Abstract
Multidrug resistance is a major barrier in the battle against tuberculosis and still a leading cause of death worldwide. In order to fight this pathogen, two routes are practicable: vaccination or drug treatment. Vaccination against Mycobacterium tuberculosis with the current vaccine Mycobacterium bovis Bacillus Calmette–Guerin is partially successful, being its efficacy variable. A few new tuberculosis vaccines are now in various phases of clinical trials. The emergence of multidrug-resistant strains of M. tuberculosis gave the impulse to discover new effective antitubercular drugs, a few of which are in clinical development. Here we focus on three different classes of very promising antitubercular drugs recently discovered (benzothiazinones, dinitrobenzamides, and benzoquinoxalines) that share the same cellular target: a subunit of the heteromeric decaprenylphosphoryl-β-d-ribose 2′-epimerase, encoded by the dprE1 (or Rv3790) gene. This enzyme is involved in the biosynthesis of d-arabinose which is crucial for the synthesis of the mycobacterial cell wall and essential for the pathogen’s survival.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Mycobacterium tuberculosis is one of the world’s most devastating human pathogens. The WHO estimated that in 2010, there were 8.8 million cases of tuberculosis (TB), 1.1 million deaths from TB among HIV-negative people, and additional 0.35 million deaths from HIV-associated TB (WHO 2011). The success of M. tuberculosis is due to its ability to persist within humans for long periods in a clinically latent state: Roughly 95 % of the people who become infected (one third of the global population) develop a latent infection (Kochi 2001). The risk of reactivation is increased in people infected with HIV to over 20 times that in HIV-negative people as immunosuppression worsens (Girardi et al. 2000; Selwyn et al. 1989). Furthermore, the currently used antitubercular drugs, mainly rifampicin, have important interactions with antiretroviral drugs (Donald and Schaaf 2007).
Vaccination is only partially successful. The current available vaccine, Mycobacterium bovis Bacillus Calmette–Guerin (BCG), is the most widely used in the world, but it has beneficial effects only in childhood (Colditz et al. 1994, 1995) for a limited number of years (Andersen and Doherty 2005). In particular, this vaccine protects only against the most severe forms of the disease and not from infection. M. tuberculosis stimulates a strong immune response, but it has evolved to resist the body’s attempts to eradicate it and even if the initial infection is successfully controlled, many individuals develop a latent infection that can persist for decades (Andersen et al. 2007; Manabe and Bishai 2000; Morrison et al. 2008; Stewart et al. 2003). Moreover, M. tuberculosis is able to interfere with almost every stage of the host’s immune response (Dietrich and Doherty 2009), and this makes difficult to design a new, effective vaccine. In fact, for more than 80 years, no new TB vaccines have been successfully developed. Presently, about 12 TB vaccines are in clinical trials (Rappuoli and Aderem 2011). Moreover, other vaccines, which show activity against the latent form of the disease, are in late preclinical stages. For example, Aagaard and collaborators have developed a multistage vaccination strategy in which the early antigens Ag85B and 6-kDa early secretory antigenic target are combined with the latency-associated protein Rv2660c (H56 vaccine). In two mouse models of latent tuberculosis, they show that H56 vaccination after exposure is able to control reactivation and significantly lower the bacterial load compared to adjuvant control mice (Aagaard et al. 2011).
Recently, the blood transcripts of individuals with active infection were compared to those of individuals who were latently infected. This investigation identified subsets of genes that correlated with the extent of the disease (Maertzdorf et al. 2011; Berry et al. 2010). The identification of the pathways associated with TB disease progression may help to define pathways that can be targeted in new vaccines.
On the other hand, the development of new drugs is urgently needed due to the increasing number of multidrug resistant (MDR), which are strains of M. tuberculosis that are resistant to both isoniazid and rifampicin, with or without resistance to other drugs, extensively drug-resistant (XDR), MDR strains also resistant to any fluoroquinolone and any of the second-line anti-TB injectable drugs (amikacin, kanamycin, or capreomycin), and totally drug-resistant (TDR) or super-XDR M. tuberculosis strains that fail to respond to first- and second-line drug treatment (Dye 2006; Gandhi Neel et al. 2006; Cegielski 2010; Riccardi et al. 2009; Rowland 2012). In particular, for TDR, there are currently no drugs available. Consequently, the demand for new and faster acting antitubercular drugs is greater than ever. In the past decade, intensive efforts have been made to discover new leads for antitubercular drugs, and a few molecules are now in clinical trials (Cole and Riccardi 2011), even if the development of new ones is still required.
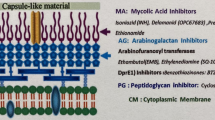
In the last 5 years, the “New Medicines for Tuberculosis” European Consortium has been working to develop new drugs for the treatment of TB through an integrated approach (http://www.sciprom.ch/nm4tb/). Within this project, it has been discovered a new effective antimycobacterial agent, belonging to the benzothiazinone (BTZ) class, with a Minimal Inhibitory Concentration (MIC) of 1 ng/ml, that quickly kills M. tuberculosis in vitro, ex vivo, and in murine models of TB (Makarov et al. 2009). The target of benzothiazinones has been identified as the DprE1 enzyme (Makarov et al. 2009), which works in concert with DprE2 to catalyze the epimerization of decaprenyl-d-ribose to decaprenyl-d-arabinose in the biosynthesis of arabinogalactan, a fundamental component of the mycobacterial cell wall (Wolucka 2008; Manina et al. 2010b).
It has been shown that DprE1 is the target also of another class of very promising drugs, the dinitrobenzamides (DNBs; Christophe et al. 2009; de Jesus Lopes Ribeiro et al. 2011). These compounds are active against mycobacteria and non-toxic for the host cells. Of particular importance is the fact that these compounds are also highly active against MDR- and XDR-TB clinical isolates (Christophe et al. 2009).
