Abstract
Terminal restriction fragment length polymorphism (T-RFLP) can be used to assess how land use management changes the dominant members of bacterial communities. We compared T-RFLP profiles obtained via amplification with forward primers (27, 63F) each coupled with the fluorescently labeled reverse primer (1392R) and multiple restriction enzymes to determine the best combination for interrogating soil bacterial populations in an agricultural soil used for potato production. Both primer pairs provide nearly universal recognition of a 1,400-bp sequence of the bacterial domain in the V1–V3 region of the 16S ribosomal RNA (rRNA) gene relative to known sequences. Labeling the reverse primer allowed for direct comparison of each forward primer and the terminal restriction fragments’ relative migration units obtained with each primer pair and restriction enzyme. Redundancy analysis (RDA) and nested multivariate analysis of variance (MANOVA) were used to assess the effects of primer pair and choice of restriction enzyme on the measured relative migration units. Our research indicates that the 63F–1392R amplimer pair provides a more complete description with respect to the bacterial communities present in this potato (Solanum tuberosum L.)–barley (Hordeum vulgare L.) rotation over seeded to crimson clover (Trifolium praense L.). Domain-specific 16S rRNA gene primers are rigorously tested to determine their ability to amplify across a target region of the gene. Yet, variability within or between T-RFLP profiles can result from factors independent of the primer pair. Therefore, researchers should use RDA and MANOVA analyses to evaluate the effects that additional laboratory and environmental variables have on bacterial diversity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Terminal restriction fragment length polymorphism (T-RFLP) is a reliable method for the generation of reproducible community profiles that can be used to determine the effects of management or physiochemical gradients on bacterial community structure as well as aid in the phylogenetic identification of members of complex microbial communities (Wick et al. 2010; Muckian et al. 2007; Marsh 1999). However, phylogenetic identification is problematic because not all species or genus of bacteria can be represented by a unique relative migration unit, and these units may differ from the standard base pair fragment length (Marsh et al. 2000). Digestion of an amplicon with multiple restriction enzymes that provide several, unique terminal restriction fragments (T-RFs) that can be compared to a robust ribosomal RNA (rRNA) gene sequence database (Kent et al. 2003; Shyu et al. 2007; Junier et al. 2008) provides increased resolution into community diversity (Clement et al. 1998). Therefore, both the primer and restriction enzymes are critical to the recovery of information with respect to the microbial community structure.
Previous studies have shown that the V1–V3 region of the 16S rRNA gene provided the greatest coverage for resolving bacteria and best represented their diversity across a range of soil systems and molecular techniques (Marchesi et al. 1998). Resolution of bacterial communities is dependent upon the bacterial primer’s ability to amplify across the entire region of the rRNA gene targeted, the restriction digests, and the number of restriction sites present in template DNA sequences (Alm et al. 1996). However, primers that target the same V1–V3 variable regions such as 27F and 63F have been shown to vary in their ability to amplify some phylogenetic groups due to primer mismatches (Frank et al. 2008; Marchesi et al. 1998; Sipos et al. 2007), GC content of primer binding sites, and “secondary structures formed by single stranded template and flanking region DNA that affect primer site accessibility or elongation efficiency” (Hansen et al. 1998). Marchesi et al. (1998) used 63F to amplify genera of coryneform that could not be previously amplified with 27F. These genera include important soil bacteria like Actinomycetes that play a role in nitrogen and carbon cycling as well as produce biocides and Arthrobacter that degrade pesticides. Therefore, shifts in community diversity of prevalent bacteria due to management and environmental factors may not be entirely resolved with a particular primer pair if the primer set excludes or poorly amplifies some of the important phylogenetic community members present (Frank et al. 2008; Sipos et al. 2007). Despite these impediments, the majority of published microbial ecology studies and the Ribosomal Database Project are principally based on a single bacterial forward primer 8-27F (Cole et al. 2005). Limitations associated with primers specific to the bacterial domain continue to be pertinent due to their adaptation to pyrosequencing platforms (Edwards et al. 2006; Kunin et al. 2009; Li et al. 2010).
Employing statistical analyses to complex T-RFLP profiles has revealed distinctly different patterns in microbial communities resulting from land applications of nutrients and waste materials (Mills et al. 2003; Widenfalk et al. 2008) across various ecosystems and land use management (Hartmann and Widmer 2006; Dunbar et al. 2000). Shifts in bacterial community patterns with treatment overtime cannot be fully assessed with diversity and similarity indices in part because these indices do not allow for repeated measures and do not account for interactions between variables that affect community structure. Measurements of analysis of variance such as multivariate analysis of variance (MANOVA) allow the user to look at the interactions among variables that are typically of greater significance to bacterial community composition than a single variable. The disadvantage of multivariate analysis is that the data must be parametric or normally distributed. In contrast, explanatory data analyses such as redundancy analysis focus on the graphical interpretation of quantitative or qualitative data and the shape and distribution of variables within a data set that can be nonparametric enabling the user to determine how redundant one set of variables is relative to another (www.statsoft.com/textbook/elementary-concepts-in-statistics/).
