Abstract
To test the dose effect of ammonium (NH4 +) fertilization on soil methane (CH4) oxidation by methanotrophic communities, batch incubations were conducted at a wide scale of NH4 + amendments: 0, 100, 250, 500, and 1,000 mg N kgdry soil −1. Denaturing gradient gel electrophoresis and real-time quantitative PCR analysis were conducted to investigate the correlation between the CH4 oxidation capacity and methanotrophic communities. Immediately after the addition of NH4 +, temporal inhibition of CH4 oxidation occurred, and this might have been due to the non-specific salt effect (osmotic stress). After a lag phase, the CH4 oxidation rates of the soils with NH4 + fertilization were promoted to levels higher than those of the controls. More than 100 mg N kgdry soil −1 of NH4 + addition resulted in the reduction of type II/type I MOB ratios and an obvious evolution of type II MOB communities, while less than 100 mg N kgdry soil −1 of NH4 + addition induced nearly no change of methanotrophic community compositions. The NH4 +-derived stimulation after the lag phase was attributed to the improvement of N availability for type I MOB. Compared with the controls, 100 mg N kgdry soil −1 of NH4 + addition doubled the CH4 oxidation peak value to more than 20 mg CH4 kgdry soil −1 h−1. Therefore, an appropriate amount of leachate irrigation on the landfill cover layer might efficiently mitigate the CH4 emissions.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Methane (CH4) is an important trace gas in atmospheric chemistry for its high global warming potential. The atmospheric concentration of CH4 has increased from a pre-industrial value of 715 to 1,774 ppb recently, mainly due to human activity (Fletcher et al. 2004). In the presence of oxygen (O2), soil can act as a sink for CH4 through the metabolism of methane-oxidizing bacteria (MOB), which use CH4 as sole carbon and energy source. According to the taxonomy and physiology characteristics, MOB were generally separated into two groups (Hanson and Hanson 1996): type I, including the genera Methylococcus, Methylocaldum, Methylomonas, Methylomicrobium, Methylobacter, Methylosphaera, and Methylosarcina, and type II, including the genera Methylocystis, Methylosinus, Methylocella, and Methylocapsa, belonging to gamma- and alpha-subdivision of the phylum Proteobacteria. Type I MOB assimilate the formaldehyde for biosynthesis by using the ribulose monophosphate (RuMP) pathway, while type II MOB are using the serine pathway. Since the RuMP pathway was more efficient than the serine one, type I MOB can yield more cells than type II MOB in pure and mixed cultures (de Viries et al. 1990; Graham et al. 1993). Compared with type I, type II MOB can survive in a nitrogen- and copper-limited environment (Graham et al. 1993; Murrell 1994), which may be due to the nitrogen fixation capacity of type II MOB. Type I MOB outcompete type II MOB under high O2 and low CH4 conditions, whereas the latter favor the inverse conditions (Graham et al. 1993). In addition, CH4 can be co-oxidized by ammonia-oxidizing bacteria (AOB) as well (Hanson and Hanson 1996) due to the broad substrate specificity of ammonia monooxygenase.
As one of the necessary nutrients for microorganisms, nitrogen (N) can affect methanotrophic activities and subsequently interfere with the capacity of soil CH4 oxidation. Nevertheless, the effect of ammonium (NH4 +)-based fertilization on CH4 oxidation is still far from conclusively understood. On one hand, inhibition has been observed in various environments and is generally attributed to the substrate competition between NH4 + and CH4 for the active site of methane monooxygenase (MMO) and the product toxicity of nitrite generated from NH4 + oxidation (Steudler et al. 1989; King 1990; King and Schnell 1994). On the other hand, NH4 + application can result in stimulation as reported in rice fields (Bodelier et al. 2000b; Kruger and Frenzel 2003; Noll et al. 2008), landfill cover soils (de Visscher et al. 1999), as well as in spruce forest soils (Rigler and Zechmeister-Boltenstern 1999). The metabolism of stimulation was still unclear, and the most accepted explanation was that added NH4 + relieved the N limitation status of the investigated methanotrophs. Different amounts of NH4 + fertilization contradict the effects on CH4 oxidation, i.e., 10–60 mM of NH4 +, corresponding to 14–62 mg N kgdry soil −1, stimulated the CH4 oxidation in soil slurry, but those higher NH4 + concentrations were inhibitory (Bender and Conrad 1995). Rigler and Zechmeister-Boltenstern (1999) investigated the effects of NH4 + fertilization on two kinds of forest soils. Their results showed that the CH4 oxidation rate in the deciduous forest soil was negatively correlated with the amount of NH4 + added (from 0 to 500 mg N kgdry soil −1), while for spruce forest soils, a progressive increase of stimulation of CH4 oxidation along with an enhanced NH4 + concentration (from 0, 10 to 100 mg N kgdry soil −1) was observed.
