Abstract
This study examined the hypothesis that different inorganic carbon (IC) conditions enrich different ammonia-oxidizing bacteria (AOB) and nitrite-oxidizing bacteria (NOB) populations by operating two laboratory-scale continuous-flow bioreactors fed with 15 and 100 mg IC/L, respectively. During this study, both bioreactors maintained satisfactory nitrification performance and stably oxidized 250 mg N/L of influent ammonium without nitrite accumulation. Based on results of cloning/sequencing and terminal restriction fragment length polymorphism targeting on the ammonia monooxygenase subunit A (amoA) gene, Nitrosomonas nitrosa lineage was identified as the dominant AOB population in the high-IC bioreactor, while Nitrosomonas europaea and Nitrosomonas nitrosa lineage AOB were dominant in the low-IC bioreactor. Results of real-time polymerase chain reactions for Nitrobacter and Nitrospira 16S rRNA genes indicated that Nitrospira was the predominant NOB population in the high-IC bioreactor, while Nitrobacter was the dominant NOB in the low-IC bioreactor. Furthermore, batch experiment results suggest that N. europaea and Nitrobacter populations are proliferated in the low-IC bioreactor due to their higher rates under low IC conditions despite the fact that these two populations have been identified as weak competitors, compared with N. nitrosa and Nitrospira, under low ammonium/nitrite environments. This study revealed that in addition to ammonium/nitrite concentrations, limited IC conditions may also be important in selecting dominant AOB/NOB communities of nitrifying bioreactors.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Nitrogen removal from wastewater is extremely important to the protection of water resources from pollution discharges as release of untreated wastewater can result in eutrophication of the receiving water bodies. Nitrification, an efficient nitrogen control strategy for wastewater treatment, is a series of microbial oxidation processes performed by nitrifiers, namely, two different bacterial groups involving ammonia-oxidizing bacteria (AOB) and nitrite-oxidizing bacteria (NOB). AOB is responsible for the oxidization of ammonia to nitrite and NOB converts nitrite to nitrate. The distribution patterns of distinct AOB/NOB species in the environments reflect the physiological properties of AOB/NOB. Among these, ammonium and nitrite concentrations are considered to be important factors for the selection of distinct AOB and NOB species, respectively. According to their physiological properties on ammonium affinity, members of Nitrosospira spp. and/or Nitrosomonas oligotropha clusters are the prevailing AOB in the environment with low ammonium, whereas Nitrosomonas europaea cluster is dominant in the environment that is rich in ammonium (Koops et al. 1991; Koops and Pommerening-Roser 2001; Siripong and Rittmann 2007). With respect to NOB, Nitrobacter species represent r-strategists with a low nitrite affinity and develop large populations at a high nitrite environment, while Nitrospira species, as K-strategists, have a high nitrite affinity and adapt to low nitrite concentrations (Schramm et al. 1999; Gieseke et al. 2003; Siripong and Rittmann 2007; Huang et al. 2010). In early studies, Nitrosomonas and Nitrobacter are considered to be the most important nitrifying bacteria responsible for ammonium and nitrite oxidation, respectively. However, recent observations, based on tools and analyses at a molecular level, indicate that Nitrosospira also are present along with well-known Nitrosomonas (Schramm et al. 1998; Schramm et al. 1999), and Nitrospira often are the dominant NOB in activated sludge systems (Schramm et al. 1998; Schramm et al. 1999; Daims et al. 2001; Dionisi et al. 2002). Although a number of studies have investigated nitrifying bacteria populations in wastewater treatment processes (Daims et al. 2001; Dionisi et al. 2002; Limpiyakorn et al. 2005; Siripong and Rittmann 2007; Whang et al. 2009; Huang et al. 2010), our understanding of linking their presence to nitrification performance at a full-scale level is still limited (Koops et al. 2006).
Previous studies have demonstrated that nitrification is feasible in full-scale wastewater treatment processes even at high ammonia levels, but nitrification failure has been frequently observed since nitrifiers can be inhibited by several environmental and engineering factors. These include low temperature, extreme pH, low dissolved oxygen concentrations, and a wide variety of chemical inhibitors (Prosser 1989). In addition to the negative effects mentioned earlier, the importance of alkalinity (Biesterfeld et al. 2003) and inorganic carbon (IC) limitations (Guisasola et al. 2007; Wett and Rauch 2003) on nitrification has been discussed. IC limitations are linked to the alkalinity requirements for buffering the acidification due to the nitrification process (Biesterfeld et al. 2003; Guisasola et al. 2007; US EPA 1993) and for cellular synthesis and growth of nitrifying bacteria (US EPA 1993). The stoichiometric relations show that per mole of ammonia removed, a substantial amount of alkalinity is consumed by the production of hydrogen ions. The accepted stoichiometric ratio for alkalinity destruction is 7.07 g of alkalinity consumed per gram of ammonia (NH3–N) oxidized to nitrate (NO −3 –N) in a closed system (US EPA 1993). Most nitrifying bacteria are autotrophic, and insufficient IC could result in a decrease of the nitrification performance. It has been demonstrated that carbon limitation is the main cause for the dramatic decrease in growth and activity of nitrifying bacteria below neutrality (Wett and Rauch 2003). Yet, IC limitations are often ignored due presumably to its abundance in full-scale systems generated during the biological oxidation of organic matters. Recently, wastewaters with the characteristic of low organic carbon to nitrogen ratio such as rejection waters of digested sludges have been studied for the development of optimized nitrogen removal systems (Carrera et al. 2003; Fux et al. 2002; Janus and van der Roest 1997; Lai et al. 2004; Wett and Rauch 2003), but little is known regarding the effects of IC limitation on nitrifying communities in such systems. Our previous study has observed that the addition of different buffering chemicals, NaOH or Na2CO3, in a bioreactor treating petrochemical wastewater may influence AOB communities (Whang et al. 2009). It is reasonable to speculate that the response to IC concentration among nitrifying populations is different, and such information is important to develop and operate a stable nitrogen removal system for wastewaters with a low carbon-to-nitrogen ratio characteristic.