This study assesses shifts in the bacterial diversity of soil from a potato barley rotation over seeded to crimson clover using T-RFLP profiles amplified with two sets of V1–V3 region primers commonly used to amplify the 16S rRNA gene and three restriction enzymes. Due to the inherent biases involved in primer selection, we chose to compare two nearly universal bacterial forward primers (27F, 63F) (Marchesi et al. 1998). Each forward primer was coupled with a fluorescently labeled reverse primer 1392R (Amann et al. 1995) and three restriction enzymes to determine which forward primer and enzyme combinations provided the best resolution of bacterial community structure in this potato production system. Restriction enzymes were selected based on their ability to provide a range of T-RFs of varying sizes and relative migration units (rmu) that covered different sections of amplicon sequences. The authors found no previous studies that compared T-RFLP profiles obtained from genomic DNA extracted from pure cultures of bacteria or environmental samples amplified with 27F–1392R and 63F–1392R.
This research will improve our ability to resolve shifts in bacterial diversity and biological processes resulting from a decade of potato small grain management. An additional goal was to determine the ability of redundancy analysis and multivariate analysis of variance to resolve differences in bacterial community diversity of dominant community members relative to standard ecological diversity indices. Diversity indices are commonly used to evaluate bacterial communities represented by shifts in the number, size, and rmu of T-RFs in and among T-RFLP profiles. However, shifts in T-RFLP profiles are often the result of variations in several factors independent of primer pair that may include restriction enzyme, laboratory, and field variability. As a result, indices that do not allow users to evaluate several variables simultaneously are far less sensitive to shifts in community diversity than multivariate analyses (Blackwood et al. 2007). Not all data are randomly distributed disallowing the choice of MANOVA designs in some instances. Therefore, a combination of primer pairs and restriction enzymes coupled with multivariate analyses provides the best means to interrogate and resolve shifts in microbial community structure across a wide range of environments and land use management.
Materials and methods
Soil sampling, DNA extraction, and amplification
Field plots were located at the University of Maine’s Aroostook Research Farm, Presque Isle, ME, USA on the Potato Ecosystem Project in an unamended potato–barley rotation over seeded to crimson clover. The plots had been in rotation for 11 years and were located on a Caribou loam (fine-loamy, mixed, frigid, Typic Haplorthod). Twelve soil samples were randomly taken from each of the four field plot replicates and composited providing one composite soil sample from each replicate field plot that measured, 14.6 m × 41.0 m. The soil samples were collected with a soil probe 2 cm in diameter to a 15-cm depth in 2002. Soils were sieved through a 2-mm screen and frozen at −20°C. The four field plot replicates sampled represent one crop rotation and fertility regime contained within a randomized complete block split plot design. Cropping system and fertility treatments included an unamended potato–barley rotation over seeded to crimson clover (used in this experiment) and amended potato rotations with a mixture of oat (Avena sativa L.), pea (Pisum sativum L.), and vetch (Vicia villosa L. Roth) (Erich et al. 2002). Unamended soils (included in this experiment) received inorganic fertilizer and amended soils received a mixture of cattle manure (Bos taurus), composted cull potatoes, green manure, and inorganic fertilizer.
Genomic DNA was extracted from 0.25 g of soil using an Ultraclean soil DNA extraction kit (Mo Bio Laboratories, Solana Beach, CA, USA). Yield and purity of genomic DNA were calculated from the A 260/A 280 ratio measured using an Agilent 8453 DAD UV/visible spectrophotometer (Agilent Technologies, Wilmington, DE, USA).
Bacterial 16S rRNA genes were amplified using two degenerate forward primers, 27F (AGA GTT TGA TCM TGG CTC AG) and 63F (CAG GCC TAA YAC ATG CAA GTC), each paired with a fluorescently labeled degenerate reverse primer, 1392R (/56-FAM/ACG GGC GGT GTG TRC) (manufactured by Integrated DNA Technologies, Coralville, IA, USA). Genomic DNA from each of the four field plot replicates was amplified in triplicate. The optimal concentration of genomic DNA amplified from environmental samples was 4 ng per 100 μl PCR reaction. Ten nanograms of Ralstonia eutropha genomic DNA was added to each control PCR reaction.