It is hypothesized that the diverse methanotrophic compositions in different soils and the changes of methanotrophic compositions affected by NH4 + fertilization induced the above-mentioned contradictions (Mohanty et al. 2006). Based on denaturing gradient gel electrophoresis (DGGE) analysis, Seghers et al. (2003) investigated the methanotrophic community structures in the soils receiving organic (compost) and mineral (NH4NO3) fertilizer, and Zheng et al. (2008) studied the soils without and with N (urea), NK (KCl), NP (superphosphate) K, and NPK+C (recycled crop residues) fertilizer. In their studies, the contradicting effects were in consistent with the distinct methanotrophic community compositions with different types of fertilization. Zheng et al. (2008) also used quantitative polymerase chain reaction (PCR) to detect the MOB abundance during different N fertilization and found that NK and NPK+C fertilizations stimulated the increase of MOB abundance in comparison with the inhibition effect with only N (urea) fertilization. However, few studies have been done to investigate the evolvement of methanotrophic communities in the soils receiving different doses of NH4 + fertilizer.
To test the dose effect of NH4 + fertilization on soil CH4 oxidation capacity, we cultured soil samples at a wide range of NH4 + concentrations in batch incubation. In the course of incubation, the CH4 oxidation rates were monitored to represent methanotrophic activities, and real-time quantitative PCR and PCR-DGGE targeting the 16S rDNA gene were applied to evaluate the abundances and community compositions of methanotrophs.
Material and methods
Batch incubation experiments
Soil samples used in this study were collected from the top cover soil (10–30 cm) of a landfill site with subsurface irrigation of leachate (referring to the condition B in Yu et al. 2009) in eastern China in May 2009. After sampling, the soils were air-dried and stored at room temperature until usage. The soil was composed of 32.8% sand, 27.9% silt, and 39.3% clay (sandy loam according to USDA classification). Other properties were as follows: Soil pH was 4.9 (1:2.5 soil–water extraction), organic matter 0.82 ± 0.05% (organic carbon content multiplied by 1.74), total N 0.08% (Kjeldahl method), and 8.2 and 17.9 mg N kgdry soil −1 of NH4 + and nitrate (NO3 −), respectively (both extracted by 2 mol L−1 of KCl followed by standard analysis methods).
The soils were sieved through 2-mm screens to remove large particulate matters; then, 0, 100, 250, 500 to 1,000 mg N kgdry soil −1 of NH4 + were applied to the soil samples (denoted as CON, N100, N250, N500, and N1000, respectively) by rewetting them to water content of 16% (w w −1) with appropriate (NH4)2SO4 solution. Three hundred grams of soil samples was transferred to 1-L serum bottles, which were sealed by rubber stoppers and injected with CH4 to obtain 5% in the headspace. The headspace of incubation bottles was flushed daily with fresh air and re-injected with CH4. When the CH4 oxidation rate was higher than 12 mg CH4 kgdry soil −1 h−1, the flush-reinjection process was conducted every 12 h. All experiments were carried out in duplicate at 25°C. Meanwhile, to test the abiotic methane oxidation, a negative control experiment was set up with autoclaved soils in parallel.
The CH4 oxidation rates were measured to represent methanotrophic activities using the method modified from Seghers et al. (2003). In brief, after the injection of CH4, the CH4 concentrations in the headspace of the serum bottles were monitored five times during the next 24 h, and when the CH4 oxidation rates were higher than 12 mg CH4 kgdry soil −1 h−1, the CH4 concentrations were monitored four times during the next 12 h. Then the CH4 oxidation rates were determined by the regression of concentrations against time. The concentrations of CH4 were measured with a gas chromatograph (GC112A, Shanghai Precision & Scientific Instrument Co., Ltd, Shanghai, China) equipped with a flame ionization detector and a 2-m stainless steel column packed with Porapak Q (60/80 mesh). The temperatures of the injector, oven and detector were set at 80°C, 60°C, and 180°C, respectively. The carrier gas was N2 with a flow rate of 40 mL min−1.
After each test of the CH4 oxidation rate, 3–5-g soil samples were obtained from the incubation bottles and used to detect the concentrations of NH4 + and NO3 −. The pH of the soil was determined on the 1st day and 72nd day and slightly reduction (about 0.1) was observed, which could be considered stable during the incubation period.
DNA extraction and quantification analysis by real-time PCR
Soil DNA was extracted from 0.4 g of soil samples by the modified phenol chloroform and ethanol precipitation methods (Ye et al. 2007). The abundances of type I MOB and type II MOB and AOB were analyzed by real-time PCR. Real-time PCR amplification was performed on a Realplex thermo-cycler (Eppendorf, Hamburg, Germany) in 20-μL reaction mixtures using SYBR® Premix Ex Tay™ as described by the suppliers (Takara Bio, Otsu, Shiga, Japan). Two reverse primers of MethT1bR (5′-GAT TCY MTG SAT GTC AAG G-3′) and MethT2bR (5′-CAT CTC TGR CSA YCA TAC CGG-3′) (Wise et al. 1999) and their common forward primer 533f (5′- GTG CCA GCA GCC GCG GTA A-3′) (Weisburg et al. 1991) were used to determine the 16S rDNA gene copy numbers of type I and type II MOB, respectively. As to AOB, the primers amoA-1F (5′-GGG GTT TCT ACT GGT GGT-3′) and amoA-2R (5′-CCC CTC KGS AAA GCC TTC TTC-3′) (Rotthauwe et al. 1997) targeting amoA gene were used. Real-time PCR assay was carried out with the protocol as follows: 95°C for 2 min; 40 cycles of 15 s at 95°C; 20 s at 57°C, 60°C, and 58°C for type I MOB, type II MOB, and AOB, respectively; 15 s at 72°C for reading the plate, followed by a melting curve analysis to confirm PCR product specificity by measuring fluorescence continuously over the temperature increase of 55–95°C. All tests were done in triplicate and data analysis was carried out with the Realplex software (version 2.2), in which the parameter C t (threshold cycle) was determined as the cycle number at which a statistically significant increase in the reporter fluorescence was detected.