This study was motivated with the goal of better understanding the influence of IC conditions on AOB and NOB community structures. Two laboratory-scale continuous-flow bioreactors were operated under IC-sufficient (high IC) and IC-limited (low IC) conditions for more than 700 days, and AOB/NOB community structures in the two bioreactors were investigated using terminal restriction fragment length polymorphism (T-RFLP) and real-time polymerase chain reaction (real-time PCR). In accordance with our previous study (Whang et al. 2009), Na2CO3 was added to adjust IC concentration. T-RFLP analysis for monitoring of AOB community changes has been well established with a reasonably good database for fingerprint analysis (Park and Noguera 2004). On the other hand, limitations of T-RFLP analysis for NOB community have been pointed out in a previous study (Regan et al. 2002). Therefore, real-time PCR analyses with designed primer sets were applied for NOB community. Finally, batch experiments were also performed to evaluate the dependence of nitrification kinetics on ammonium, nitrite, and IC concentrations for dominant AOB/NOB communities.
Materials and methods
Bioreactor operation
Two 5-L lab-scale continuous-flow bioreactors were operated to evaluate the nitrification performance and microbial communities of AOB and NOB acclimated under IC-sufficient (high IC) and IC-limited (low IC) conditions. For each bioreactor, 2 L of seed microorganisms was inoculated on day 1. The seed microorganisms for the bioreactors were obtained from a full-scale petrochemical wastewater bioreactor with a combining treatment mechanism of powder-activated carbon (PAC) adsorption and biodegradation by biofilm growth onto PAC (Whang et al. 2009). A total of 15 and 100 mg IC/L of sodium carbonate (Na2CO3) was applied to simulate low- and high-IC conditions, respectively. Ammonium chloride (NH4Cl) was added to maintain an influent concentration of 250 mg N/L for both bioreactors. The concentrations of other nutrients were identical to the ATCC 1573 medium (Park and Noguera 2004). The influent flow rate for both bioreactors was set at 0.5 L/day to maintain a 10-day hydraulic retention time. The pH was maintained at around 7 by phosphate buffer in the media. A DO concentration above 2 mg/L was maintained through aeration using diffusers. In order to minimize the introduction of IC in bioreactors through aeration, CO2 in the air was absorbed by 1 M NaOH solution before aeration. The bioreactor was mixed using a magnetic stirrer to avoid settling. The temperature was initially maintained at room temperature without additional control, but it was maintained at 30 °C using a thermal tape system after 190 days of operation. Samples were collected from the bioreactors twice a week for chemical analyses and every other week for microbial community analyses.
DNA extraction
Mixed liquor samples for DNA extraction were taken from the bioreactor and stored at −20 °C until DNA extraction. DNA was extracted using an UltraClean Soil DNA Isolation Kit (Mo Bio Laboratories, Solana Beach, CA, USA) in accordance with the manufacturer’s instructions.
Cloning and sequencing
Primers amoA-1 F and amoA-2R (Rotthauwe et al. 1997) were used to amplify a 491-bp fragment of the bacterial amoA gene. Primers 8 F and 1492R (Lane 1991) were used to amplify around 1,500 bp fragment of the bacterial 16S rRNA gene. Taq DNA Polymerase Master Mix RED (Ampliqon, Copenhagen, Denmark) was used. Cycling conditions for amoA gene included a pre-incubation step of 10 min at 95 °C, followed by 35 cycles of 95 °C for 1 min, 60 °C for 1.5 min, and 72 °C for 1.5 min, and followed by 72 °C for 10 min. Cycling conditions for bacterial 16S rRNA gene included a pre-incubation step of 5 min at 94 °C, followed by 30 cycles of 94 °C for 1 min, 55 °C for 30 s, and 72 °C for 1.5 min, and followed by 72 °C for 10 min. PCR products were ligated to the pGEM-T Easy Vector (Promega, Madison, WI, USA) and transformed into DH5α cells. Clones were randomly selected and re-amplified with amoA-1 F/amoA-2R (for amoA gene) or M13F/M13R (for 16S rRNA gene) primer sets. PCR products were digested with HhaI and HpaII restriction endonucleases. The resulting fragments were analyzed on ethidium bromide-stained agarose gels. Clones were resolved into operational taxonomic units (OTUs) based on restriction fragment length polymorphism (RFLP) patterns.
Plasmids of selected clones were extracted by the Wizard® Plus minipreps DNA purification system (Promega). DNA sequencing reactions were performed using ABI Prism 3100 and 3730 genetic analyzer (Perkin-Elmer Corp., Wellesley, MA, USA). Sequence homology searches were performed at the NCBI GenBank database using the BLAST algorithm (Altschul et al. 1997). A phylogenetic tree was constructed using Molecular Evolutionary Genetics Analysis software version 4.0.2 (Tamura et al. 2007). An unrooted phylogenetic tree was generated using the neighbor-joining method, and bootstrap tests were performed with 1,000 replicates.