Moreover, another class of antitubercular drugs, the benzoquinoxalines, has been shown to have DprE1 as its target. The lead compound of this class is VI-9376, a molecule structurally related to benzothiazinones (Magnet et al. 2010).
The scope of this mini-review is to summarize the main features of these three recently described classes of compounds which target the enzyme DprE1. This protein represents a proven vulnerable antimycobacterial drug target that could turn out magic for TB treatment (Manina et al. 2010b).
The decaprenyl-phosphoribose 2′-epimerase
The heteromeric enzyme decaprenyl-phospho-ribose 2′-epimerase catalyzes the epimerization reaction of decaprenylphosphoryl-d-ribose (DPR) into decaprenylphosphoryl-d-arabinose (DPA). This reaction occurs via a sequential oxidation–reduction mechanism involving an intermediate (decaprenylphosphoryl-2-keto-β-d-erythro-pentofuranose, DPX), which is a product of DPR oxidation and a precursor of DPA (Trefzer et al. 2012) (Fig. 1). This enzyme is composed of two proteins encoded by the dprE1 and dprE2 genes (Makarov et al. 2009; Mikusova et al. 2005). DprE1 and DprE2 have been suggested to act as decaprenylphosphoryl-β-d-ribose oxidase and decaprenylphosphoryl-d-2-keto erythro pentose reductase, respectively (Makarov et al. 2009). Recently, Trefzer and collaborators reported the in vitro characterization of the enzymatic activities of purified recombinant DprE1 and DprE2 orthologous proteins from Mycobacterium smegmatis and demonstrated that DprE1 acts as an oxidase and DprE2 as a reductase (Trefzer et al. 2012). For epimerase activity, a simultaneous expression of both polypeptides is required (Mikusova et al. 2005). The DprE1 protein contains a FAD-binding N-terminal and a C-terminal d-arabinono-1,4-lactone oxidase-like enzyme domains (Wolucka 2008).
It is noteworthy that without DPA, a complete mycobacterial cell wall cannot be produced. DPA is synthesized from phosphoribose diphosphate through a series of three successive reactions (Meniche et al. 2008): (a) transfer of phosphoribose diphosphate to decaprenyl phosphate to form decaprenylphosphoryl-5-phosphoribose, (b) removal of the 5′ phosphate to give DPR, and (c) epimerization of DPR into DPA. This sugar is used as the only donor of Araf residues in the biosynthesis of both mycobacteria and corynebacteria cell wall (Mikusova et al. 2005) which is composed of three main portions: a highly impermeable layer of mycolic acids, the complex polysaccharide arabinogalactan, and a peptidoglycan layer. Sequential additions of Araf residues to the galactan domain lead to the synthesis of arabinan domain of arabinogalactan, while lipoarabinogalactan originates from the addition of Araf units on the mannan domain by specialized arabinosyltransferases (Wolucka 2008). Arabinogalactan is covalently attached to peptidoglycan by a phosphodiester linkage and is esterified by mycolic acids. Mycobacteria cell envelope plays a crucial role in intrinsic drug resistance as well as in survival in the macrophage under stress conditions and is considered an outstanding cellular source for target discovery.
Both the dprE1 and dprE2 genes were predicted to be essential by Himar1-based transposon mutagenesis in M. tuberculosis H37Rv (Sassetti and Rubin 2003), thus validating both enzymes as targets for drug development. Moreover, the Corynebacterium glutamicum ortholog (NCgl0187) could not be deleted by Meniche et al. (2008). Recently, the construction of a conditional gene knockout strain targeting the ortholog of dprE1 in M. smegmatis, MSMEG_6382, has been reported (Crellin et al. 2011). Crellin and co-workers attempted to disrupt the MSMEG_6382 gene by insertional inactivation with a drug resistance cassette. Despite several attempts, they failed to generate this mutant. The inability to generate a viable mutant raised the possibility that the enzyme might be essential to the organism (Crellin et al. 2011). Disruption of the chromosomal copy of MSMEG_6382 was only possible in the presence of a plasmid-encoded copy of MSMEG_6382. Curing of this “rescue” plasmid from the bacterial population resulted in a cessation of growth, further demonstrating gene essentiality. This study provides the first direct experimental evidence for the essentiality of DprE1 in mycobacteria. Moreover, the essentiality of DprE1 in M. smegmatis, combined with its conservation in all sequenced mycobacterial genomes, suggests that DPA synthesis is essential in all mycobacteria (Crellin et al. 2011).
The essentiality of DprE1 in M. smegmatis also suggests that there are no alternative arabinose donors that can compensate for a lack of DPA. A possible alternative arabinose donor has been described in M. smegmatis (Wolucka 2008). However, until now, DPA is the only known Araf donor in the biogenesis of arabinogalactan and lipoarabinomannan in mycobacteria cell wall biosynthesis and is thus an essential precursor.
Another evidence on dprE1 essentiality comes from Carroll and co-workers who utilized the tetracycline-inducible system to generate knockdown strains of a number of genes/operons in M. tuberculosis to determine essentiality and vulnerability simultaneously (Carroll et al. 2011). They analyzed growth of the recombinant strains and obtained a range of phenotypes which reflect cellular requirements for viability. In particular, growth of the dprE1 knockdown strain (Ptet dprE1) was severely reduced without tetracycline, and also expression levels were shown to be reduced. The Ptet dprE1 strain demonstrated increased sensitivity to compounds targeting the cell wall; the MIC of benzothiazinones was reduced 2-fold and that of ethambutol was reduced 4-fold (Carroll et al. 2011). At present, a fundamental goal is the production of large amounts of soluble DprE1 protein in order to develop an enzymatic assay suitable for high throughput screenings and to solve the DprE1 structure to design new molecules affecting the arabinogalactan biosynthesis.