One standard curve was generated for each assay, using serial 10-fold dilutions of purified DNA solutions in sterilized water, for which DNA fragments were the PCR products obtained by using the same primers. The concentrations of the DNA solutions were determined spectrophotometrically. The slopes of standard curves were −3.435, −3.148, and −3.294, corresponding to the efficiencies of the PCR reaction of 0.95, 1.08, and 1.01 for type I MOB, type II MOB, and AOB, respectively. The corresponding regression coefficient values (R 2) were 1.000, 0.995, and 0.998.
PCR-DGGE analysis
For DGGE analysis of type II MOB, a GC clamp (CGC CCG CCG CGC CCC GCG CCC GGC CCG CCG CCC CCG CCC C) was attached to the 5′-end of the forward primer. PCR amplification was performed in a total 50-μL volume containing 0.25 μmol L−1 of each primer, 200 μmol L−1 of each dNTP, 5 μL 10× PCR buffer, 2 μmol L−1 of MgCl2, 5 U of Taq polymerase, and 5 μL of template DNA. PCR amplification conditions were as follows: pre-denaturation at 94°C for 5 min; 40 cycles of denaturation at 94°C for 30 s, annealing at 60°C for 45 s, and extension at 72°C for 90 s; and a final extension at 72°C for 5 min ended the amplification cycle.
DGGE was performed essentially as described by Muyzer et al. (1993): 40 μL of PCR product and 15 μL of 6× loading buffer were loaded onto 6% polyacrylamide gels (acrylamide/bisacrylamide, 37.5:1) with denaturing gradients of range 40–60%. The gel was electrophoresed in 1× TAE buffer at 100 V for 17 h at 60°C. After electrophoresis, the gels were stained by ethidium bromide for 15 min, followed by photography using UV transillumination.
Sequencing and sequence analysis
Dominant bands were excised from DGGE gels and transferred to 100 μL of Tris-EDTA buffer for 8 h at 4°C for DNA elution. After that, the eluted DNA was re-amplified using the original PCR primers and programs and confirmed by another DGGE. Only pure bands were amplified with primers without GC clamp and then sequenced on an ABILR-377 DNA sequencer (Applied Biosystems, Foster City, CA, USA). Sequences were aligned to related sequences available in the NCBI databases using the BLAST tool (http://www.ncbi.nlm.nih.gov/BLAST). Phylogenetic trees were calculated and drawn using the Neighbor-Joining algorithm and Mega 3.0 software. The sequences have been deposited at GenBank with accession numbers: HM209447 to HM209451 and JF769887 (16S rDNA gene sequences).
Shannon diversity index
The community diversity was assessed by Shannon diversity index (H), which was calculated based on DGGE data. Each detected band was defined as a specific phylotype and labeled on Fig. 3a. The pixel intensity for the phylotype was detected by the QUANTITY ONE® software (Version 4.6.2, Bio-Rad, USA). The Shannon diversity index was calculated by Eq. 1.
in which N i is the pixel intensity of the ith phylotype, N is the total pixel intensity of all phylotypes, and S is the number of phylotypes.
Statistical analysis
All statistic analyses were performed utilizing SPSS 16.0 (SPSS Inc., Chicago, IL, USA). The differences presented were confirmed by t test at a 95% confidence level.
Results
Effect of ammonium addition on soil methane oxidation capacities
As shown in Fig. 1, the CH4 oxidation peak values of the soils with the treatments of CON, N100, N250, N500, and N1000 were 12.2, 19.8, 16.6, 23.5, and 15.5 mg CH4 kgdry soil −1 h−1, respectively. The CH4 oxidation rates were similar to the results previously reported in mineral landfill soils (15–26 mg CH4 kgdry soil −1 h−1) but lower than that in organic-rich cover soils (128–400 mg CH4 kgdry soil −1 h−1) (Borjesson et al. 2004).