Terminal restriction fragment length polymorphism analysis
The 5′ ends of amoA-1 F and amoA-2R were labeled with 6-carboxyfluorescein (6-FAM) and tetrachlorofluorescein (TET), respectively (Park and Noguera 2004). PCR products were purified using a PCR product purification kit (Montage PCR Centrifugal Filter Devices, Millipore) and digested with TaqI restriction endonuclease for 2 h at 65 °C. Digested samples were analyzed on ABI Prism 377 genetic analyzer with GeneScan version 3.1 (Perkin-Elmer Corp.). The relative abundance of each terminal restriction fragment (T-RF) was determined by calculating the ratio between the height of each peak and the summed height from all peaks in one sample. The peaks with relative abundance of <1 % or smaller T-RF length (<50 bp) were neglected in this study due to high background noise.
Real-time polymerase chain reaction
A LightCycler 2.0 system (Roche Diagnostics, Mannheim, Germany) was employed for the real-time PCR to quantify Nitrospira and Nitrobacter 16S rRNA genes and bacterial and archaeal amoA genes. Primers EUB338f and Ntspa0685 M were used to quantify the Nitrospira 16S rRNA gene (Regan et al. 2002). Primers EUB338f and NIT3 were used for the Nitrobacter 16S rRNA gene (Regan et al. 2002). The PCR mixture consisted of 10 μl of 2× SYBR® Premix Ex Taq™ (Perfect Real Time; TaKaRa Bio, Otsu, Japan), 0.2 (Nitrospira) or 0.1 (Nitrobacter) μM of each primer, and template DNA in a final volume of 20 μl. Cycling conditions for the Nitrospira 16S rRNA gene were as follows: 95 °C for 20 s , 40 cycles consisting of 95 °C for 10 s, 58 °C for 20 s, and 72 °C for 20 s (detection), followed by the melting curve analysis (65 to 95 °C with a heating rate of 0.1 °C/s). Cycling conditions for the Nitrobacter 16S rRNA gene were as follows: 95 °C for 20 s, 40 cycles consisting of 95 °C for 7 s, 60 °C for 20 s, and 72 °C for 40 s (detection), followed by the melting curve analysis. The contents of PCR mixture and cycling conditions for bacterial and archaeal amoA genes were the same as described previously (Fukushima et al. 2012). PCR products obtained from the cloned Nitrospira (accession no. AB738418) and Nitrobacter (AB738419) 16S rRNA genes and bacterial (HM753264) and archaeal (HM753263) amoA genes were used as quantitative standard. The specificity of real-time PCR for Nitrobacter and Nitrospira was evaluated based on the melting curve analysis and agarose gel electrophoresis. The specificity of real-time PCR for bacterial and archaeal amoA genes was evaluated based on the melting curve analysis.
Batch experiments
In order to evaluate the dependences of nitrification kinetics on ammonium, nitrite, and IC concentrations, a series of batch tests was performed using activated sludge samples taken from the two bioreactors. For each batch experiment, 330 mL of biomass and 70 mL of medium were mixed in a 1-L serum bottle. Initial concentrations of IC and a nitrogen compound (ammonium or nitrite) for each batch experiment are shown in Table 1. Initial concentrations of mixed liquor volatile suspended solids were measured as biomass concentration. Other medium components were the same as those used for the bioreactor operation. After aerating with pure oxygen for 2 min, the bottles were immediately sealed with caps. The batch tests were performed in an incubator maintained at 30 °C and 125 rpm. Samples were taken frequently for nitrogen compounds. The pH and DO were monitored during experiments. The specific rates of ammonium or nitrite oxidation for batch experiments were calculated and their dependences on the nitrogen compound or IC concentrations were evaluated.
Chemical analyses
Ammonium (NH +4 –N) and nitrate (NO –3 –N) were measured by ionic chromatography (DX-120, Dionex, Sunnyvale, CA, USA). Nitrite (NO –2 –N) was measured using the colorimetric method (APHA et al. 1995). MLVSS was measured according to standard procedures (APHA et al. 1995). Total inorganic carbon was determined by Model 1020A TOC Analyzer (OI Analytical, College Station, TX, USA).
Nucleotide sequence accession numbers
The amoA gene sequences determined in this study were submitted to the EMBL/GenBank/DDBJ databases under accession numbers AB622973 to AB622983. The 16S rRNA gene sequences were also submitted to the database under accession numbers AB738418, AB738419, and AB740393 to AB740404.
Results
Performance of bioreactors
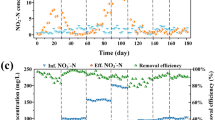
The two bioreactors, high- and low-IC, were operated for a period of more than 700 days. Influent and effluent concentrations of nitrogen species for both bioreactors are presented in Fig. 1. After approximately 90 days of operation, the activated sludge in both bioreactors achieved more than 95 % complete nitrification with occasionally observed nitrite accumulation. During day 170 to 190, nitrification performance of the high-IC bioreactor gradually declined due presumably to a decrease in room temperature from around 30 °C during summer time to 23 °C during winter time. Although the low-IC bioreactor experienced a temperature drop situation as well, the nitrification performance of the bioreactor, however, did not change. A thermal tape system was applied on day 190 to maintain an operational temperature of 30 °C, after which the nitrification performance of the high-IC bioreactor returned to previous levels. Unlike the high-IC bioreactor, the nitrification performance of the low-IC bioreactor was not influenced by the decreased room temperature during day 170 to 190.
AOB community structure
The bacterial and archaeal amoA gene copy numbers in the high- and low-IC bioreactors were determined using real-time PCR. The specificity of PCR products was confirmed by melting curve analysis (Fig. S1 of the “Electronic supplementary material”). The copy numbers of bacterial amoA gene in high- and low-IC bioreactors fluctuated within the range of 1.32 × 104 to 2.00 × 107 and 1.79 × 104 to 3.75 × 106 copy/mL sludge, respectively. The copy numbers of archaeal amoA gene in both bioreactors were below detection limit (2.50 × 103 copy/mL sludge) during most periods. The numbers were detected only after day 572 in high-IC reactor within the range of 3.83 × 103 to 6.65 × 105 copy/mL sludge. Thus, AOB was the dominant ammonia oxidizer in both bioreactors.