The reaction mixture for PCR contained 1× PCR buffer μl−1, 2.5 mM MgCl2 μl−1 (Invitrogen, Carlsbad, CA, USA), 0.025 U Taq DNA polymerase μl−1 (Invitrogen), 250 μM of each dNTP (Invitrogen), 50 ng of bovine serum albumin μl−1 (Promega, Madison, WI, USA), and 0.4 μM μl−1 of each primer. PCR mixture volumes were 100 μl. PCR was performed in a Perkin-Elmer 9600 or MJ Research Peltier PTC-100™ thermocycler. The cycling program consisted of an initial denaturation step of 95°C for 5 min, followed by 30 cycles of a program consisting of denaturation at 94°C for 40 s, primer annealing at 56°C for 40 s, and extension at 72°C for 1 min 30 s. An additional final extension at 72°C for 7 min was performed after completion of the 30 cycles.
Terminal restriction fragment length polymorphism
NEBcutter, a software program that simulates the cleaving of DNA with restriction enzymes, was used to select tetrameric enzymes providing restriction sites that produced the most phylogenetically informative species and genus-specific terminal restriction fragments providing a range of T-RFs of varying sizes and rmu covering different sections of amplicon sequences (Vincze et al. 2003). Each restriction enzyme chosen (Hha1, Msp1, and Rsa1) yielded T-RFs from a different section of the 1,400-bp PCR amplicons (New England Biolabs Inc., Beverly, MA, USA). The use of the same fluorescently labeled reverse primer (1392R) that annealed to identical DNA template binding sites independent of the forward primer binding sites allowed us to directly compare priming efficiencies of 27 and 63F. Labeling the reverse primer also ensured that differences in T-RF sizes were due to the choice of the forward primer.
Triplicate PCR products from each soil field replicate were combined and cleaned using a Qiagen PCR Purification Kit (Qiagen, Valencia, CA, USA). Yield and purity of amplicons were calculated from A 260/A 280 ratios obtained using an Agilent 8453 DAD UV/visible spectrophotometer (Agilent Technologies). Aliquots containing 200 ng of cleaned PCR amplicons from field replicates were added to restriction enzymatic digestions. Restriction endonuclease digestions were set up in individual reactions that contained one restriction enzyme per reaction. Amplicons were cut with 0.5 U of the following restriction enzymes Hha1 (recognition site: GCG^C), Msp1 (recognition site: C^CGG), and Rsa1 (recognition site: GT^AC). Three separate replicate digestions were processed for each restriction enzyme (three), primer combination (two), and field replicate (four).
Enzymatic digestions were incubated in an MJ Research Peltier PTC-100™ thermocycler at 37°C for 3 h followed by heating to 65°C for 5 min to terminate enzymatic activity. Enzymatic digestions were cleaned using a Qiagen Nucleotide Removal Kit (Qiagen). Two microliters of purified PCR products cut with a restriction enzyme was added to 7 μl of deionized formamide and 1 μl of the internal size standard ROX 1000 (Applied Biosystems, Foster City, CA, USA) and denatured at 95°C for 5 min. Samples were loaded onto an ABI PRISM® 3100 Genetic Analyzer for size analysis of T-RFs by fluorescence-based capillary electrophoresis (Applied Biosystems). The size of fluorescently labeled terminal restriction fragments was determined by Perkin-Elmer's ABI Genescan™ Analysis System (Applied Biosystems). A T-RF was considered noise and removed from a T-RFLP profile if the peak area was less than 50 fluorescence units.
Similarities between community profiles were calculated from tabulated data exported out off ABI Genescan™ into T-Align, a program designed to compare replicate T-RFLP profiles from different communities (Smith et al. 2005). A confidence value of 1 was used for binning T-RFs. Fragments were aligned using the moving average. A file was generated that showed the presence or absence of a T-RF, its average size and the percentage fluorescence intensity of total fluorescence in each profile. Terminal restriction fragments in profiles were considered to be the same fragment if they varied by 0.5 rmu or less.
Statistical analysis
Output files from T-Align were imported into Estimates S (Colwell 1997), NesANOVA (www.bio.umontreal.ca/legendre/indexEnglish.html), and Canoco 4.5 (Microcomputer Power, Ithaca, NY, USA) for further analyses. All T-RFLP community profiles were labeled for statistical analyses by primer type (27F or 63F), restriction enzyme, field plot replicate (114, 215, 315, or 423), and laboratory replicate (1, 2, or 3). The Shannon diversity and Jaccard similarity indices allowed for the comparison of T-RFs in T-RFLP profiles obtained with a specific primer set and restriction enzyme. The Jaccard similarity index measures differences in bacterial community diversity between samples based on the presence of T-RFs among T-RFLP profiles. The Shannon diversity index and Jaccard similarity index were calculated using Estimates S (Colwell 1997).