Compared with the CON, where the CH4 oxidation rates increased gradually from 4.2 to 12.2 mg CH4 kgdry soil −1 h−1 during the 72-day incubation, NH4 +-amended soils exhibited different patterns of CH4 oxidation along with time. On the first day, the CH4 oxidation rates of experiments N100, N250, N500, and N1000 were 2.6, 1.0, 0.7, and 0 mg CH4 kgdry soil −1 h−1, respectively, which were lower than that of CON (4.2 mg CH4 kgdry soil −1 h−1) and negatively correlated with the NH4 + concentration. After different levels of lag phase, the CH4 oxidation rates of the NH4 +-amended soils increased rapidly to the peak values with the similar increasing rates, i.e., 0.45–0.85 mg CH4 kgdry soil −1 h−1 per day, which were higher than 0.16 mg CH4 kgdry soil −1 h−1 per day for CON. The CH4 oxidation rates of N100 increased with no lag, exceeded that of CON after the 4th day with the oxidation rates both of 4.8 mg CH4 kgdry soil −1 h−1, and achieved the peak value of 19.8 mg CH4 kgdry soil −1 h−1 on the 21th day. For N250 and N500, the lag time before significant enhancement of the CH4 oxidation was 4 days, and the oxidation rate was 7.1 mg CH4 kgdry soil −1 h−1 when they overbore CON on the 12th day. The peak values of the CH4 oxidation rates were 13.5 and 18.8 mg CH4 kgdry soil −1 h−1 for N250 and N500, respectively, which were achieved on the 30th day and increased slightly afterward. The lag time of N1000 was the longest, lasting for 12 days, and the oxidation rate was 10.8 mg CH4 kgdry soil −1 h−1 when it overbore that of CON on the 27th day. After maintaining above this level for 26 days, the CH4 oxidation rate decreased rapidly to 5.5 mg CH4 kgdry soil −1 h−1 on the 72th day.
Throughout the experimental period, the NH4 + and NO3 − concentrations in the soils did not show a significant change. In the negative control experiment with autoclaved soils, the CH4 concentrations in the headspace remained constant over the entire experimental period indicating the absence of abiotic CH4 oxidation.
Effect of ammonium addition on methanotrophic abundance
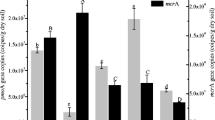
In the original soils, the 16S rDNA gene copy numbers of type I and type II MOB were 0.1 and 2.8 × 107 copies gdry soil −1, respectively, while the copy numbers of AOB were two orders of magnitude less (4.0 × 105 copies gdry soil −1) than that of type II MOB. Therefore, type II MOB were the predominant methanotrophs.
No significant trend of the 16S rDNA copy numbers of MOB was observed along with the increased amount of NH4 + added (Fig. 2a). Herein, the ratio of type II/type I MOB was used to evaluate their relative abundance. In the high NH4 +-amended samples (N250, N500, and N1000), the type II/type I MOB ratios decreased rapidly during the first 16 days (Fig. 2b) and then maintained constant in the last period, while the ratios in the other two treatments (CON and N100) fluctuated around the initial levels. In addition, Pearson correlation analysis was conducted but no significant correlation (P > 0.05) was found among the CH4 oxidation rates and the abundances of type I MOB or type II MOB or the ratios of type II/type I MOB.
Quantification of type I and type II methane-oxidizing bacteria (MOB) by real-time PCR amplification targeting 16S rDNA gene fragments (CON, N100, N250, N500, and N1000 represent the soils receiving 0, 100, 250, 500, and 1,000 mg N kgdry soil −1 of NH4 +, respectively). a Copy numbers of type I and type II MOB with incubation time (three bars in every treatment represent cell counts on the 16th, 36th, and 68th day from left to right, respectively). b The ratios of the copy numbers of type II/type I MOB. Error bars indicate the range of two measurements
During the incubation, 40% of the AOB copy numbers were undetected in all the soil samples (data not shown). In the other samples, 70% of the detected AOB copy numbers were less than 5,000 copies gdry soil −1, and the highest value was only 3.1 × 104 copies gdry soil −1. Furthermore, no correlation existed between the AOB copy numbers and the CH4 oxidation rates.
Effect of ammonium addition on the community compositions of type II methane-oxidizing bacteria
The DGGE profile (Fig. 3a) showed that the communities of type II MOB varied by the treatments. It was clear that the samples with high NH4 + amendment (N250, N500, and N1000) have distinct community compositions from the CON and N100, as the extra bands 3 and 4 were predominant but bands 1 and 2 diminished in the former ones. The CON and N100 treatments exhibited no obvious evolvements of the DGGE band patterns during the 72-day incubation. This was further supported by principal component analysis of the DGGE profile (Fig. 3b), which showed that the samples of CON and N100 at all the three sampling times clustered together with the original soil, which represented the CON sample at time 0. However, for higher NH4 +-amended samples, the methanotrophic communities evolved greatly over time with the conversion of predominant species. BAND 3 existed in the soil samples with N250 and N1000 treatment. It became predominant on the 36th day in the N250, while this time for N1000 was at least 20 days earlier. As to the N500 treatment, the only dominant specie was BAND 4 during the entire incubation period.
DGGE profile analysis of type II methane-oxidizing bacteria targeting 16S rDNA gene fragment (CON, N100, N250, N500, and N1000 represent the soils receiving 0, 100, 250, 500, and 1,000 mg N kgdry soil −1 of NH4 +, respectively). a DGGE profile. b Principal component analysis of the DGGE profile, in which only the seven labeled bands were used to do the analysis
Shown by the Shannon diversity index (Table 1), the community diversity of type II MOB experienced a reduction along with incubation time in lower NH4 + amendments (N100 and N250). Comparatively, other samples had obvious fluctuated diversities. Furthermore, Pearson correlation analysis showed that the CH4 oxidation rates were significantly negatively correlated with the Shannon diversity indexes of type II MOB (R = −0.364, P < 0.05).