Cloning and sequencing of bacterial amoA gene fragments were performed to investigate the AOB community structure of the two bioreactors. The results indicated that all of the cloned amoA sequences obtained from this study were within the genus Nitrosomonas. A neighbor-joining phylogenetic tree is presented in Fig. 2, which graphically depicts the homologies of aligned sequences as well as the expected T-RF signatures (forward/reverse). There were 29 clones randomly selected for the seeding sludge sample, and these 29 clones were classified into two OTUs including OTUs S-1 (55 % of total clones) and S-2 (45 %). Based on phylogenetical analysis, OTU S-1 was closely related to N. eutropha (T-RF 491/491) lineage and S-2 was related to N. nitrosa (48/135) lineage. For the AOB communities in the bioreactors, 46 clones were randomly selected from each sample. OTUs H143-1 (95.7 %) and H143-2 (4.3 %) were obtained from the sludge in the high-IC bioreactor on day 143. Both OTUs were related to the N. nitrosa lineage, but their T-RFs were 48/135 and 48/441, respectively. Cloning and sequencing were also performed for the sample from the high-IC bioreactor on day 273 in order to confirm the AOB community before and after temperature shock. For the day 273 sample from the high-IC bioreactor, OTUs H273-1 (95.7 %) and H273-2 (4.3 %) were obtained. OTU H273-1 was related to N. nitrosa (T-RF 48/135) lineage and H273-2 was related to N. eutropha (T-RF 491/491) lineage. This indicated that AOB community structure in high-IC bioreactor shifted back to N. nitrosa after the temperature was maintained at 30 °C. For the low-IC bioreactor sample taken on day 83, three OTUs were obtained, including L83-1 (71.7 %), L83-2 (26.1 %), and L83-3 (2.2 %). For the two major OTUs obtained, OTU L83-1 was closely related to N. europaea (T-RF 219/270) lineage and OTU L83-2 was related to N. eutropha (T-RF 491/491) lineage, leaving the minor OTU L83-3 which was related to N. nitrosa (T-RF 48/135) lineage. Cloning and sequencing of bacterial 16S rRNA gene fragments for the DNA from both bioreactors on day 442 were also performed. The total number of clones from high- and low-IC bioreactors was 86 and 112, respectively. A neighbor-joining phylogenetic tree is presented in Fig. S2 of the “Electronic supplementary material”. In the high-IC bioreactor, all AOB-related OTUs (OTU HB and HC) were related to N. nitrosa lineage. In the low-IC bioreactor, all AOB-related OTUs (OTU LA and LB) were related to N. europaea lineage.
Neighbor-joining tree based on amoA gene sequences from genus Nitrosomonas. Unrooted phylogenetic tree was generated using the neighbor-joining method and bootstrap tests were performed with 1,000 replicates. Boot-strap values (>60 %) are indicated at the branch points. The scale bar represents a 5-% sequence divergence. Accession numbers are given in parentheses. DNA from the seeding sludge (S) and sludges from high- and low-IC bioreactors (H and L) were used for cloning. OTU was decided based on RFLP analysis. Other sequences were retrieved from GenBank. The expected forward and reverse T-RF lengths after digestion with TaqI are indicated
T-RFLP analysis for the amoA gene in high- and low-IC bioreactor sludges was performed until day 760. Three major T-RFs, 491/491, 48/135, and 219/270, were observed. Based on the cloning and sequencing results, these T-RFs were closely related to N. eutropha, N. nitrosa, and N. europaea lineages, respectively. Figure 3 shows the relative abundances of major T-RFs as determined by the corresponding peak height from TET-labeled amoA-2R primer. On day 2, only N. eutropha-like AOB was observed in both bioreactors, but AOB community structures in both bioreactors shifted after day 2. In the high-IC bioreactor, N. nitrosa-like AOB became the dominant population after day 47. The dominant AOB shifted from N. nitrosa-like AOB to N. europaea-like AOB on day 196 when the temperature dropped, but N. nitrosa-like AOB came back and remained dominant after day 239. In the low-IC bioreactor, N. europaea-like AOB became the dominant population after day 47, and it continued to be dominant until day 630. After day 630, N. nitrosa-like AOB and N. europaea-like AOB were both dominant in the low-IC bioreactor at 60 and 40 %, respectively.
Dynamics of AOB populations in the high-IC (upper panel) and low-IC (lower panel) bioreactors as determined by amoA-based T-RFLP. T-RF 135 open square, T-RF 270 filled square, T-RF 491 open triangle, T-RF for others open diamond. Samples were taken for ammonium oxidation batch experiments (down arrows)
NOB community structure
The results of the phylogenetic analysis based on bacterial 16S rRNA genes (Fig. S2 of the “Electronic supplementary material”) showed that NOB-related OTUs in high-IC (OTU HA and HE) and low-IC (OTU LC) bioreactors on day 442 were related to Nitrospira and Nitrobacter, respectively. The 16S rRNA copy numbers of Nitrobacter and Nitrospira in the high- and low-IC bioreactors were determined using real-time PCR. The specificity of PCR products was confirmed by both melting curve analysis and agarose gel electrophoresis (Figs. S3 and S4 of the “Electronic supplementary material”). The results indicated that significant no-primer dimer and non-target amplicon were generated in all real-time PCR results. The 16S rRNA gene copy numbers of Nitrobacter and Nitrospira are summarized in Fig. 4. In the high-IC bioreactor, the copy number of Nitrospira was stable. The copy number of Nitrobacter increased until day 463 and then remained stable. However, the copy number of Nitrobacter was significantly lower than that of Nitrospira during this study. In the low-IC bioreactor, the copy number of Nitrospira significantly decreased from day 2 to 47. After day 47, the number remained stable, but these values were approximately ten times lower than those observed in the high-IC bioreactor. The trend for Nitrobacter copy number was similar to that observed in the high-IC bioreactor. The 16S rRNA gene copy number of total NOB was estimated by the summation of the copy numbers of Nitrospira and Nitrobacter. The total NOB copy numbers of the high-IC bioreactor were stably fluctuated between 6.61 × 103 and 4.28 × 107 copy/mL sludge. In the low-IC bioreactor, the number decreased in the first 47 days, and then it fluctuated within the range of 1.29 × 103 to 1.66 × 106 copy/mL sludge. In general, the value was approximately one order of magnitude lower than that observed in the high-IC bioreactor.
