Abstract
The suitability of three β-galactosidases as reporter enzymes for promoter expression analyses was investigated in Bacillus subtilis with respect to various temperature conditions during cultivation and assay procedures. Starting from the hypothesis that proteins derived from diverse habitats have different advantages as reporters at different growth temperatures, the beta-galactosidases from the thermophilic organism Bacillus stearothermophilus, from the mesophilic bacterium Escherichia coli and from the psychrophilic organism Pseudoalteromonas haloplanktis TAE79 were analysed under control of the constitutive B. subtilis lepA promoter. Subsequent expression of the β-galactosidase genes and determination of specific activities was performed at different cultivation and assay temperatures using B. subtilis as host. Surprisingly, the obtained results demonstrated that the highest activities over a broad cultivation temperature range were obtained using the β-galactosidase from the mesophilic bacterium E. coli whereas the enzymes from the thermophilic and psychrophilic bacteria revealed a more restricted usability in terms of cultivation temperature.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Reporter genes are important and useful tools for investigations of promoter activities and strength, translational efficiency or of localization and stability of proteins. Furthermore, reporter genes are often used to determine gene regulatory mechanisms (e.g. enhancer sequences and transcription factors). Thus, reporter genes have become indispensable for applications in various fields of research like molecular biology, biochemistry, pharmaceutical and biomedical studies (Martin et al. 2009; Debnath et al. 2010; Wang et al. 2002; Naylor 1999). The selection of a reporter gene requires the consideration of several criteria: (1) The reporter or a comparable enzymatic activity should not be present in the host organism. (2) Ideally, the reporter enzyme activity should reliably reflect the promoter strength. (3) Simple, fast, sensitive and reproducible assays should be available. (4) The physiology of the host cell should not be influenced by reporter gene expression (Kain and Ganguly 2001; Lee and Hruby 1997). Subjected to the presence of a signal peptide, reporter enzymes can be produced intracellularly or be secreted. Intracellular expression for instance offers the advantage of direct localization of cellular components, whereas cell extract preparation is avoided and gene expression profiles can be easily monitored by continuous analysis of the culture medium when the reporter protein is secreted (Alam and Cook 1990).
To date, many different reporter genes have been characterised and are used for various purposes. Among those commonly used reporter enzymes are the chloramphenicol acetyltransferase, the green fluorescent protein (GFP), the human growth hormone (hGH), an alkaline phosphatase, luciferases of Photinus pyralis and Renilla reniformis (Luc and Ruc), a β-glucuronidase (GUS) and β-galactosidases. Each of these reporter genes exhibits advantages for specific applications. Proteins like GFP, Ruc and Luc are good candidates for in vivo studies where the localisation and interactions of DNA and proteins are investigated. In vivo process studies offer the advantage that cellular processes can be visualised in real time without cell damages or excessive sampling (Kavita and Burma 2008). Another advantage for the application of GFP is that no exogenous substrates are required for fluorescence (Chalfie et al. 1994; Phillips 2001). Some of the commonly used reporter enzymes are preferentially applied in certain organisms, e.g. GUS is an excellent reporter for plant cells (Basu et al. 2004) whereas hGH is mostly used in mammalian systems (Alam and Cook 1990; Kain and Ganguly 2001).
Among the best characterised and most frequently used reporter enzymes are β-galactosidases, which are present in a variety of organisms (Hoyoux et al. 2001). β-galactosidases belong to the family of glycosylhydrolases (Coombs and Brenchley 1999) and catalyse the hydrolysis of β-galactosides into monosaccharides (Matthews 2005). In this context, the application of β-galactosidases for the hydrolysation of lactose into glucose and galactose plays a major role in dairy processes (Chen et al. 2008). Within this group of enzymes, the β-galactosidase of Escherichia coli, the product of the lacZ gene described by Jacob and Monod (1961), is the most commonly used reporter protein (Jacobson et al. 1994). In 2001, Hoyoux and coworkers isolated and characterised a β-galactosidase from the psychrophilic organism Pseudoalteromonas haloplanktis TAE79. With respect to protein folding and catalytic mechanisms this enzyme is adapted to low temperatures whereas the β-galactosidase from the thermophilic organism Bacillus stearothermophilus displays typical properties of a thermostable enzyme and is therefore characterised by an adaptation to higher temperatures (Chen et al. 2008). Based on these characteristic differences, we hypothesised that the β-galactosidases may have different advantages as reporter enzymes when subjected to variations in cultivation and assay temperatures. We compared the β-galactosidases derived from the psychrophilic marine bacterium P. haloplanktis TAE79, the mesophilic intestine bacterium E. coli and the thermophilic organism B. stearothermophilus in order to investigate their potential as reporter candidates for Bacillus subtilis expression systems. B. subtilis is an attractive host for protein production since it is well adapted to moderate and even low growth temperatures and can secrete large amounts of relevant proteins (Nijland and Kuipers 2008). Furthermore, B. subtilis is classified as GRAS organism (Generally Regarded As Safe), which is relevant with respect to production of active pharmaceutical ingredients or in the fields of food industry where the absence of pathogenic or virulence factors is essential. To approach comparable expression levels of the genes encoding the distinct β-galactosidase proteins of P. haloplanktis TAE79, E. coli and B. stearothermophilus in the host B. subtilis, the three genes were placed under control of the constitutive promoter of the B. subtilis lepA gene (Homuth et al. 1996; Hippler et al. 1997).
Materials and methods
Unless stated otherwise, all chemicals were purchased from Roth (Karlsruhe, Germany), Sigma (Steinheim, Germany) and Fluka (Buchs, Switzerland) at the highest purity available and were used without further purification. B. subtilis strain ATCC 6051 (American Type Culture Collection) was used for expression experiments. E. coli DH10b (Invitrogen, Darmstadt, Germany) [F- endA1 recA1 galE15 galK16 nupG rpsL ΔlacX74 Φ80lacZΔM15 araD139 Δ(ara,leu)7697 mcrA Δ(mrr-hsdRMS-mcrBC) λ-] was used as the host strain for all cloning procedures. All strains constructed in this study as well as the host organisms of the respective galactosidase enzymes are listed in Table 1.