Within the seven dominant bands in the DGGE profile, only six of them were successfully purified by the second round of DGGE process and subsequently sequenced and BAND 2 was excluded (Fig. 4). On the phylogenetic tree for 16S rDNA gene sequences of type II MOB, the bands 1, 3, 6, and 7 clustered with Methylocystis sp., while BAND 5 was similar to Methylosinus sp. The specie similar to BAND 1 was Methylocystis heyeri sp., which was previously isolated from acidic (pH 4.2–4.3) Sphagnum peat and tropical forest soil (Dedysh et al. 2007); at pH 4.9, our soil samples were of similar acidity. In addition, BAND 4 possessed 97–98% similarity to sequences of several species of uncultured alpha proteobacteria, which have been detected in rock biofilm from gold mines (NCBI accession numbers: FM 253641 and FN 594678) and pasture soils (NCBI accession number: AY395334).
Discussion
Effect of ammonium addition on methane oxidation
The CH4 oxidation rates of the NH4 +-amended soils were low on the 1st day but increased quickly to exceed the oxidation rates of the CON after a lag phase. Larger quantities of NH4 + fertilizations resulted in lower CH4 oxidation rates on the 1st day and longer lag time. The highest CH4 oxidation rates achieved were positively correlated with the initial NH4 + levels, except for the N500. The effect patterns of NH4 + fertilization can be described as temporal inhibition-delayed stimulation of CH4 oxidation capacity.
The positive correlation between the inhibition levels and the initial NH4 + levels made it seem that substrate competitive inhibition of the CH4 oxidation occurred. However, no increase of soil NO3 − concentrations was observed in this study, indicating that NH4 + oxidation was not significant. Similarly, Tlustos et al. (1998) found that the NH4 + concentrations in the grassland soils remained constant after the addition of (NH4)2SO4, and no NO3 − was detectable. It was generally accepted that the favorite pH for nitrification was around 6.6–8.0, and the nitrification would completely stop when the pH was lower than 4.5 (Maier et al. 2008). We attributed the insignificant NH4 + oxidation to the acidic soils used in our (pH = 4.9) and Tlustos’ (pH = 4.67) studies, under which conditions ammonium (pKa = 9.25) occurred mostly in the protonated form and was not suitable as a substrate for monooxygenase (Kowalchuk and Stephen 2001). In this case, NH4 + could not be oxidized by MOB through competing the active point of MMO with CH4, and no toxic products (e.g., nitrite) could be generated and act on MOB. The only possible explanation lasted for the temporal inhibition derived from NH4 + could be a general salt effect (osmotic stress). Saari et al. (2004) found that the osmotic stress derived from K2SO4 addition could inhibit atmospheric CH4 oxidation.
Compared with the soil samples with no fertilization (CON), the CH4 oxidation rate was promoted by NH4 + addition after a lag phase. For the NH4 +-derived stimulation of CH4 oxidation, diverse explanations can be summarized from previous literatures. Park et al. (1992) and Bodelier et al. (2000a) commented that nitrogen addition might interfere directly with the CH4 oxidizing enzyme system, as immediate CH4 consumption induced by either NH4 + or NO3 − addition. Nevertheless, this was not valid in the present study due to the delayed rather than immediate stimulation of CH4 oxidation. Cai and Mosier (2000) ascribed the promoted CH4 consumption owing to enhancement of AOB. However, in the present study, the observed stimulation effects were regarded not to be associated with AOB for two reasons: The copy numbers of AOB were not comparable with those of MOB, as the former were about 1/70 of the latter in the original soils and decreased about two magnitudes along with incubation and the CH4 oxidation rate of AOB was only 1/5 of the slowest MOB in a pure culture experiment (Hanson and Hanson 1996). Besides, Bender and Conrad (1995), de Visscher et al. (1999), and Papen et al. (2001) found that NH4 +-based fertilization could relieve the N limitation of methanotrophs rather than stimulate CH4 oxidation process. As a necessary nutrient for methanotrophs, 0.25 mol of nitrogen is required for 1 mol of carbon assimilation (Anthony 1982). If 1 mol of CH4 consumption is assumed to lead to 0.4 mol of carbon assimilation, then the ratios of NH4 +/CH4 should be >0.1 to ensure no N limitation happened (Bodelier and Laanbroek 2004). In this study, the NH4 +/CH4 ratio of the original soils was 0.096 near the N limitation threshold for methanotrophs.
We hypothesized that the mechanisms of NH4 + utilization and CH4 oxidation in this study were as follows: The bioavailable NH4 + could be inherent, mineralized from organic N and externally added as (NH4)2SO4, and was regulated by the adsorption or desorption from the high cation-exchanging capacity clay particles. The NH4 + oxidation process was impeded at low pH conditions. Considering the process of CH4 oxidation, the NH4 + in the soils could be assimilated in the cells of MOB, which could reduce the NH4 + concentrations in the soil solution. Then, the adsorbed NH4 + could be released from the clay particles to the soil solution, and this might explain the observation that the extractable NH4 + concentrations in the soils displayed no significant variation.