The 16S rRNA gene copy number is assumed to be equivalent to the bacteria cell number for Nitrobacter and Nitrospira since these species typically possess one 16S rRNA gene operon (Dionisi et al. 2002; Navarro et al. 1992). On day 2, the relative abundance of Nitrospira in both bioreactors was greater than 99 %, indicating that Nitrospira was dominant in the seeding sludge. Nitrospira, with a relative abundance over 80 %, was the dominant NOB in the high-IC bioreactor during this study. In the low-IC bioreactor, the relative abundance of Nitrospira was higher than that of Nitrobacter during day 2 to 149, but the abundance of Nitrospira became lower than Nitrobacter after day 205.
Evaluation of oxidation rates of nitrifying populations in the bioreactors
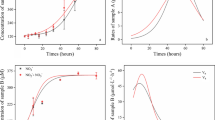
The dependences of specific ammonium oxidation rates on IC and ammonium concentrations for biomass taken from high- and low-IC bioreactors are summarized in Fig. 5. In all batch experiments, the specific ammonia oxidization rates of the low-IC bioreactor AOB were obviously higher than those of the high-IC bioreactor AOB. Among all experiments, the difference in specific ammonium oxidation rates between high- and low-IC AOB was minimum at an initial ammonium concentration of 50 mg N/L.
The dependences of specific nitrite oxidation rates on IC and nitrite concentrations for biomass taken from high- and low-IC bioreactors were evaluated with three batch experiments as summarized in Fig. 6. In the NOB-1 batch experiment with an initial nitrite concentration of 40 mg N/L, the specific nitrite oxidization rates of the low-IC bioreactor NOB were all higher than those of the high-IC bioreactor NOB, disregarding the IC concentration applied. For the NOB-2 batch experiment with an initial nitrite concentration of 5 mg N/L, NOB in both bioreactors attained similar rates. In the NOB-3 batch experiment with an initial IC concentration of 100 mg/L, the rates for the low-IC bioreactor NOB were higher than those for the high-IC bioreactor NOB, except that at an initial nitrite concentration of 5 mg N/L the high-IC bioreactor NOB attained a slightly higher rate.
Discussion
To evaluate the effects of IC concentration on the nitrifying communities and their nitrification performance, we operated two lab-scale continuous-flow bioreactors with high and low IC concentrations, respectively. Both bioreactors achieved more than 95 % of nitrification efficiency except for the high-IC bioreactor during a period of reduced ambient temperature. In this study, we demonstrated that IC concentration did not significantly affect nitrification performance, but it affected nitrifying community structures.
The dominant AOB populations in the high- and low-IC bioreactors were of the N. nitrosa and N. europaea lineage, respectively. N. nitrosa has been reported for its abundance in activated sludge treating industrial wastewaters (Cabezas et al. 2009; Dionisi et al. 2002; Kim et al. 2011; Kuo et al. 2006; Layton et al. 2005). In fact, N. nitrosa Nm90 was isolated from an industrial sewage in Germany (Koops et al. 1991). Compared with N. europaea, N. nitrosa was reported to have a higher affinity for ammonium (Koops and Pommerening-Roser 2001; Prosser 1989), suggesting its competitiveness in a low-ammonium environment like the bioreactors operated in this study. However, the details regarding the conditions that may favor the proliferation of N. nitrosa over other AOB species are limited and warrant future research.
During day 170 to 190, the nitrification performance of the high-IC bioreactor gradually declined due presumably to a decrease in room temperature, while the performance of the low-IC bioreactor did not change. Schmidt et al. (2004) reported that different temperature settings had no significant effect on the growth mode of and the proteins expressed by N. europaea. Although the reason is unclear, the dominance of N. nitrosa lineage under a higher temperature has been reported (Avrahami et al. 2011; Shore et al. 2012). Those results suggest that different AOB communities present in between high- and low-IC reactors may result in different responses to temperature drop.
N. europaea has been known to be the key AOB in high-ammonium-containing wastewater treatment systems because it is the most frequently recovered AOB pure culture in nitrogen-rich culture conditions (Koops and Pommerening-Roser 2001; Prosser 1989). Previous studies have shown that members of the N. europaea lineage are relatively abundant among AOB species in several bioreactors (Gieseke et al. 2003; Schramm et al. 2000) due to its relatively high maximum ammonium conversion rate compared to those of other AOB populations (Koops and Pommerening-Roser 2001; Prosser 1989). The low-IC biomass dominated by N. europaea attained higher specific ammonia oxidization rates in all batch experiments with initial ammonium > 50 mg N/L, indicating that the results of microbial community analysis agreed with the results of kinetic analysis. On the other hand, a previous study has reported that N. europaea was found to be dominant in a pilot-scale bioreactor buffered with NaOH under an IC-limited condition, while Nitrosospira became proliferated in the same bioreactor during the operation buffered with Na2CO3 (Whang et al. 2009). Accordingly, N. europaea was able to dominate under low-IC conditions, although the mechanism is not clear at this moment.