Nested multivariate analysis of variance with permutation testing, using two factors, primer pair and field plot replicate and a nested factor (field plot primer pair), was performed in PERMANOVA and compared to the RDA analyses. The software PERMANOVA was used to perform permutational multivariate analysis of variance on the basis of Bray–Curtis distances, using permutation procedures to obtain P values for the tests (including interactions), for a balanced multi-factorial ANOVA design (http://www.stat.auckland.ac.nz/~mja/Programs.htm). Redundancy analysis does not assume a random distribution of error and explores the relationship between sets of categorical variables (www.statsoft.com/textbook/elementary-concepts-in-statistics/). The bacterial community diversity represented by the T-RFs contained in the T-RFLP profiles was related to choice of forward primer, field replicate, and laboratory replicate by redundancy analysis. Primer type, field replicate, and laboratory replicate were coded as environmental variables and used to define the ordination axes of RDA in Canoco 4.5.
Results
Diversity indices and unique terminal restriction fragments
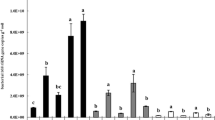
The Shannon index revealed a trend toward greater diversity when the soil bacterial biomass was amplified using the 63F–1392R primer pair, and this trend was statistically significant for T-RFs cut with Hha1 (Table 1). The Jaccard index illustrates that there is a significant (greater than 50%) overlap between T-RFs in profiles obtained from DNA amplified with a given primer pair and cut with a specific restriction enzyme (Table 1). Each primer set and restriction enzyme produced unique and shared T-RFs. More unique fragments were obtained from DNA amplified with 63F–1392R digested with Rsa1 while 27F–1392R cut with Hha1 provided greater numbers of unique T-RFs. However, the number of unique fragments does not determine whether a given primer and enzyme combination provided greater diversity of T-RFs. Profiles obtained from genomic DNA amplified using 27F– or 63F–1392R and cut with Rsa1 contained the same number of shared (8) and unique (10) T-RFs (Tables 1 and 2).
Nested multivariate analysis of variance
A nested MANOVA revealed that the main factor primer pair and nested factor, field plot, were significant for the Hha1 T-RFs (Table 3). The MANOVA analysis of Msp1-derived T-RFs indicated that primer pair (P < 0.03) was significant. The nested factor field plot primer pair was highly significant among Rsa1 T-RFs.
Redundancy analysis
The redundancy analyses grouped samples based on the presence or absence of T-RFs corrected for the percent peak area in T-RFLP profiles. Redundancy analyses like the MANOVA indicated that primer pair explained a significant portion of the variation of the T-RFs obtained from each combination of primer pair coupled with Hha1 and Msp1 restriction enzymes (Table 4 and Figs. 1 and 2). Field plot replicate explained additional variations in T-RFLP profiles produced by digestion with Hha1 and Rsa1. The primer and primer by plot variables are represented by arrows in Figs. 1, 2, and 3 that point in positive directions, indicating increases in T-RFs representing increased bacterial community diversity of the dominant community members when the 63F primer was employed. Terminal restriction fragments from each primer pair are located in separate circles on the graph.
Redundancy analysis depicting the relationship between Hha1 terminal restriction fragments, choice of forward primer, field plot, and laboratory replicate labeled as arrows. Axis 1, primer pair, explained 58% of the T-RFLP variance; axis 2, primer × plot explained a further 22.3% of the T-RFLP variance. All terminal restriction fragment length polymorphism (T-RFLP) community profiles were labeled by primer type (27F or 63F) and field plot replicate (114, 215, 315, or 423). Individual laboratory replicates are identified as circles. The laboratory replicates for each field plot are on top of one another
Redundancy analysis depicting the relationship between Msp1 terminal restriction fragments, choice of forward primer, field plot, and laboratory replicates. Axis 1, primer pair, explained 11% of the T-RFLP variance. Forward primer had a significant effect on terminal restriction fragment length polymorphism (T-RFLP) community profiles as indicated by the labeled arrow. All T-RFLP community profiles were labeled by primer type (27F or 63F). Individual laboratory replicates are identified as circles
Redundancy analyses depicting the relationship between Rsa1 terminal restriction fragments, choice of forward primer, field plot, and laboratory replicate are labeled as arrows. Axis 1, primer pair; axis 2, primer × plot, primer × field plot × laboratory replicate explained 29% of the T-RFLP variance. All terminal restriction fragment length polymorphism (T-RFLP) community profiles were labeled by primer type (27F or 63F). Individual laboratory replicates are identified as circles
Terminal restriction fragments obtained via amplification with 63F and digestion with Hha1 are located in the positive quadrant of the graph showing increased bacterial community diversity of the prevalent bacterial members (Fig. 1). Terminal restriction fragments produced by amplification with 27F and digestion with Hha1 are in the negative quadrant, indicating decreased community diversity. Primer pair explained 58% of the variability of T-RFs obtained after digestion with Hha1 and primer × plot described an additional 26% of the variability (Table 4). The primer × plot arrow indicates that field replicate variability had a significant effect on bacterial community diversity of the dominant community members (Fig. 1).