What is evident now, however, is that the decaprenyl-phosphoribose 2′-epimerase enzyme is a very important and authentic validated target for antitubercular drugs, being the target for at least three different classes of antitubercular drugs, namely benzothiazinones, dinitrobenzamides, and benzoquinoxalines (Makarov et al. 2009; Christophe et al. 2009; Magnet et al. 2010) that will be described in the paragraphs below.
Benzothiazinones
The first described class of drugs targeting DprE1 is that of BTZs, a series of sulfur-containing heterocycle compounds with antibacterial activity (Makarov et al. 2006). One of them, the 2-[2-methyl-1,4-dioxa-8-azaspiro[4.5]dec-8-yl]-8-nitro-6-(trifluoromethyl)-4H-1,3 benzothiazin-4-one (BTZ038), has been well characterized (Makarov et al. 2009). The compound was synthesized in seven steps with a yield of 36 %. BTZ038 has a single chiral center, and both enantiomers, BTZ043 (S) and BTZ044 (R), were found to be equipotent in vitro (Fig. 2).
The MICs of a variety of BTZs against different mycobacteria ranged from ∼0.1 to 80 ng/ml for fast growers and from 1 to 30 ng/ml for members of the M. tuberculosis complex. The MICs of BTZ043 against M. tuberculosis H37Rv and M. smegmatis were 1 and 4 ng/ml, respectively (Makarov et al. 2009).
BTZ043 is bactericidal, reducing viability in vitro by more than 1,000-fold in under 72 h. The uptake, intracellular killing, and potential cytotoxicity of BTZ compounds in an ex vivo model were determined. Macrophages treated with BTZ043 were protected as compared with those treated with the negative controls (Makarov et al. 2009).
The in vivo efficacy of BTZ043 was assessed in BALB/c mice in the chronic model of TB. Four weeks of treatment with BTZ043 reduced the bacterial burden in the lungs and spleens by one and two logs, respectively. Additional results suggested that BTZ efficacy is time rather than dose dependent (Makarov et al. 2009).
The fact that BTZ targets DprE1 was demonstrated by two independent genetic approaches. Firstly, cosmids bearing DNA from M. smegmatis that confer increased resistance in M. smegmatis were identified. The region responsible for resistance was subsequently pinpointed by subcloning experiments. This approach revealed that the responsible gene was MSMEG_6382 of M. smegmatis or its M. tuberculosis ortholog, dprE1. Secondly, M. smegmatis, M. bovis BCG, and M. tuberculosis mutants with a high level of BTZ resistance were isolated and characterized. All these resistant mutants harbored missense mutations in MSMEG_6382, Mb3819, and dprE1 genes, respectively (Makarov et al. 2009).
In all of the drug-resistant mutants examined, the same codon of dprE1 was affected: the cysteine at position 387 was replaced by serine or glycine codons, respectively. The BTZ resistance-determining region of dprE1 was highly conserved in orthologous genes from various Actinobacteria, except that in a few cases where Cys387 was replaced by serine or alanine. The corresponding bacteria, Mycobacterium avium and Mycobacterium aurum, were found to be naturally resistant to BTZ, thus supporting the identification of DprE1 as the BTZ target (Makarov et al. 2009).
Further corroboration was obtained biochemically by using membrane preparations from M. smegmatis to catalyze the epimerization reaction from radiolabeled DPR precursor (Mikusova et al. 2005), in the presence or absence of BTZ. Addition of BTZ abolished the production of DPA from DPR; furthermore, when the highly BTZ-resistant mutants of M. bovis BCG or M. smegmatis were used as sources of enzymes, epimerization was no longer inhibited, thereby confirming identification of the BTZ target (Makarov et al. 2009).
As early metabolic studies with bacteria or mice indicated that the BTZ nitro group could be reduced to an amino group, the S and R enantiomers of the amino derivatives and the likely hydroxylamine intermediate were synthesized and tested for antimycobacterial activity in vitro. The amino and hydroxylamine derivatives were substantially less active (500- to 5,000-fold) respect to the nitro form (Makarov et al. 2009).
Respecting this, a resistance mechanism to BTZs was described in M. smegmatis (Manina et al. 2010a). The overexpression of the nitroreductase NfnB leads to the inactivation of the drug by reduction of a critical nitro group to an amino group. Some M. smegmatis BTZ-resistant mutants which harbored neither mutations in MSMEG_6382 (dprE1) nor in MSMEG_6385 (dprE2), but in the MSMEG_6503 gene, coding for a putative transcriptional regulator from the TetR family were isolated. It was demonstrated that this regulator controls the transcription of the MSMEG_6505 gene, coding for NfnB enzyme. This mutation led to a defective repressor, causing overexpression of NfnB and, consequently, the reduction of the BTZ nitro molecule to its less active amino derivative (Manina et al. 2010a). To further confirm the direct role of NfnB in the BTZ resistance, an in-frame unmarked deletion was created in the nfnB gene and the ΔnfnB strain was sensitive to BTZ (Manina et al. 2010a).
Both wild-type M. smegmatis and one of the resistant mutants were evaluated for their ability to convert the nitro compound to the amino derivative, by high-pressure liquid chromatography analysis of culture media. The mutant transformed the nitro to the amino compound more efficiently and more rapidly compared with M. smegmatis wild-type strain (Manina et al. 2010a).
In order to assess the activity of the purified NfnB protein toward BTZ043, recombinant NfnB enzyme was added to the assay mixtures to monitor DPA synthesis. When BTZ043 was pretreated with purified NfnB prior to addition to the reaction mixture, DPA was still formed, most likely due to the conversion of the active drug to its inactive amino form (Manina et al. 2010a).