Recombinant DNA technologies, genes and vectors
All routine DNA technologies were carried out according to standard protocols (Sambrook and Russell 2001). Restriction enzymes and other DNA-modifying enzymes were used as specified by the suppliers (Roche, Mannheim, Germany, New England Biolabs, Frankfurt a. M., Germany). DNA sequencing was carried out by LGC Genomics (Berlin, Germany). Oligonuceotides were synthesised and provided by Invitrogen (Darmstadt, Germany).
To investigate the suitability of different β-galactosidases as reporter enzymes in B. subtilis expression systems, the β-galactosidase-encoding genes of E. coli (UNIPROT accession number P00722), B. stearothermophilus (UNIPROT accession number P19668) and P. haloplanktis TAE79 (UNIPROT accession number P81650) were cloned into the vector backbone of the pbgaB plasmid (Mogk et al. 1996) under control of the constitutive B. subtilis lepA promoter.
Plasmid and strain constructions
In order to construct the appropriate expression vectors with the thermophilic β-galactosidase from B. stearothermophilus, the mesophilic β-galactosidase from E. coli and the cold-adapted β-galactosidase from P. haloplanktis TAE79, the backbone of the previously described plasmid pbgaB, harbouring the coding sequence of the B. stearothermophilus β-galactosidase gene (Mogk et al. 1996), was used for all cloning procedures. For integration of the B. subtilis lepA promoter region, B. subtilis chromosomal DNA was isolated using the High Pure Template Preparation Kit (Roche, Mannheim, Germany). The promoter fragment was amplified by polymerase chain reaction (PCR) using Phusion DNA-polymerase (New England Biolabs, Frankfurt a.M., Germany) with oligonucleotides lepA-forward 5′-ACGTGGATCCAGTGCTTACTGCTTGTCACG-3′ and lepA-reverse 5′-GAATTCCATAATCTATCACTCCTACTATT-3′ (restriction sites are underlined). PCR product digestion with BamHI and EcoRI and subsequent purification of the promoter fragment was carried out using the Qiaquick Gel Extraction Kit (Qiagen, Hilden, Germany). The purified promoter fragment was then ligated with the BamHI- and EcoRI-restricted vector pbgaB, and E. coli DH10B was transformed with the resulting recombinant plasmid (Fig. 1), yielding strain E. coli B.st.bgaB (Table 1). Correct insertion of the lepA promoter fragment was verified by sequencing. The resulting plasmid was designated pP lepA -B.st.bgaB.
The nucleotide sequence of the β-galactosidase gene from P. haloplanktis TAE79 was synthesised and provided by GENEART GmbH (Regensburg, Germany). For this purpose, the amino acid sequence was obtained from the UNIPROT database (accession number P81650), and the synthetic gene was codon-optimised for B. subtilis using the GeneOptimizer® software (GENEART). The algorithm utilises a matrix for the most frequently occurring codons in B. subtilis. The construct was assembled from synthetic oligonucleotides and cloned into the pP lepA -B.st.bgaB backbone using the EcoRI and XbaI restriction sites, thereby replacing the bgaB coding sequence with the synthetic gene construct of the β-galactosidase gene from P. haloplanktis TAE79. The recombinant plasmid was used for transformation of E. coli DH10B, yielding strain E. coli P.h.lacZ, and the sequence identity was validated. The resulting plasmid was designated pP lepA -P.h.lacZ.
For the E. coli lacZ construct, the E. coli lacZ gene was amplified from the pAC7 vector (Weinrauch et al. 1991) by PCR using Phusion DNA-polymerase (New England Biolabs, Frankfurt a.M., Germany) with oligonucleotides lacZ-forward 5′-ACGTATGAATTCGTCGTTTTACAACGTCGTG-3′ and lacZ-reverse 5′-ACGTAGTCTAGATTATTTTTGACACCAGACCAAC-3′ (restriction sites are underlined). After digestion of the PCR product with EcoRI and XbaI, the E. coli lacZ gene was gel-purified and ligated with the EcoRI- and XbaI-restricted vector pP lepA -B.st.bgaB. With this construction, the vector-based bgaB coding sequence was replaced with the coding sequence of the E. coli lacZ gene. After transformation of E. coli DH10B with the recombinant plasmid, yielding strain E. coli E.c.lacZ, the correct insertion of the lacZ gene was verified by sequencing. Finally, all three expression plasmids pP lepA -B.st.bgaB, pP lepA -P.h.lacZ and pP lepA -E.c.lacZ shared the same vector backbone with the identical promoter region and transcriptional terminator sequence and differed solely in the coding β-galactosidase sequences.
The resulting plasmids were isolated from the appropriate E. coli host strain, linearised with ScaI and integrated at the amyE locus of the B. subtilis ATCC 6051 chromosome by a double-crossover event. Successful integration was verified by screening the resulting neomycin-resistant clones for an alpha-amylase negative phenotype on agar plates containing 1.0% (w/v) starch and by PCR.
Calculation of B. subtilis codon adaptation index
The codon adaptation index (CAI) is a measure of codon usage within a gene relative to a reference set of genes. Values were calculated using the CAI Calculator (Puigbo et al. 2008 ; http://genomes.urv.es/CAIcal/) with the ß-galactosidase sequences described above and a codon usage table for B. subtilis (www.kazusa.or.jp).
Cell cultivation and harvesting
Cells were cultivated in Luria Bertani medium [yeast extract (5 g L−1), peptone (10 g L−1) and NaCl (10 g L−1)] supplemented with the appropriate antibiotics. For constitutive expression of β-galactosidases, all strains were grown continuously at 15°C, 20°C, 37°C and 50°C. Samples for β-galactosidase assays were collected at an optical density (OD578nm) of ~0.5 and 15, 30 and 60 min later. The harvested cells were subsequently pelleted at 13,000 rpm for 5 min at 4°C; the supernatant was discarded, and the cell pellets were stored at −20°C until further analysis. Samples for protein preparation were collected at the same time points, normalised by 5/OD and centrifuged for 5 min at 4°C and 8,500 rpm. The supernatant was discarded, and the pellet was washed with 10 mL of lysis buffer I (50 mM NaH2PO4; 300 mM NaCl, pH 8.0) and centrifuged. The cell pellet was then resuspended in 300 μL of Lysis buffer I and stored at −20 °C until further analysis. Samples for RNA isolation were collected at OD 0.5 (t 0) and 60 minutes later (t 60). Cells were harvested in half of the sample volume ice-cold killing buffer (20 mM NaN3; 20 mM Tris–HCL, pH 7.5; 5 mM MgCl2) and centrifuged for 5 min and 8,500 rpm at 4°C. The supernatant was discarded, and the cell pellets were immediately frozen in liquid nitrogen and stored at −70°C.