Terminal restriction fragments obtained from PCR products amplified with each primer pair (27F and 63F–1392R) and digested with Msp1 and Rsa1 contained less variability among field plots and sample replicates (Table 4, Figs. 2 and 3). The effect of primer pair was significant in determining the T-RFs obtained from Msp1 digests but accounted for only 11% of the total variability among T-RFs (Tables 3 and 4). T-RFLP community profiles obtained by amplification with 63F and digestion with Msp1 are located in the positive quadrant of Fig. 2, indicating increased bacterial community diversity of dominant members, relative to T-RFs amplified with 27F and cut with Msp (located in the negative quadrant). The effect of primer pair, field plot, and replicate explained 29% of the variation among T-RFs obtained via digestion with Rsa1 (Table 4). The T-RFLP profiles obtained from PCR products amplified with 63F–1392R and cut with Rsa1 increased in community diversity as a result of primer pair, field plot, and laboratory replicate (Fig. 3).
Discussion
Amplimer pairs
Few primers targeting the bacterial domain have been employed consistently across a range of ecosystems and managements. The primer pair 27F–1492R has been used to amplify the majority of sequences contained in the Ribosomal Database Project. While this primer set and 27F in particular provides sufficient resolution for what is believed to be the majority of sequences in the V1–V3 region of the 16S rRNA gene, 27F has failed to amplify template from some environments and phylogenetic groups that other primer pairs amplify (Marchesi et al. 1998). As new technologies come on-line that include pyrosequencing and PostLight™ semiconductor-based sequencing technologies (Ion Torrent Systems, Inc., San Francisco, CA, USA), researchers will continue to employ existing primers like 27F that target the 16S rRNA gene (Edwards et al. 2006; Kunin et al.; 2009; Li et al. 2010). The consistent use of 27F stems from the primer’s robustness, general adaption across a range of environments, the previously existing database of published sequences amplified using 27F, and the need to compare sequences generated from older technologies with new sequencing platforms. Therefore, our results add to a wider body of knowledge that extends beyond T-RFLP analysis.
Both 27F and 63F forward primers amplify the V1–V3 region of the 16S rRNA gene of the bacterial domain (Baker et al. 2003). Each primer contains mismatches with several phylogenetic groups of soil bacteria (Frank et al. 2008; Marchesi et al. 1998; Sipos et al. 2007). The 27F primer has mismatches with binding site sequences in Actinobacteria, Proteobacteria, Campylobacterales, Sphingomonadales, Gardnerella, and Borrelia spp. that could lead to bias in PCR amplicons (Frank et al. 2008). The primer set 63F–1392R has also failed to amplify some phylogenetic groups such as the bacterial division Verrucomicrobia (Marchesi et al. 1998). However, 63F forward is known to have more matches with sequences contained in the Ribosomal Database Project than 8-27F (Huws et al. 2007).
Mismatches associated with both forward primers are minimal at low annealing temperatures but exponentially increase preferential amplification due to 63F primer mismatches at temperatures between 59.9°C and 61.0°C (Sipos et al. 2007). Our annealing temperatures were below this reported range. Previous research has demonstrated that at lower annealing temperatures, 27F and 63F amplified a majority of pure culture bacterial strains and provided similar T-RFs after restriction enzyme digests were performed and T-RFLP profiles analyzed (Sipos et al. 2007). While the presence of mismatches is problematic and inherent to all primers, binding sites containing no primer mismatches may fail to amplify or lower amplification of genomic DNA template if DNA sequences flanking the binding site interfere with annealing (Hansen et al. 1998).
The forward primers, 27 and 63F, in our study were 39 bp apart relative to the 16S rRNA gene sequenced from Escherichia coli (Brosius et al. 1981). Despite a significant overlap between the two primers and “universal” specificity for the bacterial domain of each, the 63F–1392R pair allowed for greater resolution of bacterial community diversity as revealed by the MANOVA and RDA analyses as well as the diversity indices. An additional benefit of the 63F–1392R primer pair is that 63F minimizes amplification of sequences across kingdoms relative to 27F (Galkiewicz and Kellogg 2008).
A previous study by Marchesi et al. (1998) employed two similar but distinct reverse primers (1387R, 1392R) to mixtures of cultured bacteria and found no clear explanation for greater template amplification and specificity of 63F–1387R relative to 27F–1392R. The 63F–1387R primer pair was more effective in amplifying 16S rRNA genes from species showing higher levels of theoretical 5′ mismatches than 27F coupled with 1392R. Our experiment utilized primers 27F and 63F coupled with the same fluorescently labeled reverse primer (1392R) to evaluate bacterial populations in a potato–barley rotation over seeded to crimson clover. Profiles derived from our experiment revealed that the 63F–1392R amplimer pair repeatedly contained greater richness of T-RFs than profiles derived from 27F–1392R indicating that 63F may provide more complete and additional information with respect to bacterial populations present in these soil community profiles. The conserved area of the 16S rRNA gene that 63F was designed to anneal to may have greater potential to form base pairs with a complementary region of a DNA strand than the conserved region of a complementary region of DNA targeted by 27F (Brunk et al. 1996; Suzuki and Giovannoni 1996). It is also possible that 27F may form an intramolecular duplex with a 5′ overhang that could result in unfavorable intramolecular complementarity between 27F and 1392R causing binding on the 3′ end (Marchesi et al. 1998).