It has also been observed that the nitro BTZ compounds are transformed into the corresponding amino derivatives not only in M. smegmatis cultures overexpressing NfnB but also in blood and urine from treated mice (V. Makarov, unpublished data), strongly suggesting that one or more nitroreductases, either mammalian or from the intestinal microbial flora, could carry out such a conversion (Roldán et al. 2008). Interestingly, M. tuberculosis most probably lacks enzymes able to inactivate BTZ043, consistent to the low MIC values of BTZ043 in M. tuberculosis and to the fact that all the BTZ-resistant mutants isolated so far in this species harbored mutations in the target gene dprE1 (Manina et al. 2010a). However, this finding is useful for the design of new BTZ molecules or new antitubercular drugs, which may be more effective in vivo.
On the other hand, interesting information about the binding between the BTZs and their target came from Trefzer and collaborators who demonstrated that BTZs are activated by reduction of an essential nitro group to a nitroso derivative, which then specifically reacts with a cysteine residue in the active site of DprE1 (Trefzer et al. 2010) (Fig. 3). Electron-deficient nitroaromatic compounds are known to be reduced in biological systems (Spain 1995) and to readily react with thiols (Ellis et al. 1992) and also to covalently modify cysteine residues in proteins (Reeve et al. 2002; Callan et al. 2009). As the mutation of Cys387 in DprE1 leads to BTZ resistance, Trefzer et al. (2010) speculated that the nitroso derivatives of BTZs form a semimercaptal with Cys387. To test this hypothesis, they studied the reaction of a nitroso derivative of BTZs with thiols. They incubated M. smegmatis coexpressing DprE1 and DprE2 from M. tuberculosis with BTZ043 and isolated the proteins. Subsequent analysis by mass spectrometry permitted the detection of a protein with a mass which corresponds to a semimercaptal formed from the nitroso derivative of BTZ043 and DprE1. To demonstrate that the observed labeling depends on Cys387, they mutated Cys387 in DprE1 to glycine. Coexpression of this gene with dprE2 in M. smegmatis resulted in high level resistance to BTZ043, indicating that also the mutated DprE1 from M. tuberculosis is functional. Analysis of mutated DprE1 by mass spectrometry after incubation of M. smegmatis with BTZ043 allowed only the detection of unmodified DprE1. Together, these experiments support the hypothesis of the formation of a semimercaptal between Cys387 and a BTZ nitroso derivative (Trefzer et al. 2010). To independently confirm the observed covalent modification of DprE1 by BTZs, a 14C-labeled BTZ derivative was synthesized. The degree of labeling of DprE1 after purification was determined to be 25 %. When these experiments were repeated with the mutated DprE1, the degree of labeling was 20-fold lower (Trefzer et al. 2010). To gain further insight into the interaction of BTZs with DprE1, Trefzer and co-workers built a structural model of DprE1 by homology modeling with alditol oxidase, which possesses the highest sequence identity to DprE1 (18 %). In their model, Cys387 points into the substrate binding pocket of DprE1 (Trefzer et al. 2010). Thus, modification of Cys387 should block the substrate binding site of the enzyme.
An amino acid sequence alignment between NfnB and DprE1 showed a common amino acid stretch of 30 amino acids between NfnB (residues 86–115) and DprE1 (residues 386–417). It is noteworthy that this amino acid stretch is located at the C-terminal end of DprE1, the same region in which spontaneous mutations conferring resistance to BTZ were identified (Makarov et al. 2009), suggesting that this polypeptide portion might also play a key role in defining the relative specificity of NfnB toward the nitro molecules from the BTZ class (Manina et al. 2010a).
Recently, it has been demonstrated that BTZs act as suicide inhibitors of DprE1 (Trefzer et al. 2012). BTZs are reduced by DprE1 to an electrophile, which then reacts in a near quantitative manner with an active-site cysteine of DprE1, thus providing a rationale for the extraordinary potency of BTZs (Trefzer et al. 2012).
The pharmaceutical industry usually estimates the likelihood of the development of resistance against a new drug by focusing on the mutation resistance rate, assuming that this rate is a major determinant of resistance development in clinical settings (Andersson 2006). It is noteworthy that M. tuberculosis BTZ-resistant mutants isolated in vitro were rare, arising at a frequency of 10−8 (Makarov et al. 2009), and they did not show a good fitness (M.R. Pasca, unpublished data).
Moreover, a total of 240 M. tuberculosis clinical isolates from four European hospitals were surveyed for the presence of mutations in the dprE1 gene and for BTZ susceptibility (Pasca et al. 2010). These M. tuberculosis clinical isolates included MDR- and XDR-TB strains and were screened for mutations in the Cys387 codon of dprE1 and for sensitivity to BTZ. None of the M. tuberculosis isolates showed mutations in the dprE1 gene, and hence, these isolates were presumably sensitive to BTZ. This was confirmed by MIC determination, with values ranging from 0.75 to 30 ng/ml (Pasca et al. 2010). This result confirms that the currently circulating strains of M. tuberculosis do not have mutations in the BTZ target, thus confirming BTZs as very promising drugs, active against both sensitive and resistant strains, including MDR-TB and XDR-TB (Pasca et al. 2010). These results will have very important implications for future clinical trials. Moreover, it will be possible to perform an easy and rapid diagnostic test for BTZ resistance in clinical isolates simply by sequencing the dprE1 gene around the Cys387 codon or by using a real-time PCR assay. In summary, the mechanistic insights presented in this work represent an important step in the further development of BTZs as antitubercular drug candidates.
Dinitrobenzamides
A few months after the publication of BTZs as promising antitubercular drugs targeting DprE1 another new class, the DNB derivatives (Fig. 4) were identified through a screening of chemicals which interfere with M. tuberculosis replication within macrophages (Christophe et al. 2009). The assay employed is based on the use of automated confocal fluorescent microscopy to monitor intracellular growth of green fluorescent protein-expressing M. tuberculosis H37Rv in Raw264.7 macrophages (Christophe et al. 2009). The screening of a library of more than 50,000 small molecules led to the identification of 135 active and non-toxic compounds. These hit compounds had an MIC of about 70 ng/ml, which is similar to that of isoniazid (Christophe et al. 2009).