For further investigation of temperature profiles of recombinant β-galactosidases, all strains were cultivated close to the respective identified optimal conditions of the individual β-galactosidases to an optical density (OD578nm) of ~1.0. Samples for protein preparation were collected and treated as described above.
Protein extract preparation and enzyme assays
Total cellular protein was isolated by mechanical disruption of the cells using the RiboLyser Fast Prep Fp 120 (Thermo Electron, Dreieich, Germany) in lysis buffer I for 45 s at 4.5 m s−1. Afterwards, the samples were centrifuged twice at 4°C for 15 min and 13,000 rpm. The supernatant was transferred into a new Eppendorf tube, and the β-galactosidase assay was carried out immediately. For each single measurement, 50 μL of the protein extract were mixed with 384 μL of Z-buffer (60 mM Na2HPO4; 40 mM NaH2PO4; 10 mM KCl; 1 mM MgSO4, pH 7.5) and incubated for 10 min at the respective assay temperature. The reaction was started by addition of 66 μL of the substrate ONPG (ortho-nitrophenyl-β-galactopyranoside) and stopped with 500 μL of 1 M Na2CO3. Absorption at 420 nm was measured using the Ultrospec 3000 (GE Healthcare, Freiburg, Germany), and the specific β-galactosidase activities were calculated on the basis of an extinction coefficient for ONPG at pH 10 of 4.86 mM−1 cm−1. One unit of β-galactosidase activity was defined as the amount of enzyme that hydrolyzes 1 μmol of ONPG per minute, and specific β-galactosidase activities were expressed in units per milligram of cellular protein. Protein concentrations were determined using the Bradford method with BSA (0.1 mg mL-1) as the standard (Bradford 1976). In parallel, β-galactosidase activities were carried out using the cell pellets as described earlier (Prágai and Harwood 2002). All analyses were done in triplicate.
RNA isolation
Total RNA of B. subtilis was isolated using the KingFisher mL™ pipetting robot (Thermo Electron, Dreieich, Germany) and the MagNa Pure LC RNA Isolation Kit (Roche, Mannheim, Germany). For this purpose, cell pellets were resuspended in lysis buffer II (3 mM EDTA; 200 mM NaCl), and mechanical cell disruption was performed using the RiboLyser at 6.5 m s−1 for 45 s. The ribolyser tubes contained glass beads (0.10–0.11 mm diameter, Sartorius, Göttingen, Germany) and acidic phenol chloroform isoamylalcohol (50:48:2). RNA was separated from cell debris by centrifugation of the tubes for 5 min and 13,000 rpm at room temperature. The upper RNA-containing phase was transferred into a new Eppendorf tube for subsequent isolation and purification with the KingFisher mL™ as described earlier (Jürgen et al. 2005). Purity and concentration of the RNA preparations were determined using the Nanodrop ND 1000 UV/Vis spectrophotometer (Peqlab, Erlangen, Germany) as recommended by the manufacturers.
Northern blot analysis
Northern blot analysis was performed as described by Homuth et al. (1996). Complete and even RNA transfer onto the membrane was verified by visualisation of both the ethidiumbromide-stained RNA gel and the membrane after blotting. After hybridisation of the blotted RNA samples with the appropriate RNA probes, chemiluminescence was detected using the Vilber Lourmat Fusion-SL-3500-WL (Eberhardzell, Germany). Labeling of the gene-specific probes was done using the DIG RNA Labeling Kit (SP6/T7) from Roche (Mannheim, Germany) following the manufacturer´s instructions. The following oligonucleotides were used for PCR synthesis. For E. coli lacZ: lacZ-probe-for 5′-AGGAAGGCCAGACGCGAATT-3′ and lacZ-probe-rev 5′-CTAATACGACTCACTATAGGGAGAAACCACCACGCTCATCGATA-3′, for B. stearothermophilus bgaB: bgaB-probe-for 5′-ATTCTCTCGTTTGGCAGTAG-3′ and bgaB-probe-rev 5′-CTAATACGACTCACTATAGGGAGATGAAATTAGTTGATACTGG-3′, and for P. haloplanktis lacZ: TAE-probe-for 5′-CAGAATTTGATCTGTCAGAACTT-3′ and TAE-probe-rev 5′-CTAATACGACTCACTATAGGGAGAATCCAGCAGTGAAACAACGCATC-3′ (the sequence of the T7 promoter is underlined).
Results
In order to investigate the applicability of distinct β-galactosidases as reporter enzymes in B. subtilis expression systems, we integrated the genes encoding for β-galactosidases from three different organisms into the chromosome of the host strain B. subtilis ATCC 6051. The β-galactosidase genes from E. coli and B. stearothermophilus were expressed with their original nucleotide sequences whereas the P. haloplanktis gene was codon-optimised before chemical synthesis. Finally, all expression constructs used in this study were preceded by the constitutive promoter of the B. subtilis lepA gene and differed solely in the coding sequence for the specific β-galactosidase genes. The resulting codon adaptation indexes (referring to the most frequently used codons in B. subtilis) were 0.98 for P. haloplanktis lacZ, 0.80 for E. coli lacZ and 0.76 for bgaB.
mRNA analysis and comparison with β-galactosidase activities
Since the intracellular level of a given bacterial protein is predicted to be at least partially reflected by the amounts of the corresponding specific mRNA, we performed Northern analyses to investigate the transcript levels specified by the three different β-galactosidase genes. The samples were taken at 15°C, 20°C, 37°C and 50°C at an optical density of 0.5 (t 0) and 1 h later (t 60) as shown in Fig. 2. For this purpose, electrophoretically separated RNA samples that were blotted to nylon membranes were hybridised with the respective gene-specific β-galactosidase probes. As illustrated in Fig. 2a, chemiluminographs for all three ectopically inserted genes could be detected at all expression temperatures for the samples taken at an optical density of 0.5 (t 0). However, the detected signal intensities exhibited diverse patterns within each strain at different cultivation temperatures, which did not correlate in all cases with the measured β-galactosidase activities summarised in Fig. 3.