Comparison of bacterial communities in environmental samples
The use of the same reverse primer labeled at the 5′ end ensured that differences in T-RF sizes were comparable between primer sets, and biases associated with the choice of the reverse primer were consistent across primer pair and enabled us to directly measure the ability of the 27F and 63F primers to amplify the template. The use of several enzymes that cut at different base pair sequences insured that T-RFs of distinct size and rmu were derived from all regions of the 1,400-bp amplicons produced in our study. Labeling of the 5′ end of the 1392R reverse primer resulted in fewer and in some instances smaller terminal restriction fragments due to the greater number of restriction sites of greater length at the 3′ end of DNA sequences. Many of the Rsa1 restriction sites were at the 5′ end of the approximately 1,400-bp 16S rRNA gene amplicons to be digested. A study that paired 27F with a reverse primer (536R) followed by endonuclease digestion with Rsa1 or Msp1 produced approximately a quarter of the fragments when the 3′ end was fluorescently labeled in place of the 5′ end (Liu et al. 1997). Therefore, it is likely that samples from this experiment also contained fewer T-RFs in each profile due to labeling of the reverse primer, but comparisons of the relative diversity inherent in T-RFLP profiles representing combinations of primer pairs and restriction enzymes were conserved.
Amplicons cut with Msp1 had the least variability associated with field and laboratory sampling based on the redundancy analyses. A comparison of 16S rRNA gene restriction endonucleases used in generating T-RFLP profiles revealed that Msp1 has high fidelity in resolving T-RFs (Engebretson and Moyer 2003). Our study revealed that additional information with respect to bacterial community diversity was obtained when PCR templates were amplified with 63F–1392R and cut with Hha1. An evaluation of 16S rRNA gene restriction endonucleases used to obtain T-RFLP profiles ranked Hha1 second in resolving the greatest number of T-RFs above Msp1 (Engebretson and Moyer 2003).
Diversity indices, redundancy analyses, and nested multivariate analysis of variance
A standardized T-RFLP protocol including multiple enzymatic digestions and explanatory data analysis has been shown to provide data that allows for rapid comparison of bacterial communities in environmental samples relative to denaturing gel electrophoresis and single strand conformation polymorphism (Dunbar et al. 2000; Smalla et al. 2007). Diversity indices are commonly used to interpret T-RFLP profiles but terminal restriction fragments may represent more than one species or phylotype leading to a lack of resolution in biodiversity measurements. The Jaccard similarity index and redundancy analyses measure differences in bacterial community diversity between samples based on the presence of T-RFs among T-RFLP profiles. Diversity and similarity indices are limited because they explain the variance of one dependant variable at a time. This more basic approach is sufficient for some applications. But most environmental applications of T-RFLP analysis require interpretation of multiple variables within the laboratory and environment of interest. In contrast, RDA allows for resolution of multiple environmental variables that are nonparametric (Blackwood et al. 2007). Multivariate analyses were critical for the interpretation of our T-RFLP profiles that varied due to primer pair, restriction enzyme, and field plot. Our data were parametric allowing us to use MANOVA and RDA analyses. All of the statistical analyses in this experiment employed distance measures based on the presence of a T-RF in all profiles. The absence of a T-RF could be due to primer mismatches or other limitations associated with PCR reactions that are unrelated to environment conditions (Schütte et al. 2008). The diversity indices used in this experiment corroborated the explanatory data analysis revealing greater overall measures of diversity in sequences amplified with the 63F–1392R primer pair.
Nested multivariate analysis of variance and redundancy analyses allowed us to resolve differences in T-RFLP profiles resulting from biological and physiochemical gradients among field plots. This insured that variation from factors other than primer pair and restriction enzymes was not equated with the primer pair employed. Both multivariate analyses revealed that there were shifts in prevalent bacterial members on each of the field plots. Eleven years of consistent land use management in the Potato Ecosystem Project was expected to limit the variability of bacterial communities measured among our field plots. However, redundancy analyses revealed that diversity of prevalent bacterial community members, measured via the number and size of T-RFs, varied significantly among field crop replicates. A number of studies have utilized ordination analyses to relate changes in environmental variables such as hydrocarbons, pesticides, bioremediation, and soil properties to shifts in bacterial community diversity (Muckian et al. 2007; Schütte et al. 2008; Wick et al. 2010; Widenfalk et al. 2008).