To identify the chemical substituents necessary for benzamide antibacterial activity, over 155 additional derivatives were synthesized and their structure–activity relationship was analyzed using both intracellular and in vitro growth assays. The two major compounds from this series [N-(2-(4-methoxyphenoxy) ethyl)-3,5-dinitrobenzamide] and [N-(2-(benzyloxy) ethyl)-3,5-dinitrobenzamide] named DNB1 and DNB2, respectively, were pursued further since their activities on intracellular and extracellular M. tuberculosis were particularly favorable (Fig. 4). No cell toxicity was noted for these compounds using conventional cytotoxicity assays of uninfected cells (Christophe et al. 2009). Analysis of the broad antimicrobial spectrum revealed that the effect of these DNB derivatives was mainly restricted to Actinomycetes, with the most potent activity observed against Mycobacterium with an MIC of 75 ng/ml. DNB1 and DNB2 were also highly active against MDR and XDR-TB clinical isolates. Moreover, these two compounds were also associated with low levels of spontaneous resistance. The bactericidal effect on M. tuberculosis of DNB1 and DNB2 was found to be time dependent and to require several days to reach bacterial clearance, implying that they could interfere with de novo mycobacterial component biosynthesis. This was further corroborated by the fact that the DNB compounds lost their activity in a non-replicating M. tuberculosis system (Christophe et al. 2009).
To gain insight into the possible targets of DNBs, the effect of DNB1 and DNB2 on the lipid composition of the cell envelope of M. tuberculosis was investigated; no effects on the biosynthesis of fatty acids, mycolic acids, and/or other lipids were noted. By contrast, DNB1 and DNB2 showed a clear-cut effect on the synthesis of the arabinan domains of arabinogalactan and lipoarabinomannan. The effects of DNB in the inhibition of the synthesis of DPA were tested. Analyses revealed complete inhibition of DPA formation in the DNB-treated extracts, concurrent with the accumulation of DPR, indicating that the target of DNBs could be the heteromeric decaprenylphospho-ribose epimerase encoded by the dprE1/dprE2 genes in M. tuberculosis H37Rv. Moreover, BTZ-resistant mutants of M. smegmatis and M. bovis BCG, having a mutation in dprE1 gene, were also resistant to DNBs (Christophe et al. 2009).
This hypothesis has been recently confirmed showing that the DNBs and the BTZs have not only the same target but also the same mechanisms of resistance (de Jesus Lopes Ribeiro et al. 2011). In particular, to better understand the mechanism of resistance to DNBs, several spontaneous M. smegmatis mutants resistant to N-(2-(3-chlorobenzyloxy)ethyl)-3,5-dinitrobenzamide (DNB3) were isolated. DNB3 was chosen because of its higher solubility in solid medium in respect to the other DNBs derivatives. The spontaneous mutants exhibited two different resistance levels to DNB3. The first series of M. smegmatis mutants showed a high level of resistance to DNBs (800-fold the MIC for the wild-type strain) and the second series a lower level of resistance (64–128-fold the MIC). The possible cross-resistance between DNBs and BTZs was checked and demonstrated for all M. smegmatis mutants. All the DNB-resistant mutants were screened for the presence of mutations in MSMEG_6382 gene (M. tuberculosis dprE1 ortholog). All the M. smegmatis mutants with a higher level of resistance to DNB had a point mutation at codon 394 of MSMEG_6382 gene, as previously observed for M. smegmatis mutants resistant to BTZ (Makarov et al. 2009). This position (Cys394) corresponds to Cys387 position in M. tuberculosis dprE1 gene. All mutants turned out to have the substitution Cys394Gly or Cys394Ser in DprE1. These data support the idea that Cys394 is involved in the binding of DNB, as previously suggested for BTZ (Makarov et al. 2009). To obtain direct evidence on the binding of DNBs to DprE1, the interactions between the two counterparts were investigated monitoring the change in fluorescence of the enzyme bound FAD, in the presence of different ligand concentrations. The intensity of the fluorescence was enhanced by the ligand, showing a saturation curve. Moreover, no alteration in fluorescence was found when dimethyl sulfoxide, the solvent of DNBs, was used alone. This experiment showed that DprE1 binds DNB compounds with high affinity, thus demonstrating unambiguously that DprE1 is the target of DNBs (de Jesus Lopes Ribeiro et al. 2011).
The mutants less resistant to DNB did not show mutations nor in MSMEG_6382 or in MSMEG_6385 (dprE2 ortholog), thus suggesting that their resistance should be due to a different molecular mechanism. All these mutants showed a mutation in the MSMEG_6503 gene, encoding the nfnB regulator (see the previous paragraph) (de Jesus Lopes Ribeiro et al. 2011). The overexpression of the nitroreductase NfnB, caused by the altered repressor, accounts for the resistance to this new class of antitubercular drugs, as shown by real-time PCR experiments. In fact, a statistically significant increase in transcription of nfnB was observed in these mutants with respect to the wild-type strain (de Jesus Lopes Ribeiro et al. 2011). Moreover, the purified enzyme was evaluated for its ability to convert the nitro form of DNB1 into the corresponding hydroxylamine and/or amino derivatives by LC/MS2 analysis. Indeed NfnB was able to convert DNB1 into the corresponding hydroxylamine derivative, but the DNB amino derivatives were not detected in these experimental settings (de Jesus Lopes Ribeiro et al. 2011). On the other hand, previous studies performed on M. smegmatis extracts incubated with DNBs evidenced the production of both amino and hydroxylamine derivative forms of the drug, as in the case of BTZ compounds (Christophe et al. 2009; Manina et al. 2010a). Moreover, the in vitro synthesis of the DNB amino derivative and hydroxylamine-form was successful, and these forms of DNB were not effective against mycobacteria, similarly to BTZ derivatives (Makarov et al. 2009; Christophe et al. 2009; Manina et al. 2010a). In conclusion, de Jesus Lopes Ribeiro et al. (2011) showed that DNBs and BTZs have the same target and the same mechanisms of resistance.