The difference in the mechanisms and characteristics of carbon fixation among AOB species may provide possible explanations for their different responses to IC concentrations. AOB fix CO2 through the Calvin–Benson–Bassham (CBB) cycle, and one of the key enzymes is the ribulose bisphosphate carboxylase (RubisCO) (Schramm et al. 1998; Utaker et al. 2002). It has been reported for N. europaea that the transcription of cbb genes, which encode carbon-fixing enzyme RubisCO, was upregulated when the carbon source was limited (Wei et al. 2004). This property suggests that N. europaea may primarily distribute available resources towards key components for carbon assimilation under limited IC conditions, resulting in its competitiveness and dominance in the low-IC bioreactor. Another possibility is that N. nitrosa has carboxysomes, while N. europaea does not (Koops and Pommerening-Roser 2001). It is suggested that CO2, rather than bicarbonate, is used as a substrate by RubisCO for microbial CO2 fixation, and the conversion of bicarbonate to CO2 requires another enzyme, carbonic anhydrase (Tabita 1999). Carboxysome is a microbial microcompartment that performs CO2 fixation by co-localizing the two enzymes RuBisCO and carbonic anhydrase within the carboxysome (Yeates et al. 2008). Compared to AOB species with the carboxysome, it is likely that N. europaea, without the carboxysome, can fix CO2 more efficiently, especially under an IC-limited environment. Furthermore, N. europaea Nm50 had a “green-like” type I RubisCO (similar to RubisCO genes found in green algae), whereas Nitrosospira spp. generally has a “red-like” type I RubisCO (Utaker et al. 2002). The differences in affinity of the RubisCO enzyme for NaHCO3 among the AOB species (Hatayama et al. 2000) can also be attributed to their different responses to IC concentrations. Nevertheless, the exact mechanisms are not clear at this time, but the answer to whether those differences cause the different responses to varying IC concentrations among AOB species awaits future investigations.
For the NOB population, Nitrospira and Nitrobacter were dominant in the high- and low-IC bioreactors, respectively. Nitrobacter, an r-strategist, has been reported to be a superior competitor when nitrite is abundant, while Nitrospira, a K-strategist, thrives under conditions with nitrite scarcity (Blackburne et al. 2007; Huang et al. 2010; Nogueira and Melo 2006). Under high-nitrite conditions in the batch experiments, the low-IC sludge dominated by Nitrobacter attained higher specific nitrite oxidization rates compared to the Nitrospira-dominated high-IC sludge, indicating that the results of microbial community analysis agreed with the results of kinetic analysis. Kim and Kim (2006) reported that specific nitrite oxidization activities of Nitrobacter (93.8 mg/gNOB h) were nine times higher than that of Nitrospira (10.5 mg/g NOB/h). In addition, higher accumulated nitrate concentration might present an inhibitory effect to the Nitrospira. These may explain why low-IC biomass showed a higher activity in batch experiments even if the total NOB copy number of the low-IC biomass was lower than that of the high-IC biomass. Under low nitrite together with IC-limited conditions, the Nitrobacter-dominant sludge maintained similar specific nitrite oxidization rates compared to the Nitrospira-dominated sludge despite the fact that Nitrospira was considered a K-strategist. It is likely that IC limitation may favor the selection of Nitrobacter over Nitrospira even at a low-nitrite condition like the bioreactors operated in this study. By evaluating oxygen uptake rates, Guisasola et al. (2007) also observed no IC limitation effect on the nitratation process for a nitrifying sludge containing Nitrobacter at 5 % of the total bacterial populations, although detailed information regarding Nitrospira population was not mentioned in that study.
Metagenomic analyses on Nitrobacter winogradskyi (Starkenburg et al. 2006), Nitrobacter hamburgensis X14 (Starkenburg et al. 2008), and “Candidatus Nitrospira defluvii” (Lucker et al. 2010) indicated that different NOB populations may perform different carbon fixation mechanisms. Genes for all of the enzymes of a functional CBB cycle are present in Nitrobacter but not in Candidatus Nitrospira defluvii, suggesting that the CBB cycle does not operate in Candidatus Nitrospira defluvii. All genes of the reductive tricarboxylic acid cycle are instead present in Candidatus Nitrospira defluvii, suggesting that a different carbon fixation pathway is employed for Nitrospira. The difference in carbon fixation mechanisms between Nitrobacter and Nitrospira is likely attributed to their different responses to IC concentrations.
In summary, under high ammonium/nitrite conditions, N. europaea/Nitrobacter was a dominant AOB/NOB disregarding the IC concentration. Compared with N. nitrosa/Nitrospira, N. europaea/Nitrobacter attained higher specific oxidation rates in such environments even at low IC conditions. Under low-ammonium/nitrite conditions, IC availability became important in selecting dominant nitrifying populations. With sufficient IC conditions like the high-IC bioreactor, N. nitrosa/Nitrospira was dominant since it was a K-strategist in terms of ammonium/nitrite, while in the low-IC bioreactor, when available IC became limited, N. europaea/Nitrobacter became important due to its efficient utilization of IC compared with N. nitrosa/Nitrospira.
References
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25(17):3389–3402
APHA/AWWA/WEF (1995) Standard methods for the examination of water and wastewater, 19th edn. Washington, DC: American Public Health Association/American Water Works Association/Water Environment Federation.