Northern blot analyses (a) of B. subtilis strains expressing the cold-adapted β-galactosidase from P. haloplanktis TAE79 (P.h.lacZ), the mesophilic β-galactosidase from E. coli (E.c.lacZ) and the thermostable β-galactosidase from B. stearothermophilus (B.st.bgaB) using gene-specific RNA probes. RNA preparations were electrophoretically separated in a 1.5% agarose gel (b). Each lane contained 10 μg of total RNA, and samples were taken at OD578nm of 0.5 (Co, left lane) and 60 min later (60 min, right lane) at the respective cultivation temperature
Influence of cultivation- and assay-temperatures on specific β-galactosidase activities of the cold-adapted β-galactosidase from P. haloplanktis TAE79, the mesophilic β-galactosidase from E. coli and the thermophilic β-galactosidase from B. stearothermophilus. Cultivation of the strains was done continuously at 15°C, 20°C, 37°C and 50°C. Samples for determination of enzyme activity were taken at OD578nm 0.5 and 15, 30 and 60 min later. The enzyme assay was performed at 15°C, 37°C and 50°C. Specific β-galactosidase activities are drawn as average values from three independent cultivations; standard deviations are presented as error bars
The mRNA specified by the E.c.lacZ gene was detected in highest amounts at the low growth temperature of 20°C at t 0 as well as 60 min later, whereas the transcript was present in clearly decreased amounts at t 0 when the cells were cultivated at 37°C and 50°C and was barely detectable 60 min later (Fig. 2a). In contrast to that, the highest specific β-galactosidase activities—even significantly higher compared with cells cultivated at 20°C—were measured for both time points when the cells were grown at 37°C (Fig. 3). At 50°C cultivation temperature, there was a dramatic drop in β-galactosidase activity, although the mRNA pattern was identical with the one observed at 37°C, which co-occurred with the described high activities.
The measured specific β-galactosidase activities of P.h.LacZ were clearly higher at 15°C than at 20°C at all time points, which is also contradictory to the observed transcriptional pattern: While the largest amounts of P.h.lacZ specific mRNA were detected at t 0 and t 60 when cells were cultivated at 20°C, the amount of P.h.lacZ transcript was clearly lower at 15°C growth temperature, whereby the signal slightly increased between t 0 and t 60. At 37°C and 50°C, the mRNA pattern was very similar to the one observed for E.c.lacZ at these temperatures: The P.h.lacZ transcript was present in clearly decreased amounts as compared with cells grown at 20°C at t 0 and was barely detectable 60 min later (Fig. 2a). There was nearly no measurable β-galactosidase activity at 37°C and 50°C.
The Northern analysis of the bgaB expression revealed distinct signals at all time points and all cultivation temperatures, with the exception of a slight increase in the mRNA amount between t 0 and t 60 at 15°C and a pronounced decrease of the mRNA amount between t 0 and t 60 at 37°C. However, the measured β-galactosidase activities at 15°C and 20°C were very low, and only moderately higher when cells were grown at 37°C, whereas the clearly highest activities were measured at 50°C cultivation temperature.
For all Northern analyses, the uniform loading of equal amounts of total RNA per gel lane and the absence of degradation of the RNA samples was validated by ethidium bromide staining of the RNA gels before the vacuum blotting. One representative example is shown in Fig. 2b.
Influence of cultivation and assay temperatures on measured β-galactosidase activities
Cultivation of the three expression strains at all four growth temperatures resulted in measurable reporter enzyme activities, indicating that all three enzymes where produced in an active conformation in the host B. subtilis (Fig. 3). The determined maximal β-galactosidase activities at the respective strain-specific optimal cultivation and assay temperatures amounted to 43.1 U mg−1 for the cold-adapted β-galactosidase P.h.LacZ, 627.5 U mg−1 for the mesophilic enzyme E.c.LacZ and 61.0 U mg−1 for the thermostable enzyme BgaB.
Our investigations on the influence of the cultivation temperature upon the measured activities exhibited significant differences between the three expression strains. As illustrated in Fig. 3, the highest β-galactosidase activity in the strain producing the enzyme from the psychrophilic organism P. haloplanktis TAE79 was obtained when the strain was cultivated at 15°C (43.1 U mg−1) and, slightly lower, at 20°C (28.3 U mg−1) whereas the activity dropped dramatically at higher growth temperatures (<2.5 U mg−1 at 37°C and 50°C). For the strain producing the β-galactosidase from the mesophilic organism E. coli, we observed the maximum activity values at a cultivation temperature of 37°C (627.5 U mg−1) with a slight decrease at lower cultivation temperatures (469.3 U mg−1 at 20°C and 290.1 U mg−1 at 15°C) and a quite pronounced drop at a cultivation temperature of 50°C (119.5 U mg−1). The producer strain of the B. stearothermophilus β-galactosidase displayed the highest activities (61.0 U mg−1) during cultivation at 50°C which was the maximum temperature applied in this study. The activity decreased continuously at lower growth temperatures (29.0 U mg−1 at 37°C, 12.4 U mg−1 at 20°C and 9.3 U mg−1 at 15°C). Determination of specific β-galactosidase activities using the method of Prágai and Harwood (2002) resulted in highly similar measured enzyme activities (data not shown).
Besides the cultivation temperatures, the applied assay temperatures also strongly affected the measured activities of the β-galactosidases produced by the three strains investigated in this study (Fig. 3). For the strain producing the β-galactosidase of P. haloplanktis TAE79, the highest specific activities were obtained at assay temperatures of 37°C and, significantly reduced, at 15°C, whereas the β-galactosidase activity drastically dropped when the enzyme assay was performed at 50°C. In the case of the E. coli LacZ producer strain, the most suitable assay temperature was 37°C, but very similar results were obtained when an assay temperature of 50°C was applied. The latter observation was in sharp contrast to the described strong reduction in β-galactosidase activity at 50°C assay temperature observed for the strain producing the cold-adapted enzyme of P. haloplanktis TAE79. Clearly, lower activities were measured when the assays were performed at 15°C. The highest values for the strain producing the BgaB enzyme were obtained at an assay temperature of 50°C. It became clear that temperatures of 15°C and 37°C are not suitable for assaying the specific activity of this enzyme since a very pronounced reduction in activity at these temperatures was observed.