Benzoquinoxalines
The last class of compounds known to target the M. tuberculosis DprE1 is the benzoquinoxalines. In this case, to find antimycobacterial scaffolds, a kinase inhibitor library of more than 12,000 compounds from Vichem Chemie Ltd. was screened using an integrated strategy involving whole-cell-based assays and a target-based assay with the protein kinase PknA (Magnet et al. 2010). In fact, signaling pathways in Prokaryotes are also controlled by protein kinases. Moreover, a few examples of compounds derived from protein kinase pharmacophores have been shown to inhibit non-kinase antibacterial targets, such as d-alanine–d-alanine ligase (Triola et al. 2009) or biotin carboxylase kinase (Miller et al. 2009). Thus, kinase inhibitor libraries can potentially be a source of inhibitors for a wide range of bacterial enzymes.
Of the 12,100 compounds tested, more than 200 showed activity against C. glutamicum of which 17 also displayed activity against M. tuberculosis. These 17 compounds were tested for inhibition of PknA and PknB. None of them specifically inhibited the serine/threonine protein kinases activity. They were further examined for their potential genotoxic and cytotoxic activities, and their MIC against M. tuberculosis H37Rv was determined. Among these, only three compounds were found to be non-mutagenic, non-cytotoxic, and displayed an MIC <10 mM against M. tuberculosis H37Rv (Magnet et al. 2010).
Among the compounds active on M. tuberculosis, the structure of VI-9376 (2-methyl-3-phenyl-5-nitro-7-bromoquinoxaline, Fig. 5) partly resembled that of the BTZs and DNBs. For this reason, VI-9376 was tested against several BTZ-resistant mutants of mycobacteria and C. glutamicum. The MIC results revealed cross-resistance between the BTZ lead compound, BTZ043, and VI-9376. Forty derivatives of VI-9376 were subsequently synthesized and tested against M. tuberculosis H37Rv. The structure–activity relationship and MIC data obtained for the derivatives showed that the nitro group at the fifth position of the quinoxaline scaffold is absolutely required for activity. Replacement of the bromine at the fourth position by a trifluoromethyl group increased the potency of the scaffold, while modifications at position 2 or position 3 did not lead to a significant improvement of activity against M. tuberculosis (Magnet et al. 2010).
In conclusion, three independent whole-cell-based screens have now identified the compounds targeting DprE1 recently both in vitro and ex vivo (Makarov et al. 2009; Christophe et al. 2009; Magnet et al. 2010). On the whole, these results highlight the vulnerability of DprE1 and its importance as an antimycobacterial target and show that whole-cell screens can overcome the difficulties associated with the development of high throughput assays using membrane-bound proteins or enzymes that use complex substrates, which until now are not commercially available, as is the case with DprE1.
Conclusions
The inexorable rise in cases of TB worldwide highlights the need for new drugs and, in particular, for those that can shorten the duration of treatment. In fact, vaccination is only partially successful, being the only current available vaccine against M. tuberculosis and M. bovis BCG, effective only in childhood (Colditz et al. 1994, 1995) for a limited number of years (Andersen and Doherty 2005). The aim of the Stop TB Partnership is to reduce the annual incidence of new TB cases to less than one per million people by 2050. In fact, these ambitious goals will only be achieved by the combined introduction of new diagnostics, drugs, and vaccines (Dye and Williams 2008).
The development of new drugs is urgently needed due to the increasing number of M. tuberculosis MDR, XDR, and TDR strains that fail to respond to first- and second-line drug treatment (Dye 2006; Gandhi Neel et al. 2006; Cegielski 2010). Recently, three new promising antitubercular drugs were discovered: the benzothiazinones, the dinitrobenzamides, and the benzoquinoxalines (Makarov et al. 2009; Christophe et al. 2009; Magnet et al. 2010). All these drugs are in preclinical studies (Cole and Riccardi 2011) and have the same cellular target, the DprE1 enzyme from M. tuberculosis. A Cys residue, probably near the active site, is covalently modified by these drugs, and this may prevent substrate access to the enzyme. Mutants or natural variants lacking the cysteine residue are resistant to these compounds. This highlights the vulnerability of DprE1 and its importance as an antitubercular target which can be exploited in structure-assisted drug design to find new pharmacophores which, hopefully, could result essential for the treatment of this disease.