Our research indicates that the 63F–1392R primer pair provided more complete information with respect to the bacterial communities present in this potato–barley rotation over seeded to crimson clover. Variations in land use management lead to changes in biological and physiochemical gradients that select for different bacterial community members. In this study, agronomic management resulted in the selection of soil bacterial community members that could not be resolved when the 27F primer pair was employed with multiple restriction enzymes. Therefore, we recommend initially utilizing two different sets of primer pairs specific to the V1–V3 region of the bacterial domain in conjunction with multiple restriction enzymes to provide a greater number and diversity of sequences that generate fragments of varying sizes and rmu across the entire length of amplicon sequences. This approach will facilitate the determination of which primer pair and restriction enzymes provide the greatest coverage of soil bacterial populations for a given set of management and environmental conditions. T-RFLP profiles coupled with redundancy analyses can be used as a tool to aid in resolving the efficacy of current and future domain-specific 16S rRNA gene primers and determine the impact of environment variables on bacterial community diversity. The adaptation of existing domain-specific bacterial primers like 27F to pyrosequencing ensures that our research is pertinent to fields outside of T-RFLP analysis.
References
Alm EW, Oerther DB, Larsen N, Stahl DA, Raskin L (1996) The oligonucleotide probe database. Appl Environ Microbiol 62:3557–3559
Amann RI, Ludwig W, Schleifer K-H (1995) Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev 59:143–169
Baker GC, Smith JJ, Cowan DA (2003) Review and re-analysis of domain-specific 16S primers. J Microbiol Meth 55:541–555
Blackwood CB, Hudleston D, Zak DR, Buyer JS (2007) Interpreting ecological diversity indices applied to terminal restriction fragment length polymorphism data: insights from simulated microbial communities. Appl Environ Microbiol 73:5276–5283
Brosius J, Dull TL, Sleeter DD, Noller HF (1981) Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol 148:107–127
Brunk CF, Avaniss-Aghjani E, Brunk CA (1996) A computer analysis of primer and probe hybridization potential with bacterial small-subunit rRNA sequences. Appl Environ Microbiol 62:872–879
Clement BG, Kehl LE, DeBord KL, Kitts CL (1998) Terminal restriction fragment patterns (T-RFPs), a rapid, PCR-based method for the comparison of complex bacterial communities. J Microbiol Meth 31:135–142
Cole JR, Chai B, Farris RJ, Wang Q, Kulam SA, McGarrell DM, Garrity GM, Tiedje JM (2005) The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res 33:294–296
Colwell, RK (1997) User’s guide to estimate S5. Statistical estimation of species richness and shared species from samples. (http://www.viceroy.eeb.uconn.edu/estimates). Accessed 7 Jun 2011 and 28 Jan 2011
Dunbar J, Ticknor LO, Kuske CR (2000) Assessment of microbial diversity in four southwestern United States soils by 16S rRNA gene terminal restriction fragment analysis. Appl Environ Microbiol 66:2943–2950
Edwards RA, Rodriguez-Brito B, Wegley L, Haynes M, Breitbart M, Peterson DM, Saar MO, Alexander S, Alexander EC, Rohwer F (2006) Using pyrosequencing to shed light on deep mine microbial ecology. MC Genomics 7:1471–2164
Engebretson JJ, Moyer CL (2003) Fidelity of select restriction endonuclease in determining microbial diversity by terminal-restriction fragment length polymorphism. Appl Environ Microbiol 69:4823–4829
Erich MS, Fitzgerald CB, Porter GA (2002) The effect of organic amendments on phosphorus chemistry in a potato cropping system. Agric Ecosyst Environ 88:79–88
Frank JA, Reich CI, Sharma S, Weisbaum JS, Wilson BA, Olsen GJ (2008) Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl Environ Microbiol 74:2461–2470
Galkiewicz JP, Kellogg CA (2008) Cross-kingdom amplification using bacteria-specific primers: complications for coral microbial ecology. Appl Environ Microbiol 74:7828–7831
Hansen MC, Tolker-Nielsen T, Givskov M, Molin S (1998) 16S rDNA PCR amplification caused by interference from DNA flanking the template region. FEMS Microbiol Ecol 26:141–149
Hartmann M, Widmer F (2006) Community structure analyses are more sensitive to differences in soil bacterial communities than anonymous diversity indices. Appl Environ Microbiol 72:7804–7812
Huws SA, Edwards JE, Kim EJ, Scollan ND (2007) Specificity and sensitivity of eubacterial primers utilized for molecular profiling of bacteria within complex microbial ecosystems. J Microbiol Methods 70:565–569