Derived temperature profiles of recombinant β-galactosidases
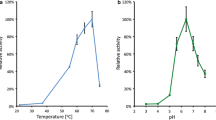
In an independent experiment, temperature profiles in the range from 5°C to 50°C where recorded using protein extracts prepared from the three β-galactosidase producer strains close to the respective individual enzyme-specific optimal cultivation temperatures. The applied cultivation temperatures were 20°C for the cold-adapted enzyme of P. haloplanktis, 37 °C for the E. coli β-galactosidase and 50°C for the BgaB enzyme of B. stearothermophilus. The profiles obtained for the strains producing the P.h.LacZ and the E.c.LacZ enzymes show bell-shaped curve characteristics, whereas the profile from BgaB reflects a maximum of activity beyond the investigated range. The temperature profile obtained with the protein extract from the strain producing the cold-adapted enzyme P.h.LacZ was characterised by a continuous increase of the specific β-galactosidase activity up to the maximum of 33.8 U mg−1 at 35°C and by a rapid drop at 50°C (Fig. 4). The residual activity at 50°C amounted to only 9.9% of the activity measured at 35°C, whereas the corresponding percentage at 45°C still amounted to 81.5%. The profile generated using the extract prepared from the strain producing the LacZ enzyme of E. coli exhibited a temperature optimum at 40°C and showed a linear increase starting from 5°C. The decrease of the activity to a residual value of 72.3% at 50°C as compared with the maximum temperature measured at 40°C was much less distinctive than in the case of the extracts containing the cold-adapted enzyme of P. haloplanktis TAE79. The extract prepared from the BgaB enzyme exhibited completely different profile characteristics. Here, the activity increased continuously at temperatures above 30°C. Below this temperature, the enzyme is obviously significantly less active than the cold-adapted and the mesophilic enzyme. The maximum activity measured with BgaB-containing extracts was obtained at 50°C in our experiments. However, as this was the highest temperature applied, no clear temperature optimum could be determined.
Temperature profile of the β-galactosidases from P. haloplanktis TAE79 (a), E. coli (b) and B. stearothermophilus (c). Specific β-galactosidase activity is expressed in units per milligram, whereas the relative activity refers to the respective maximum activity of each β-galactosidase in the investigated temperature range from 5°C to 50°C. Specific enzyme activities are drawn as average values from three independent cultivations; standard deviations are presented as error bars
Discussion
In this study, we compared three β-galactosidase genes originating from a psychrophilic, a mesophilic and a thermophilic host in order to investigate their usability as reporter enzymes for expression analyses at different cultivation temperatures. For this purpose, the genes were placed under the control of the promoter of the B. subtilis lepA gene that has been described to be constitutively active under several different conditions during the exponential growth phase (Hippler et al. 1997). As the transcriptional termination of all three genes is mediated by the same downstream terminator structure present in the pbgaB vector backbone and as all three genes share, besides the lepA promoter, also the 5´ leader sequence of the B. subtilis lepA gene including the Shine-Dalgarno sequence, the three constructs exclusively differ in their coding sequences. The three β-galactosidases originated from the psychrophilic marine bacterium P. haloplanktis TAE79, the mesophilic bacterium E. coli and the thermophilic bacterium B. stearothermophilus. The natural habitats of these organisms differ significantly with respect to their surrounding temperatures: The ambient temperature in the cold marine environment of P. haloplanktis TAE79 averages −1°C (Hoyoux et al. 2001), whereas the habitat for B. stearothermophilus with a growth temperature between 50°C and 60°C is thermophilic (Nazina et al. 2001). The intestinal bacterium E. coli grows well at moderate temperatures around 37°C as a consequence of adaptation to its mammalian host. Due to the respective specific natural habitat, individual enzyme properties like overall folding state, topology and stability differ significantly, and adaptations in the enzyme structure are necessary to ensure activity under defined conditions (Coombs and Brenchley 1999). As suggested by Brenchley (1996), enzymes may have evolved within limited temperature ranges and, as a consequence, catalytic activity is provided either at high or low temperatures, but not at both. These features may influence the ability of the investigated β-galactosidases as reporter enzymes, especially at different cultivation temperatures. The amino acid sequence identity between the β-galactosidases of P. haloplanktis TAE79 (1,038 aa) and of E. coli (1,023 aa) is 51% (Hoyoux et al. 2001). For the BgaB enzyme (672 aa), there is no significant overall amino acid homology when compared with β-galactosidases from E. coli, P. haloplanktis, Klebsiella pneumoniae or Kluyveromyces lactis (Hirata et al. 1986). However, the highest partial homology (21% in a 52 aa overlap) is observed when the amino acid sequence is compared with the β-galactosidase of E. coli. The reporter enzymes investigated in this study could also be predicted to demonstrate diversity on the DNA adaptation level since different organisms exhibit a specific individual codon usage and are equipped with an adequate corresponding tRNA population. As a consequence, the codon usage of heterologously expressed genes often differs significantly from the codon usage of the host (Kurland and Gallant 1996). To solve this problem, codon optimisation is a general approach to improve heterologous gene expression in foreign hosts (Wu et al. 2006). For expression of the β-galactosidase of P. haloplanktis TAE79, we used a synthetic gene, which was optimised for expression in B. subtilis. Hereby, the original CAI of the P. haloplanktis galactosidase was increased from 0.77 to 0.98, which is supposed to facilitate higher translation rates in B. subtilis. The calculated CAI values for E. coli lacZ and bgaB were slightly lower (0.80 and 076), but still in a proper range.