References
Aagaard C, Hoang T, Dietrich J, Cardona PJ, Izzo A, Dolganov G, Schoolnik GK, Cassidy JP, Billeskov R, Andersen P (2011) A multistage tuberculosis vaccine that confers efficient protection before and after exposure. Nature Med 17:189–194
Andersen P, Doherty TM (2005) The success and failure of BCG—implications for a novel tuberculosis vaccine. Nature Rev Microbiol 3:656–662
Andersen P, Doherty TM, Pai M, Weldingh K (2007) The prognosis of latent tuberculosis: can disease be predicted? Trends Mol Med 13:175–182
Andersson DI (2006) The biological cost of mutational antibiotic resistance: any practical conclusions? Curr Opin Microbiol 9:461–465
Berry MP, Graham CM, McNab FW, Xu Z, Bloch SA, Oni T, Wilkinson KA, Banchereau R, Skinner J, Wilkinson RJ, Quinn C, Blankenship D, Dhawan R, Cush JJ, Mejias A, Ramilo O, Kon OM, Pascual V, Banchereau J, Chaussabel D, O’Garra A (2010) An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature 466:973–977
Callan HE, Jenkins RE, Maggs JL, Lavergne SN, Clarke SE, Naisbitt DJ, Park BK (2009) Multiple adduction reactions of nitroso sulfamethoxazole with cysteinyl residues of peptides and proteins: implications for hapten formation. Chem Res Toxicol 22:937–948
Carroll P, Faray-Kele M-C, Parish T (2011) Identifying vulnerable pathways in Mycobacterium tuberculosis by using a knockdown approach. Appl Environ Microbiol 77:5040–5043
Cegielski JP (2010) Extensively drug-resistant tuberculosis: “there must be some kind of way out of here”. Clin Infect Dis 50:S195–S200
Christophe T, Jackson M, Jeon HK, Fenistein D, Contreras-Dominguez M, Kim J, Genovesio A, Carralot JP, Ewann F, Kim EH, Lee SY, Kang S, Seo MJ, Park EJ, Skovierová H, Pham H, Riccardi G, Nam JY, Marsollier L, Kempf M, Joly-Guillou ML, Oh T, Shin WK, No Z, Nehrbass U, Brosch R, Cole ST, Brodin P (2009) High content screening identifies decaprenyl-phosphoribose 2′ epimerase as a target for intracellular antimycobacterial inhibitors. PLoS Pathogens 5:e1000645
Colditz GA, Brewer TF, Berkey CS, Wilson ME, Burdick E, Fineberg HV, Mosteller F (1994) Efficacy of BCG vaccine in the prevention of tuberculosis. Meta-analysis of the published literature. JAMA 271:698–702
Colditz GA, Berkey CS, Mosteller F, Brewer TF, Wilson ME, Burdick E, Fineberg HV (1995) The efficacy of Bacillus Calmette–Guerin vaccination of newborns and infants in the prevention of tuberculosis: meta-analyses of the published literature. Pediatrics 96:29–35
Cole ST, Riccardi G (2011) New tuberculosis drugs on the horizon. Curr Op Microbiol 14:570–576
Crellin PK, Brammananth R, Coppel RL (2011) Decaprenylphosphoryl-β-D-ribose 2′-epimerase, the target of benzothiazinones and dinitrobenzamides, is an essential enzyme in Mycobacterium smegmatis. PLoS One 6:e16869
de Jesus Lopes Ribeiro AL, Degiacomi G, Ewann F, Buroni S, Incandela ML, Chiarelli LR, Mori G, Kim J, Contreras-Dominguez M, Park Y-S, Han S-J, Brodin P, Valentini G, Rizzi M, Riccardi G, Pasca MR (2011) Analogous mechanisms of resistance to benzothiazinones and dinitrobenzamides in Mycobacterium smegmatis. PLoS One 6:e26675
Dietrich J, Doherty TM (2009) Interaction of Mycobacterium tuberculosis with the host: consequences for vaccine development. APMIS 117:440–457
Donald PR, Schaaf HS (2007) Old and new drugs for the treatment of tuberculosis in children. Paediatr Resp Rev 8:134–141
Dye C (2006) Global epidemiology of tuberculosis. Lancet 367:938–940
Dye C, Williams BG (2008) Eliminating human tuberculosis in the twenty-first century. J R Soc Interface 5:653–662
Ellis MK, Hill S, Foster PMD (1992) Reactions of nitrosonitrobenzenes with biological thiols: identification and reactivity of glutathione-S-yl conjugates. Chem Biol Interact 82:151–163
Gandhi Neel R, Moll A, Sturm AW, Pawinski R, Govender T, Lalloo U, Zeller K, Andrews J, Friedland G (2006) Extensively drug-resistant tuberculosis as a cause of death in patients co-infected with tuberculosis and HIV in a rural area of South Africa. Lancet 368:1575–1580
Girardi E, Raviglione MC, Antonucci G, Godfrey-Faussett P, Ippolito G (2000) Impact of the HIV epidemic on the spread of other diseases: the case of tuberculosis. AIDS 14:S47–S56
Kochi A (2001) The global tuberculosis situation and the new control strategy of the World Health Organization 1991. Bull World Health Organ 79:71–75
Maertzdorf J, Repsilber D, Parida SK, Stanley K, Roberts T, Black G, Walzl G, Kaufmann SH (2011) Human gene expression profiles of susceptibility and resistance in tuberculosis. Genes Immun 12:15–22
Magnet S, Hartkoorn RC, Székely R, Pató J, Triccas JA, Schneider P, Szántai-Kis C, Orfi L, Chambon M, Banfi D, Bueno M, Turcatti G, Kéri G, Cole ST (2010) Leads for antitubercular compounds from kinase inhibitor library screens. Tuberculosis (Edinb) 90:354–360
Makarov V, Riabova OB, Yuschenko A, Urlyapova N, Daudova A, Zipfel PF, Möllmann U (2006) Synthesis and antileprosy activity of some dialkyldithiocarbamates. J Antimicrob Chemother 57:1134–1138
Makarov V, Manina G, Mikušová K, Möllmann U, Ryabova O, Saint-Joanis B, Dhar N, Pasca MR, Buroni S, Lucarelli AP, Milano A, De Rossi E, Belanova M, Bobovska A, Dianišková P, Kordulakova J, Sala C, Fullam E, Schneider P, McKinney JD, Brodin P, Christophe T, Waddell S, Butcher P, Albrethsen J, Rosenkrands I, Brosch R, Nandi V, Bharath S, Gaonkar S, Shandil RK, Balasubramanian V, Balganesh T, Tyagi S, Grosset J, Riccardi G, Cole ST (2009) Benzothiazinones kill Mycobacterium tuberculosis by blocking arabinan synthesis. Science 324:801–804