Junier P, Junier T, Witzel K-P (2008) TRiFLe, a program for in silico terminal restriction fragment length polymorphism analysis with user-defined sequence sets. Appl Environ Microbiol 74:6452–6456
Kent AD, Smith DJ, Benson BJ, Triplett EW (2003) Web-based phylogenetic assignment tool for analysis of terminal restriction fragment length polymorphism profiles of microbial communities. Appl Environ Microbiol 69:6768–6776
Kunin V, Engelbrektson A, Ochman H, Hugenholtz P (2009) Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ Microbiol 12(1):118–123
Li L, Hsiao WWL, Nandakumar R, Barbuto SM, Mongodin EF, Paster BJ, Fraser-Liggett CM, Fouad AF (2010) Analyzing endodontic infections by deep coverage pyrosequencing. J Dent Res 89:980–984
Liu WT, Marsh TL, Cheng H, Forney LJ (1997) Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl Environ Microbiol 63:4516–4522
Marchesi JR, Sato T, Weightman AJ, Martin TA, Fry JC, Hiom SJ, Wade WG (1998) Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl Environ Microbiol 64:795–799
Marsh TL (1999) Terminal restriction fragment length polymorphism (T-RFLP): an emerging method for characterizing diversity among homologous populations of amplification products. Curr Opin Microbiol 2:323–327
Marsh TL, Saxman P, Cole J, Tiedje JM (2000) Terminal restriction fragment length polymorphism analysis program, a web-based research tool for microbial community analysis. Appl Environ Microbiol 66:3616–3620
Mills DK, Fitzgerald K, Litchfield CD, Gillevet PM (2003) A comparison of DNA profiling techniques for monitoring nutrient impact on microbial community composition during bioremediation of petroleum-contaminated soils. J Microbiol Methods 54(1):57–74
Muckian L, Grant R, Doyle E, Clipson N (2007) Bacterial community structure in soils contaminated by polycyclic aromatic hydrocarbons. Chemosphere 68:1535–1541
Schütte UME, Abdo Z, Bent SJ, Shyu C, Williams CJ, Pierson JD, Forney LJ (2008) Advances in the use of terminal restriction fragment length polymorphism (T-RFLP) analysis of 16S rRNA genes to characterize microbial communities. Appl Microbiol Biotechnol 80:365–380
Shyu C, Soule T, Bent SJ, Foster JA, Forney LJ (2007) MiCA: a web-based tool for the analysis of microbial communities based on terminal-restriction fragment length polymorphisms of 16S and 18S rRNA genes. J Microbial Ecol 53:562–570
Sipos R, Székely AJ, Palatinszky M, Révész S, Márialigeti K, Nikolausz M (2007) Effect of primer mismatch, annealing temperature and PCR cycle number on 16S rRNA gene-targeting bacterial community analysis. FEMS Microbiol Ecol 60(2):1574–6941
Smalla K, Oros-Sichler M, Milling A, Heuer H, Baumgarte S, Becker R, Neuber G, Kropf S, Ulrich A, Tebbe CC (2007) Bacterial diversity of soils assessed by DGGE, T-RFLP and SSCP fingerprints of PCR-amplified 16S rRNA gene fragments: do the different methods provide similar results? J Microbiol Meth 69(3):470–479
Smith CJ, Danoilowicz BS, Clear AK, Costello FJ, Wilson B, Meijer WG (2005) T-Align, a web-based tool for comparison of multiple terminal restriction fragment length polymorphism profiles. FEMS Microbiol Ecol 54:375–380
Suzuki MT, Giovannoni SJ (1996) Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol 62:625–630
Vincze T, Posfai J, Roberts RJ (2003) NEBcutter: a program to cleave DNA with restriction enzymes. Nucleic Acids Res 31:3688–3691
Wick LY, Buchholz F, Fetzer I, Kleinsteuber S, Hartig C, Shi L, Miltner A, Harms H, Pucci GN (2010) Responses of soil microbial communities to weak electric fields. J Sci Tot 408(20):4886–4893
Widenfalk A, Bertilsson S, Sundh I, Goedkoop W (2008) Effects of pesticides on community composition and activity of sediment microbes—response at various levels of microbial community organization. Environ Pollut 152(3):576–584
Acknowledgments
Funding for this project was provided from Congressional appropriations to the United States Department of Agriculture, Agriculture Research Service. The authors would like to thank Dr. Ronald Turco of Purdue University for his editorial advice and constructive suggestions.
Author information
Authors and Affiliations
Corresponding author
Additional information
Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
Rights and permissions
About this article
Cite this article
Fortuna, AM., Marsh, T.L., Honeycutt, C.W. et al. Use of primer selection and restriction enzymes to assess bacterial community diversity in an agricultural soil used for potato production via terminal restriction fragment length polymorphism. Appl Microbiol Biotechnol 91, 1193–1202 (2011). https://doi.org/10.1007/s00253-011-3363-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-011-3363-7