The transcript patterns of all three β-galactosidase-specifying mRNAs as determined by Northern analysis were generally similar: The highest mRNA amounts were observed at 15°C and 20°C cultivation temperature, as well at t 0 (Optical Density (OD)578nm ~ 0.5) as at t 60 (1 h later). At the higher cultivation temperatures of 37°C and 50°C, there were pronounced differences between the strains: In the case of those expressing the lacZ genes of P. haloplanktis TAE79 and E. coli, there was much less mRNA present at t 0 as compared with 20°C cultivation temperature, and this amount even strongly dropped at t 60. In the strain expressing the B. stearothermophilus bgaB gene, the mRNA amount at t 0 and 37°C was comparable to the amounts observed at 20°C; however, it also clearly decreased at t 60. Most surprisingly, there was no difference between the bgaB mRNA amounts at t 0 and t 60 at 50°C cultivation temperature, in this case, a drop in the mRNA amount at t 60 was not observed. The cellular net amount of a given mRNA molecule is determined by two major factors: The mRNA synthesis rate mediated primarily by the strength of the particular promoter and the specific mRNA stability modulated primarily by the accessibility of the mRNA 5′- and 3′-ends for ribonucleases (Bechhofer and Zen 1989; Bechhofer 2009; Makrides 1996). As already mentioned, the three recombinant β-galactosidase genes used in this study differed solely in their coding sequences, which means that their expression was driven by the same promoter (the lepA promoter) and that they shared the same 5′- and 3′-leader regions. Thereby, it is tempting to speculate that the partially similar transcript profiles of the mRNAs specifying the three different β-galactosidases are caused by the combined effects of varying lepA promoter activities and varying mRNA stabilities.
It has to be emphasised that, at 37°C and at 50°C, cells were fully exponentially growing at t 0 (OD578nm ~ 0.5) whereas, at t 60, they already approached the transition state. At 15°C and at 20°C, there were no growth rate differences between t 0 and t 60 because, at these lower cultivation temperatures and resulting slower growth rates, cells did not yet approach the transition state at t 60 (Fig. 5). Therefore, the drastic drop in the mRNA amount observed for all three transcripts between t 0 and t 60 at 37°C is most probably caused by decreased lepA promoter activity or/and decreased mRNA stability as consequences of the physiological changes associated with the beginning transition from exponential growth to the stationary phase. The lepA gene encodes a ribosomal elongation factor that recognises ribosomes after a defective translocation reaction and induces a back-translocation, thus giving EF-G a second chance to translocate the tRNAs correctly (Qin et al. 2006). Therefore, downregulation of the lepA expression might be part of an adaptation to the stationary phase when the complete cellular protein synthesis machinery is strongly curtailed and might be mediated by reduced activity of the lepA promoter. Interestingly, at 50°C, the bgaB-specific mRNA amount did not exhibit the dramatic decrease at t 60. It can be speculated that the protein coding sequence of this particular mRNA might be adapted to this high temperature as a consequence of the evolution of B. stearothermophilus in its original habitat characterised by high temperatures and therefore exhibits a significantly higher stability at this temperature. Specific structural properties of the resulting mRNA might for instance impede its recognition by specific ribonucleases. The relatively large amounts of mRNA at 20°C could be explained by increased mRNA stability at this lower cultivation temperature, where exoribonuclease-impeding secondary structures, like the one located downstream of the β-galactosidase-coding sequences, are stabilised, and the enzymatic activity of ribonucleases is generally decreased, whereas the lepA promoter might still be fairly active. The mRNA amounts at 15°C that are reduced compared with those detected at 20°C might primarily result from a strongly reduced lepA promoter activity at this low temperature as an adaptation to the slow growth, which exceeds the effect of the increased mRNA stability that might cause the increased net amount in mRNA at 20°C. Of course, a final clear definition of the precise contributions of mRNA stability and promoter strength at the different cultivation temperatures and time points would require the determination of the respective mRNA stabilities. However, such a comprehensive analysis was beyond the scope of this study where the major question concerned the practical usability of the different reporter enzymes under the different conditions tested.
Growth characteristics of B. subtilis E.c.lacZ at 15°C (black circles), 20°C (white circles), 37°C (black triangles) and 50°C (white triangles). Start and end points of sampling for each applied temperature are indicated by t 0 and t 60, respectively. Growth curve characteristics are representative for the strains B. subtilis P.h.lacZ and B. subtilis B.st.bgaB as well
For the β-galactosidase of P. haloplanktis TAE79, the highest activities were measured when the enzyme assay was performed at 37°C, whereas they were clearly lower when the assay was performed at higher (50°C) or lower (15°C) temperatures (Fig. 6). This finding is somewhat surprising in view of the fact that the habitat of P. haloplanktis TAE79 is characterised by an average temperature of −1°C (Hoyoux et al. 2001). The result was independent from the respective cultivation temperature of the corresponding expression strain from which the protein extracts were prepared. However, an optimal temperature of around 40°C for enzyme activity was also reported for a cold-adapted β-galactosidase synthesised by another psychrophilic P. haloplanktis strain (isolate 22b), and this enzyme shares 90% sequence identity and similar properties to the investigated enzyme of P. haloplanktis TAE79 (Cieśliński et al. 2005). On the other hand, comparison of the different cultivation temperatures revealed that the maximal enzyme activities were obtained when the expression strain was grown at 15°C or, slightly higher, at 20°C, without significant differences between t 0 and t 60, respectively. The amount of P. haloplanktis TAE79 β-galactosidase-specific mRNA was clearly lower at 15°C as compared with 20°C. Apparently, the relatively low amount of mRNA present at 15°C corresponded to a very high enzyme activity that nearly reached the highest detected P. haloplanktis TAE79 β-galactosidase activity at 20°C, where the mRNA amount was clearly higher. These observations strongly indicate that the enzyme is maximally active in the host B. subtilis when synthesised at 15°C.