Manabe YC, Bishai WR (2000) Latent Mycobacterium tuberculosis—persistence, patience, and winning by waiting. Nature Med 6:1327–1329
Manina G, Bellinzoni M, Pasca MR, Neres J, Milano A, Ribeiro AL, Buroni S, Skovierová H, Dianišková P, Mikušová K, Marák J, Makarov V, Giganti D, Haouz A, Lucarelli AP, Degiacomi G, Piazza A, Chiarelli LR, De Rossi E, Salina E, Cole ST, Alzari PM, Riccardi G (2010a) Biological and structural characterization of the Mycobacterium smegmatis nitroreductase NfnB, and its role in benzothiazinone resistance. Mol Microbiol 77:1172–1185
Manina G, Pasca MR, Buroni S, De Rossi E, Riccardi G (2010b) Decaprenylphosphoryl-β-D-ribose 2′-epimerase from Mycobacterium tuberculosis is a magic drug target. Curr Med Chem 17:3099–3108
Meniche X, de Sousa-d’Auria C, Van-der-Rest B, Bhamidi S, Huc E, Huang H, De Paepe D, Tropis M, McNeil M, Daffé M, Houssin C (2008) Partial redundancy in the synthesis of the D-arabinose incorporated in the cell wall arabinan of Corynebacterineae. Microbiology 154:2315–2326
Mikusova K, Huang H, Yagi T, Holsters M, Vereecke D, D’Haeze W, Scherman MS, Brennan PJ, McNeil MR, Crick DC (2005) Decaprenylphosphoryl arabinofuranose, the donor of the D-arabinofuranosyl residues of mycobacterial arabinan, is formed via a two-step epimerization of decaprenylphosphoryl ribose. J Bacteriol 187:8020–8025
Miller JR, Dunham S, Mochalkin I, Banotai C, Bowman M, Buist S, Dunkle B, Hanna D, Harwood HJ, Huband MD, Karnovsky A, Kuhn M, Limberakis C, Liu JY, Mehrens S, Mueller WT, Narasimhan L, Ogden A, Ohren J, Prasad JV, Shelly JA, Skerlos L, Sulavik M, Thomas VH, VanderRoest S, Wang L, Wang Z, Whitton A, Zhu T, Stover CK (2009) A class of selective antibacterials derived from a protein kinase inhibitor pharmacophore. Proc Natl Acad Sci USA 106:1737–1742
Morrison J, Pai M, Hopewell PC (2008) Tuberculosis and latent tuberculosis infection in close contacts of people with pulmonary tuberculosis in low-income and middle-income countries: a systematic review and meta-analysis. Lancet Infect Dis 8:359–368
Pasca MR, Degiacomi G, Ribeiro AL, Zara F, De Mori P, Heym B, Mirrione M, Brerra R, Pagani L, Pucillo L, Troupioti P, Makarov V, Cole ST, Riccardi G (2010) Clinical isolates of Mycobacterium tuberculosis in four European hospitals are uniformly susceptible to benzothiazinones. Antimicrob Agents Chemother 54:1616–1618
Rappuoli R, Aderem AA (2011) 2020 vision for vaccines against HIV, tuberculosis and malaria. Nature 26:463–469
Reeve IT, Voss JC, Miller MG (2002) 1,3-Dinitrobenzene metabolism and GSH depletivo. Chem Res Toxicol 15:361–366
Riccardi G, Pasca MR, Buroni S (2009) Mycobacterium tuberculosis: drug resistance and future perspectives. Fut Microbiol 4:597–614
Roldán MD, Pérez-Reinado E, Castillo F, Moreno-Vivián C (2008) Reduction of polynitroaromatic compounds: the bacterial nitroreductases. FEMS Microbiol Rev 32:474–500
Rowland K (2012) Totally drug-resistant TB emerges in India. Nature. doi:10.1038/nature.2012.9797
Sassetti CM, Rubin EJ (2003) Genetic requirements for mycobacterial survival during infection. Proc Natl Acad Sci USA 100:12989–12994
Selwyn PA, Hartel D, Wasserman W, Drucker E (1989) Impact of the AIDS epidemic on morbidity and mortality among intravenous drug users in a New York City methadone maintenance program. Am J Public Health 79:1358–1362
Spain JC (1995) Biodegradation of nitroaromatic compounds. Ann Rev Microbiol 49:523–555
Stewart GR, Robertson BD, Young DB (2003) Tuberculosis: a problem with persistence. Nature Rev Microbiol 1:97–105
Trefzer C, Rengifo-Gonzalez M, Hinner MJ, Schneider P, Makarov V, Cole ST, Johnsson K (2010) Benzothiazinones: prodrugs that covalently modify the decaprenylphosphoryl-β-D-ribose 2′-epimerase DprE1 of Mycobacterium tuberculosis. J Am Chem Soc 132:13663–13665
Trefzer C, Skovierová H, Buroni S, Bobovská A, Nenci S, Molteni E, Pojer F, Pasca MR, Makarov V, Cole ST, Riccardi G, Mikušová K, Johnsson K (2012) Benzothiazinones are suicide inhibitors of mycobacterial decaprenylphosphoryl-β-d-ribofuranose 2′-oxidase DprE1. J Am Chem Soc 134:912–915
Triola G, Wetzel S, Ellinger B, Koch MA, Hubel K, Rauh D, Waldmann H (2009) ATP competitive inhibitors of D-alanine-D-alanine ligase based on protein kinase inhibitor scaffolds. Bioorg Med Chem 17:1079–1087
Wolucka BA (2008) Biosynthesis of D-arabinose in mycobacteria—a novel bacterial pathway with implications for antimycobacterial therapy. FEBS J 275:2691–2711
WHO (2011) www.who.int/mediacentre/factsheets/fs104/en/ Accessed March 2012
Acknowledgments
We thank the European Commission for support (VI Framework, contract no. LSHP-CT 2005-018923 and VII Framework, contract no. 260872).
Author information
Authors and Affiliations
Corresponding author
Additional information
This article has been retracted due to copyright issues that cannot be resolved.
About this article
Cite this article
Buroni, S., Pasca, M.R., de Jesus Lopes Ribeiro, A.L. et al. RETRACTED ARTICLE: Antituberculars which target decaprenylphosphoryl-β-D-ribofuranose 2′-oxidase DprE1: state of art. Appl Microbiol Biotechnol 94, 907–916 (2012). https://doi.org/10.1007/s00253-012-4013-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-012-4013-4