Avrahami S, Jia Z, Neufeld JD, Murrell JC, Conrad R, Küsel K (2011) Active autotrophic ammonia-oxidizing bacteria in biofilm enrichments from simulated creek ecosystems at two ammonium concentrations respond to temperature manipulation. Appl Environ Microbiol 77:7329–7338
Biesterfeld S, Farmer G, Russell P, Figueroa L (2003) Effect of alkalinity type and concentration on nitrifying biofilm activity. Water Environ Res 75(3):196–204
Blackburne R, Vadivelu VM, Yuan Z, Keller J (2007) Kinetic characterisation of an enriched Nitrospira culture with comparison to Nitrobacter. Water Res 41(14):3033–3042
Cabezas A, Draper P, Etchebehere C (2009) Fluctuation of microbial activities after influent load variations in a full-scale SBR: recovery of the biomass after starvation. Appl Microbiol Biotechnol 84(6):1191–1202
Carrera J, Baeza JA, Vicent T, Lafuente J (2003) Biological nitrogen removal of high-strength ammonium industrial wastewater with two-sludge system. Water Res 37(17):4211–4221
Daims H, Nielsen JL, Nielsen PH, Schleifer K-H, Wagner M (2001) In situ characterization of Nitrospira-like nitriteoxidizing bacteria active in wastewater treatment plants. Appl Environ Microbiol 67:5273–5284
Dionisi HM, Layton AC, Harms G, Gregory IR, Robinson KG, Sayler GS (2002) Quantification of Nitrosomonas oligotropha-like ammonia-oxidizing bacteria and Nitrospira spp. from full-scale wastewater treatment plants by competitive PCR. Appl Environ Microbiol 68:245–253
Fukushima T, Wu YJ, Whang LM (2012) The influence of salinity and ammonium levels on amoA mRNA expression of ammonia-oxidizing prokaryotes. Water Sci Technol 65(12):2228–2235
Fux C, Boehler M, Huber P, Brunner I, Siegrist H (2002) Biological treatment of ammonium-rich wastewater by partial nitritation and subsequent anaerobic ammonium oxidation (anammox) in a pilot plant. J Biotechnol 99(3):295–306
Gieseke A, Bjerrum L, Wagner M, Amann R (2003) Structure and activity of multiple nitrifying bacterial populations co-existing in a biofilm. Environ Microbiol 5(5):355–369
Guisasola A, Petzet S, Baeza JA, Carrera J, Lafuente J (2007) Inorganic carbon limitations on nitrification: experimental assessment and modelling. Water Res 41(2):277–286
Hatayama R, Takahashi R, Ohshima M, Shibasaki R, Tokuyama T (2000) Ribulose-1,5-bisphosphate carboxylase/oxygenase from an ammonia-oxidizing bacterium, Nitrosomonas sp K1: purification and properties. J Biosci Bioeng 90(4):426–430
Huang Z, Gedalanga PB, Asvapathanagul P, Olson BH (2010) Influence of physicochemical and operational parameters on Nitrobacter and Nitrospira communities in an aerobic activated sludge bioreactor. Water Res 44(15):4351–4358
Janus HM, van der Roest HF (1997) Don't reject the idea of treating reject water. Water Sci Technol 35(10):27–34
Kim DJ, Kim SH (2006) Effect of nitrite concentration on the distribution and competition of nitrite-oxidizing bacteria in nitratation reactor systems and their kinetic characteristics. Water Res 40:887–894
Kim YM, Lee DS, Park C, Park D, Park JM (2011) Effects of free cyanide on microbial communities and biological carbon and nitrogen removal performance in the industrial activated sludge process. Water Res 45(3):1267–1279
Koops H, Bottcher B, Moller U, Pommerening-Roser A, Stehr G (1991) Classification of eight new species of ammonia-oxidizing bacteria: Nitrosomonas communis sp. nov., Nitrosomonas ureae sp. nov., Nitrosomonas aestuarii sp. nov., Nitrosomonas marina sp. nov., Nitrosomonas nitrosa sp. nov., Nitrosomonas eutropha sp. nov., Nitrosomonas oligotropha sp. nov. and Nitrosomonas halophila sp. nov. Microbiol 137(7):1689–1699
Koops HP, Pommerening-Roser A (2001) Distribution and ecophysiology of the nitrifying bacteria emphasizing cultured species. FEMS Microbiol Ecol 37:1–9
Koops H, Purkhold U, Pommerening-R A, Timmermann G, Wagner M (2006) The lithoautotrophic ammonia-oxidizing bacteria. The Prokaryotes 5:778–811
Kuo DHW, Robinson KG, Layton AC, Meyers AJ, Sayler GS (2006) Real-time PCR quantification of ammonia-oxidizing bacteria (AOB): solids retention time (SRT) impacts during activated sludge treatment of industrial wastewater. Environ Eng Sci 23(3):507–520
Lai E, Senkpiel S, Solley D, Keller J (2004) Nitrogen removal of high strength wastewater via nitritation/denitritation using a sequencing batch reactor. Water Sci Technol 50(10):27–33
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. Wiley, New York, pp 115–175
Layton AC, Dionisi H, Kuo HW, Robinson KG, Garrett VM, Meyers A, Sayler GS (2005) Emergence of competitive dominant ammonia-oxidizing bacterial populations in a full-scale industrial wastewater treatment plant. Appl Environ Microbiol 71(2):1105–1108
Limpiyakorn T, Shinihara Y, Kurisu F, Yagi O (2005) Communities of ammonia-oxidizing bacteria in activated sludge of various sewage treatment plants in Tokyo. FEMS Microbiol Ecol 54:205–117