Dependency of enzyme activities on cultivation and assay temperatures. The optimal conditions for specific β-galactosidase activities of the cold-adapted β-galactosidase from P. haloplanktis TAE79 (P.h.LacZ), the mesophilic β-galactosidase from E. coli (E.c.LacZ) and the thermophilic β-galactosidase from B. stearothermophilus (B.st.BgaB) are illustrated in red colours. Suboptimal cultivation and assay conditions are characterised by transition into green and blue colours
As illustrated in Fig. 6, the β-galactosidase encoded by the lacZ gene of E. coli exhibited the highest activities when the enzyme assay was carried out at 37°C and 50°C, whereas the activities were clearly lower at 15°C. When the different cultivation temperatures were compared, it turned out that maximal enzyme activities were measured when the expression strain was grown at 37°C. The activities were slightly lower at 20°C cultivation temperature and exhibited a more pronounced decrease at 15°C. Most impressive was a drastic drop observed at a cultivation temperature of 50°C. Because the measured enzyme activities obtained with the protein extracts prepared from the lacZ expression strain cultivated at 37°C were very similar when the enzyme assay was performed at 37°C or 50°C, it can be concluded that the breakdown of the β-galactosidase activity observed at 50°C cultivation temperature is not caused by thermal denaturation (i.e. unfolding) of the E. coli enzyme. Most probably, cultivation at 50°C results in the induction of a heat-inducible proteolytic activity mediating the degradation of the E. coli β-galactosidase in B. subtilis, which is in accordance with other findings described in the literature (Zuber and Schumann 1994). The highest enzyme activity, without significant differences between t 0 and t 60, was measured at a temperature (37°C) where the amount of E. coli lacZ-specific mRNA was much lower than at 15°C and 20°C. This suggests that the enzyme is indeed maximally active in the host B. subtilis when synthesised at 37°C. Considering the comparable mRNA profile and the similar enzyme activities measured when the protein extract prepared from cells cultivated at 37°C was assayed at 50°C, the initial enzyme activity might be similar at 37°C and 50°C cultivation temperature. However, due to the obvious heat-induced degradation of the enzyme, this remains unclear.
For the BgaB enzyme, the β-galactosidase of B. stearothermophilus, the clearly highest activities were measured when the enzyme assay was performed at 50°C (Fig. 6). The activities were drastically reduced when the assays were carried out at 37°C and even more pronounced at 15°C, underscoring the thermophilic origin of the BgaB enzyme. Similarly, the highest activities were measured when the expression strain was cultivated at 50°C, whereas activity strongly decreased at all lower temperatures. Since temperature profiles were recorded from 5–50°C the optimal temperature for enzyme activity was outside the investigated range. However, a temperature optimum of 70°C was recently proposed for BgaB, with the measured β-galactosidase activity amounting values twice as high compared with those at an assay temperature of 55°C (Chen et al. 2008). The bgaB-specific mRNA amounts did not show strong variations between the t 0 time points at the different cultivation temperatures, although the corresponding measured enzyme activities varied dramatically, with the described optimum at 50°C cultivation temperature. It can therefore be presumed that the BgaB enzyme is maximally active in the host B. subtilis when synthesised at 50°C, which was the maximum temperature applied in this study.
In summary, simultaneously considering the associated specific mRNA levels, the β-galactosidases of P. haloplanktis TAE79, E. coli and B. stearothermophilus exhibited maximal activities in the heterogeneous host B. subtilis when synthesised at 15°C, 37°C and 50°C, respectively and when assayed at 37°C (P.h.lacZ), 37°C or 50°C (E.c.lacZ), and 50°C (B.st.bgaB), respectively. However, even under the optimal conditions of the P. haloplanktis TAE79 enzyme (15°C cultivation temperature and 37°C assay temperature), its maximally measured activities (around 40 U mg−1) were clearly lower than the corresponding activities of the E. coli β-galactosidase (>200 U mg−1), which unambiguously demonstrates that the E. coli enzyme represents a much better suited reporter system for low and intermediate temperatures in the host B. subtilis than the P. haloplanktis TAE79 enzyme. At intermediate cultivation temperature (37°C) and when assayed at 37°C or 50°C, the E. coli β-galactosidase also exhibited much higher activities (around 400 U mg−1) than the enzyme from B. stearothermophilus (maximally around 30 U mg−1 when assayed at 50°C). However, cultivation of the lacZ expression strain at 50°C resulted in the described drastic near-total loss of β-galactosidase activity, whereas the B. stearothermophilus enzyme reached its maximal activities (40–60 U mg–1) in the B. subtilis host under these conditions. Using purified enzymes in in vitro assays, maximal β-galactosidase activities of around 1,200, 138, and 125 U mg–1 were measured for the enzymes from E. coli (Sigma Aldrich protein catalogue no. G5635), P. haloplanktis TAE79 (Hoyoux et al. 2001) and B. stearothermophilus (Chen et al. 2008), respectively. The corresponding values in our study amounted to around 600, 40 and 60 U mg–1. The observation that in all cases expression in B. subtilis resulted in lower maximal activities is supposedly primarily based on the fact that in our study non-purified enzymes were used to determine β-galactosidase activities.
Finally, it can be concluded that, for the host B. subtilis, the E. coli β-galactosidase might represent the best suited reporter enzyme for low and intermediate cultivation temperatures, whereas, at high temperatures, as described earlier (Chen et al. 2008; Hirata et al. 1985), the B. stearothermophilus enzyme indeed represents the first choice.