Effect of ammonium addition on the methanotrophic communities
The CH4 oxidation rates were negatively correlated with the Shannon diversity indexes of type II MOB, but were not correlated with methanotrophic abundances. Compared with the DGGE profile, when the CH4 oxidation rates of high NH4 +-amended samples reached the peak values on the 36th day (Fig. 1), the predominant species correspondingly appeared. This was consistent with the work of Seghers et al. (2005), who observed that long-term herbicide application altered the methanotrophic community structures, but not significantly affected either the soil CH4 oxidation rates or methanotrophic populations. It was demonstrated that methanotrophic community structures rather than the methanotrophic quantities were more sensitive to outside environmental factors.
Considering the responses of methanotrophs on NH4 + addition, two kinds of effect patterns can be summarized: (1) Lower NH4 + amendments (CON and N100) induced relatively constant community compositions and (2) higher NH4 + application (N250, N500, and N1000) posed decreased ratios of type II/type I MOB and significant community evolutions of type II MOB. It was generally believed that type II MOB can survive better in nutrient-limited environments, but type I MOB were more competitive than type II MOB in nutrient-rich environments (Amaral and Knowles 1995; Borjesson et al. 1998; Wise et al. 1999). Microarray analysis (Lee et al. 2009) of methanotrophs in landfill soils revealed that 16.5 mg N kgdry soil −1 of NH4 + amendment led to increased signals of type I MOB but relatively constant signals of type II MOB. Thus, we hypothesized that for the N-limited soils (as in this study), lower NH4 + application (≤100 mg N kgdry soil −1) can relieve the N limitation status of MOB and subsequently promote the activities of all MOB with no significant competitions among methanotrophic species. Whereas, >100 mg N kgdry soil −1 of NH4 + fertilization was higher enough to favor the growth of part of the MOB species, i.e., type I MOB, kinds of Methylocystis-like species (BAND 3 and BAND 4 in the DGGE profile, Fig. 3a), and altered the methanotrophic community structures. The only Methylosinus-like species (BAND 5 in the DGGE profile, Fig. 3a) detected in this study diminished under high NH4 + amendments.
Possible threshold level distinguishing ammonium-derived inhibition or stimulation
Many researchers have found that NH4 + application could stimulate CH4 oxidation at high CH4 mixing ratios, while at low CH4 concentrations, NH4 + acted as an inhibitor. For example, de Visscher et al. (2001) incubated landfill cover soils with 3% CH4 and found that the CH4 uptake rates of the soils were initially stimulated when mixed with sugar beet leaves rich in ammonium, while Boeckx et al. (1996) observed an inhibition of CH4 oxidation derived from NH4 + application in a similar experiment at 10 ppm of CH4. Bodelier et al. (2000a, b) and Mohanty et al. (2006) have done many studies to investigate the influence of NH4 + fertilization on CH4 oxidation in rice fields, either in situ or in lab scale, and most of their studies showed stimulation effects. From their experimental conditions, with 1% CH4 and 40–400 kg N ha−1 of NH4 + added, we calculated that the NH4 +/CH4 ratios during the experiments were 0.4–4, within the above-mentioned N limitation threshold of 0.1 and the maximum value of 9.6 in our study. As to the upland soil, fed with atmospheric CH4 (about 1.8 ppm), NH4 + fertilization always inhibited CH4 oxidation. With the NH4 + fertilization of 0.1–20 mg N kgdry soil −1 (Gulledge et al. 1997), the NH4 +/CH4 ratios in upland soils were 30–600, much greater than the value of 9.6 in the present study and the thresholds of N inhibition observed in littoral sediments (Bosse et al. 1993) and planted intertidal marshes (van der Nat et al. 1997) at 190 and 30, respectively.
Accordingly, we attribute the influence patterns of NH4 + application on CH4 oxidation to the ratios of NH4 +/CH4. For the environment with high NH4 +/CH4 ratio (>30 and mainly at the atmospheric CH4 concentrations), nitrogen content is sufficient for assimilation and added NH4 + would compete with CH4 or introduce more toxic byproducts during its oxidation. However, at lower NH4 +/CH4 ratios (<0.1 and mainly at percent magnitude CH4 mixing ratios), inherent N is not sufficient for the higher requirement of more CH4 assimilation and appropriate NH4 + application can promote the CH4 oxidation rate. The latter situation often occurs in rice fields, wetlands, and landfill cover soils. Considering the adjustability, landfill may be one of the most practical places to enhance the CH4 oxidation capacity by the addition of NH4 +.
Ammonium addition to reduce methane emission from waste landfill
As discussed before, in N-limited environment, an appropriate amount of NH4 + addition could stimulate the CH4 oxidation capacity. In this study, the CH4 oxidation peak value of the landfill cover soils nearly doubled from 12.2 mg CH4 kgdry soil −1 h−1 in CON to 19.8 mg CH4 m−2 h−1 in N100, equaling 3,800 to 6,200 mg CH4 m−2 h−1 in field scale. Considering the sampling site with the largest CH4 flux of about 4500 mg CH4 m−2 h−1 (Zhang et al. 2008), if we assume that other environmental conditions are as favorable as in this study and the highest CH4 oxidation rate can be maintained, appropriate amount of NH4 + addition could convert the CH4 emission to a net sink. Compared with costly commercial mineral NH4 +-based fertilization, landfill leachate irrigation, a process to reduce the liquid quantity and the pollutants by solar evaporation and soil absorption, respectively (Jones et al. 2006), may be a good option for NH4 + application. By investigating CH4 emissions from landfill sites with or without landfill leachate irrigation, Shao et al. (2009) demonstrated that the average CH4 flux from test sites with 1 mm day−1 leachate irrigation (1,000‒1,340 mg N L−1 of NH4 +) was 1/10 of that with no irrigation. However, besides the mitigation of CH4 emission, leachate irrigation may lead to some other environmental problems owing to increased NH4 + and water content in the cover soils, such as larger emissions of nitrous oxide (Watzinger et al. 2005), which should always be kept in mind.