Lucker S, Wagner M, Maixner F, Pelletier E, Koch H, Vacherie B, Rattei T, Damste JS, Spieck E, Le Paslier D, Daims H (2010) A Nitrospira metagenome illuminates the physiology and evolution of globally important nitrite-oxidizing bacteria. Proc Natl Acad Sci USA 107(30):13479–13484
Navarro E, Fernandez MP, Grimont F, Claysjosserand A, Bardin R (1992) Genomic heterogeneity of the genus Nitrobacter. Int J Syst Bacteriol 42(4):554–560
Nogueira R, Melo LF (2006) Competition between Nitrospira spp. and Nitrobacter spp. in nitrite-oxidizing bioreactors. Biotechnol Bioeng 95(1):169–175
Park H-D, Noguera DR (2004) Evaluating the effect of dissolved oxygen on ammonia-oxidizing bacterial communities in activated sludge. Water Res 38(14–15):3275–3286
Prosser JI (1989) Autotrophic nitrification in bacteria. Adv Microb Physiol 30:125–181
Regan JM, Harrington GW, Noguera DR (2002) Ammonia- and nitrite-oxidizing bacterial communities in a pilot-scale chloraminated drinking water distribution system. Appl Environ Microbiol 68(1):73–81
Rotthauwe JH, Witzel KP, Liesack W (1997) The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol 63(12):4704–4712
Schramm A, de Beer D, Wagner M, Amann R (1998) Identification and activities in situ of Nitrosospira and Nitrospira spp. as dominant populations in a nitrifying fluidized bed reactor. Appl Environ Microbiol 64(9):3480–3485
Schramm A, de Beer D, van den Heuvel J, Ottengraf S, Amann R (1999) Microscale distribution of populations and activities of Nitrosospira and Nitrospira spp. along a macroscale gradient in a nitrifying bioreactor: quantification by in situ hybridization and the use of microsensors. Appl Environ Microbiol 65(8):3690–3696
Schramm A, De Beer D, Gieseke A, Amann R (2000) Microenvironments and distribution of nitrifying bacteria in a membrane-bound biofilm. Environ Microbiol 2(6):680–686
Schmidt I, Steenbakker PJM, Op den Camp HJM, Schmidt K, Jetten MSM (2004) Physiologic and proteomic evidence for a role of nitric oxide in biofilm formation by Nitrosomonas europaea and other ammonia oxidizers. J Bacteriol 186(9):2781–2788
Shore JL, M’Coy WS, Gunsch CK, Deshusses MA (2012) Application of a moving bed biofilm reactor for tertiary ammonia treatment in high temperature industrial wastewater. Biores Technol 122:51–60
Siripong S, Rittmann B (2007) Diversity study of nitrifying bacteria in full-scale municipal wastewater treatment plants. Water Res 41(5):1110–1120
Starkenburg SR, Chain PS, Sayavedra-Soto LA, Hauser L, Land ML, Larimer FW, Malfatti SA, Klotz MG, Bottomley PJ, Arp DJ, Hickey WJ (2006) Genome sequence of the chemolithoautotrophic nitrite-oxidizing bacterium Nitrobacter winogradskyi Nb-255. Appl Environ Microbiol 72(3):2050–2063
Starkenburg SR, Larimer FW, Stein LY, Klotz MG, Chain PS, Sayavedra-Soto LA, Poret-Peterson AT, Gentry ME, Arp DJ, Ward B, Bottomley PJ (2008) Complete genome sequence of Nitrobacter hamburgensis X14 and comparative genomic analysis of species within the genus Nitrobacter. Appl Environ Microbiol 74(9):2852–2863
Tabita FR (1999) Microbial ribulose 1,5-bisphosphate carboxylase/oxygenase: a different perspective. Photosynth Res 60(1):1–28
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24(8):1596–1599
US EPA (1993) Process design manual: nitrogen control, EPA/625/R-93/010. US EPA, Office of Research and Development, Office of Water, Washington, DC.
Utaker JB, Andersen K, Aakra A, Moen B, Nes IF (2002) Phylogeny and functional expression of ribulose 1,5-bisphosphate carboxylase/oxygenase from the autotrophic ammonia-oxidizing bacterium Nitrosospira sp. isolate 40KI. J Bacteriol 184(2):468–478
Wei X, Sayavedra-Soto LA, Arp DJ (2004) The transcription of the cbb operon in Nitrosomonas europaea. Microbiol 150(6):1869–1879
Wett B, Rauch W (2003) The role of inorganic carbon limitation in biological nitrogen removal of extremely ammonia concentrated wastewater. Water Res 37(5):1100–1110
Whang LM, Yang KH, Yang YF, Han YL, Chen YJ, Cheng SS (2009) Microbial ecology and performance of ammonia oxidizing bacteria (AOB) in biological processes treating petrochemical wastewater with high strength of ammonia: effect of Na2CO3 addition. Water Sci Technol 59(2):223–231
Yeates TO, Kerfeld CA, Heinhorst S, Cannon GC, Shively JM (2008) Protein-based organelles in bacteria: carboxysomes and related microcompartments. Nat Rev Micro 6(9):681–691
Acknowledgments
The authors would like to acknowledge the financial support from the National Science Council of Taiwan under Grant NSC 97-2221-E-006-045-MY3. We thank Meng Chung Hung of The University of Waterloo for valuable discussion.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 423 kb)
Rights and permissions
About this article
Cite this article
Fukushima, T., Whang, LM., Chiang, TY. et al. Nitrifying bacterial community structures and their nitrification performance under sufficient and limited inorganic carbon conditions. Appl Microbiol Biotechnol 97, 6513–6523 (2013). https://doi.org/10.1007/s00253-012-4436-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-012-4436-y