References
Alam J, Cook JL (1990) Reporter genes: application to the study of mammalian gene transcription. Anal Biochem 188:245–254
Basu C, Kausch AP, Chandlee JM (2004) Use of beta-glucuronidase reporter gene for gene expression analysis in turfgrasses. Biochem Biophys Res Commun 320:7–10
Bechhofer DH (2009) Messenger RNA decay and maturation in Bacillus subtilis. Prog Mol Biol Transl Sci 85:231–273
Bechhofer DH, Zen KH (1989) Mechanism of erythromycin-induced ermC mRNA stability in Bacillus subtilis. J Bacteriol 171:5803–5811
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Brenchley J (1996) Psychrophilic microorganisms and their cold-active enzymes. J Ind Microbiol Biotechnol 17:432–437
Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC (1994) Green fluorescent protein as a marker for gene expression. Science (New York, NY) 263:802–805
Chen W, Chen H, Xia Y, Zhao J, Tian F, Zhang H (2008) Production, purification, and characterization of a potential thermostable galactosidase for milk lactose hydrolysis from Bacillus stearothermophilus. J Dairy Sci 91:1751–1758
Cieśliński H, Kur J, Białkowska A, Baran I, Makowski K, Turkiewicz M (2005) Cloning, expression, and purification of a recombinant cold-adapted beta-galactosidase from Antarctic bacterium Pseudoalteromonas sp. 22b. Protein Expression Purif 39:27–34
Coombs JM, Brenchley JE (1999) Biochemical and phylogenetic analyses of a cold-active β-galactosidase from the lactic acid bacterium Carnobacterium piscicola BA. Appl Environ Microbiol 65:5443–5450
Debnath M, Prasad GBKS, Bisen PS (2010) Molecular diagnostics: promises and possibilities. Springer, New York, 520
Hippler B, Homuth G, Hoffmann T, Hungerer C, Schumann W, Jahn D (1997) Characterization of Bacillus subtilis hemN. J Bacteriol 179:7181–7185
Hirata H, Fukazawa T, Negoro S, Okada H (1986) Structure of a beta-galactosidase gene of Bacillus stearothermophilus. J Bacteriol 166:722–727
Hirata H, Negoro S, Okada H (1985) High production of thermostable {beta}-galactosidase of Bacillus stearothermophilus in Bacillus subtilis. Appl Environ Microbiol 49:1547–1549
Homuth G, Heinemann M, Zuber U, Schumann W (1996) The genes of lepA and hemN form a bicistronic operon in Bacillus subtilis. Microbiology 142(Pt 7):1641–1649
Hoyoux A, Jennes I, Dubois P, Genicot S, Dubail F, François JM, Baise E, Feller G, Gerday C (2001) Cold-adapted β-galactosidase from the Antarctic psychrophile Pseudoalteromonas haloplanktis. Appl Environ Microbiol 67:1529–1535
Jacob F, Monod J (1961) Genetic regulatory mechanisms in the synthesis of proteins. J Mol Biol 3:318–356
Jacobson RH, Zhang XJ, DuBose RF, Matthews BW (1994) Three-dimensional structure of beta-galactosidase from E. coli. Nature 369:761–766
Jürgen B, Barken KB, Tobisch S, Pioch D, Wümpelmann M, Hecker M, Schweder T (2005) Application of an electric DNA-chip for the expression analysis of bioprocess-relevant marker genes of Bacillus subtilis. Biotechnol Bioeng 92:299–307
Kain SR, Ganguly S (2001) Uses of fusion genes in mammalian transfection. In: Current protocols in molecular biology. Wiley & Sons, Inc., Hoboken, NJ, USA
Kavita P, Burma PK (2008) A comparative analysis of green fluorescent protein and beta-glucuronidase protein-encoding genes as a reporter system for studying the temporal expression profiles of promoters. J Biosci (Bangalore) 33:337–343
Kurland C, Gallant J (1996) Errors of heterologous protein expression. Curr Opin Biotechnol 7:489–493
Lee P, Hruby DE (1997) Detection of recombinant protein based on reporter enzyme activity: chloramphenicol acetyltransferase. Methods Mol Biol (Clifton, NJ) 63:31–40
Makrides SC (1996) Strategies for achieving high-level expression of genes in Escherichia coli. Microbiol Rev 60:512–538
Martin L, Che A, Endy D (2009) Gemini, a bifunctional enzymatic and fluorescent reporter of gene expression. PLoS One 4:e7569–e7569
Matthews BW (2005) The structure of E. coli beta-galactosidase. C R Biol 328:549–556
Mogk A, Hayward R, Schumann W (1996) Integrative vectors for constructing single-copy transcriptional fusions between Bacillus subtilis promoters and various reporter genes encoding heat-stable enzymes. Gene 182:33–36
Naylor LH (1999) Reporter gene technology: the future looks bright. Biochem Pharmacol 58:749–757
Nazina TN, Tourova TP, Poltaraus AB, Novikova EV, Grigoryan AA, Ivanova AE, Lysenko AM, Petrunyaka VV, Osipov GA, Belyaev SS, Ivanov MV (2001) Taxonomic study of aerobic thermophilic bacilli: descriptions of Geobacillus subterraneus gen. nov., sp. nov. and Geobacillus uzenensis sp. nov. from petroleum reservoirs and transfer of Bacillus stearothermophilus, Bacillus thermocatenulatus, Bacillus thermoleovorans, Bacillus kaustophilus, Bacillus thermodenitrificans to Geobacillus as the new combinations G. stearothermophilus, G. th. Int J Syst Evol Microbiol 51:433–446
Nijland R, Kuipers OP (2008) Optimization of protein secretion by Bacillus subtilis. Recent Patents Biotechnol 2:79–87
Phillips GJ (2001) Green fluorescent protein—a bright idea for the study of bacterial protein localization. FEMS Microbiol Lett 204:9–18
Prágai Z, Harwood CR (2002) Regulatory interactions between the Pho and sigma(B)-dependent general stress regulons of Bacillus subtilis. Microbiology 148:1593–1602
Puigbo P, Bravo I, Garcia-Vallve S (2008) CAIcal: a combined set of tools to assess codon usage adaptation. Biol Direct 3:38–38
Qin Y, Polacek N, Vesper O, Staub E, Einfeldt E, Wilson DN, Nierhaus KH (2006) The highly conserved LepA is a ribosomal elongation factor that back-translocates the ribosome. Cell 127:721–733
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual. CSHL
Wang Y, Yu YA, Shabahang S, Wang G, Szalay AA (2002) Renilla luciferase-Aequorea GFP (Ruc-GFP) fusion protein, a novel dual reporter for real-time imaging of gene expression in cell cultures and in live animals. Mol Genet Genom MGG 268:160–168
Weinrauch Y, Msadek T, Kunst F, Dubnau D (1991) Sequence and properties of comQ, a new competence regulatory gene of Bacillus subtilis. J Bacteriol 173:5685–5693
Wu G, Bashir-Bello N, Freeland SJ (2006) The synthetic gene designer: a flexible web platform to explore sequence manipulation for heterologous expression. Protein Expression Purif 47:441–445
Zuber U, Schumann W (1994) CIRCE, a novel heat shock element involved in regulation of heat shock operon dnaK of Bacillus subtilis. J Bacteriol 176:1359–1363
Acknowledgements
We thank Jana Priebe for excellent technical assistance and Stephanie Markert for critical reading of the manuscript. This work was partially supported by funds of the Competence Network “BioIndustrie 2021-Biokatalyse2021” financed by the German Federal Ministry of Education and Research (BMBF).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Welsch, N., Homuth, G. & Schweder, T. Suitability of different β-galactosidases as reporter enzymes in Bacillus subtilis . Appl Microbiol Biotechnol 93, 381–392 (2012). https://doi.org/10.1007/s00253-011-3645-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-011-3645-0