References
Amaral JA, Knowles R (1995) Growth of methanotrophs in methane and oxygen counter gradients. FEMS Microbiol Lett 126:215–220
Anthony C (1982) The biochemistry of methylotrophs. Academic, London
Bender M, Conrad R (1995) Effect of CH4 concentrations and soil conditions on the induction of CH4 oxidation activity. Soil Biol Biochem 27:1517–1527
Bodelier PLE, Laanbroek HJ (2004) Nitrogen as a regulatory factor of methane oxidation in soils and sediments. FEMS Microbiol Ecol 47:265–277
Bodelier PLE, Hahn AP, Arth IR, Frenzel P (2000a) Effects of ammonium-based fertilisation on microbial processes involved in methane emission from soils planted with rice. Biogeochemistry 51:225–257
Bodelier PLE, Roslev P, Henckel T, Frenzel P (2000b) Stimulation by ammonium-based fertilizers of methane oxidation in soil around rice roots. Nature 403:421–424
Boeckx P, van Cleemput O, Villaralvo I (1996) Methane emission from a landfill and the methane oxidising capacity of its covering soil. Soil Biol Biochem 28:1397–1405
Borjesson G, Sundh I, Tunlid A, Svensson BH (1998) Methane oxidation in landfill cover soils, as revealed by potential oxidation measurements and phospholipid fatty acid analyses. Soil Biol Biochem 30:1423–1433
Borjesson G, Sundh I, Svensson B (2004) Microbial oxidation of CH4 at different temperatures in landfill cover soils. FEMS Microbiol Ecol 48:305–312
Bosse U, Frenzel P, Conrad R (1993) Inhibition of methane oxidation by ammonium in the surface-layer of a littoral sediment. FEMS Microbiol Ecol 13:123–134
Cai ZCC, Mosier AR (2000) Effect of NH4Cl addition on methane oxidation by paddy soils. Soil Biol Biochem 32:1537–1545
de Viries GE, Kuees U, Stahl U (1990) Physiology and genetics of methylotrophic bacteria. FEMS Microbiol Rev 75:57–102
de Visscher A, Thomas D, Boeckx P, van Cleemput O (1999) Methane oxidation in stimulated landfill cover soil environments. Environ Sci Technol 33:1854–1859
de Visscher A, Schippers M, van Cleemput O (2001) Short-term kinetic response of enhanced methane oxidation in landfill cover soils to environmental factors. Biol Fertility Soils 33:231–237
Dedysh SN, Belova SE, Bodelier PLE, Smirnova KV, Khmelenina VN, Chidthaisong A, Trotsenko YA, Liesack W, Dunfield PF (2007) Methylocystis heyeri sp nov., a novel type II methanotrophic bacterium possessing ‘signature’ fatty acids of type I methanotrophs. Int J Syst Evol Microbiol 57:472–479
Fletcher SEM, Tans PP, Bruhwiler LM, Miller JB, Heimann M (2004) CH4 sources estimated from atmospheric observations of CH4 and its C-13/C-12 isotopic ratios: 1. Inverse modeling of source processes. Global Biogeochem Cycles 18:GB4004
Gulledge J, Doyle AP, Schimel JP (1997) Different NH4 +-inhibition patterns of soil CH4 consumption: a result of distinct CH4-oxidizer populations across sites? Soil Biol Biochem 29:13–21
Graham DW, Chaudhary JA, Hanson RS, Arnold RG (1993) Factors affecting competition between type I and type II methanotrophs in two-organism, continuous-flow reactors. Microb Ecol 25:1–17
Hanson RS, Hanson TE (1996) Methanotrophic bacteria. Microbiol Rev 60:439–471
Jones DL, Williamson KL, Owen AG (2006) Phytoremediation of landfill leachate. Waste Manage 26:825–837
King GM (1990) Regulation by light of methane emissions from a wetland. Nature 345:513–515
King GM, Schnell S (1994) Ammonium and nitrite inhibition of methane oxidation by Methylobacter-albus Bg8 and Methylosinus-trichosporium Ob3b at low methane concentrations. Appl Environ Microbiol 60:3508–3513
Kowalchuk GA, Stephen JR (2001) Ammonia-oxidizing bacteria: a model for molecular microbial ecology. Annu Rev Microbiol 55:485–529
Kruger M, Frenzel P (2003) Effects of N-fertilisation on CH4 oxidation and production and consequences for CH4 emissions from microcosms and rice fields. Global Change Biol 9:773–784
Lee SW, Im J, DiSpirito AA, Bodrossy L, Barcelona MJ, Semrau JD (2009) Effect of nutrient and selective inhibitor amendments on methane oxidation, nitrous oxide production, and key gene presence and expression in landfill cover soils: characterization of the role of methanotrophs, nitrifiers, and denitrifiers. Appl Microbiol Biotechnol 85:389–403
Maier R, Paepper I, Gerba C (2008) Environmental microbiology. Academic, Salt Lake
Mohanty SR, Bodelier PLE, Floris V, Conrad R (2006) Differential effects of nitrogenous fertilizers on methane-consuming microbes in rice field and forest soils. Appl Environ Microbiol 72:1346–1354
Murrell JC (1994) Molecular genetics of methane oxidation. Biodegradation 5:145–160
Muyzer G, de Waal EC, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59:695–700
Noll M, Frenzel P, Conrad R (2008) Selective stimulation of type I methanotrophs in a rice paddy soil by urea fertilization revealed by RNA-based stable isotope probing. FEMS Microbiol Ecol 65:125–132
Papen H, Daum M, Steinkamp R, Butterbach-Bahl K (2001) N2O and CH4-fluxes from soils of a N-limited and N-fertilised spruce forest ecosystem of the temperate zone. J Appl Bot-Angew Bot 74:159–163
Park S, Shah NH, Taylor RT, Droege MW (1992) Batch cultivation of Methylosinus trichosporium OB3b: II. Production of particulate methane monooxygenase. Biotechnol Bioeng 40:151–157
Rigler E, Zechmeister-Boltenstern S (1999) Oxidation of ethylene and methane in forest soils-effect of CO2 and mineral nitrogen. Geoderma 90:147–159
Rotthauwe JH, Witzel KP, Liesack W (1997) The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol 63:4704–4712
Saari A, Rinnan R, Martikainen PJ (2004) Methane oxidation in boreal forest soils: kinetics and sensitivity to pH and ammonium. Soil Biol Biochem 36:1037–1046
Seghers D, Top EM, Reheul D, Bulcke R, Boeckx P, Verstraete W, Siciliano SD (2003) Long-term effects of mineral versus organic fertilizers on activity and structure of the methanotrophic community in agricultural soils. Environ Microbiol 5:867–877
Seghers D, Siciliano SD, Top EM, Verstraete W (2005) Combined effect of fertilizer and herbicide applications on the abundance, community structure and performance of the soil methanotrophic community. Soil Biol Biochem 37:187–193
Shao LM, Zhong JS, Zhang HH, He PJ (2009) CH4 emissions from municipal solid waste landfills in spring and summer and influencing factors analysis. Res Environ Sci (in Chinese) 22:83–88
Steudler PA, Bowden RD, Melillo JM, Aber JD (1989) Influence of nitrogen fertilization on methane uptake in temperate forest soils. Nature 341:314–316
Tlustos P, Willison TW, Baker JC, Murphy DV, Pavlikova D, Goulding KWT, Powlson DS (1998) Short-term effects of nitrogen on methane oxidation in soils. Biol Fertil Soils 28:64–70
van der Nat FJWA, de Brouwer JFC, Middelburg JJ, Laanbroek HJ (1997) Spatial distribution and inhibition by ammonium of methane oxidation in intertidal freshwater marshes. Appl Environ Microbiol 63:4734–4740
Watzinger A, Reichenauer TG, Blum WEH, Gerzabek MH, Zechmeister-Boltenstern S (2005) The effect of landfill leachate irrigation on soil gas composition: methane oxidation and nitrous oxide formation. Water Air Soil Pollut 164:295–313
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703
Wise MG, McArthur JV, Shimkets LJ (1999) Methanotroph diversity in landfill soil: isolation of novel type I and type II methanotrophs whose presence was suggested by culture-independent 16S ribosomal DNA analysis. Appl Environ Microbiol 65:4887–4897
Ye NF, Lv F, Shao LM, Godon JJ, He PJ (2007) Bacterial community dynamics and product distribution during pH-adjusted fermentation of vegetable wastes. J Appl Microbiol 103:1055–1065
Yu T, He PJ, Lu F, Shao LM (2009) Mediating N2O emissions from municipal solid waste landfills: Impacts of landfill operating conditions on community structure of ammonia-oxidizing bacteria in cover soils. Ecol Eng 35:882–889
Zhang HH, He PJ, Shao LM (2008) Methane emissions from MSW landfill with sandy soil covers under leachate recirculation and subsurface irrigation. Atmos Environ 42:5579–5588
Zheng Y, Zhang LM, Zheng YM, Di HJ, He JZ (2008) Abundance and community composition of methanotrophs in a Chinese paddy soil under long-term fertilization practices. J Soils Sed 8:406–414
Acknowledgments
This work was financially supported by the National High-Tech Research and Development Program of China (No. 2003AA644020) and Natural Key Technology R&D Program of China (No. 2006BAJ04A06).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yang, N., Lü, F., He, P. et al. Response of methanotrophs and methane oxidation on ammonium application in landfill soils. Appl Microbiol Biotechnol 92, 1073–1082 (2011). https://doi.org/10.1007/s00253-011-3389-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-011-3389-x