Abstract
Aerial conidia are central dispersing structures for most fungi and represent the infectious propagule for entomopathogenic fungus Beauveria bassiana, thus the active ingredients of commercial mycoinsecticides. Although a number of formic-acid-extractable (FAE) cell wall proteins from conidia have been characterized, the functions of many such proteins remain obscure. We report that a conidial FAE protein, termed CP15, isolated from B. bassiana is related to fungal tolerance to thermal and oxidative stresses. The full-length genomic sequence of CP15 was shown to lack introns, encoding for a 131 amino acid protein (15.0 kDa) with no sequence identity to any known proteins in the NCBI database. The function of this new gene with two genomic copies was examined using the antisense-RNA method. Five transgenic strains displayed various degrees of silenced CP15 expression, resulting in significantly reduced conidial FAE protein profiles. The FAE protein contents of the strains were linearly correlated to the survival indices of their conidia when exposed to 30-min wet stress at 48°C (r 2 = 0.93). Under prolonged 75-min heat stress, the median lethal times (LT50s) of their conidia were significantly reduced by 13.6–29.5%. The CP15 silenced strains were also 20–50% less resistant to oxidative stress but were not affected with respect to UV-B or hyperosmotic stress. Our data indicate that discrete conidial proteins may mediate resistance to some abiotic stresses, and that manipulation of such proteins may be a viable approach to enhancing the environmental fitness of B. bassiana for more persisting control of insect pests in warmer climates.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Aerial conidia of Beauveria bassiana are the infectious propagule of the entomopathogenic fungus and usually form on mycosed insects but can also be readily produced on solid substrates such as agar plates and small grains in order to be formulated as a mycoinsecticide for arthropod pest control (Feng et al. 1994; Lomer et al. 2001; Ye et al. 2006). As active ingredients, the formulated conidia usually germinate and kill insect pests around an optimal temperature of 25°C, but these cells may rapidly lose viability and insect control potential under environmental stress conditions, such as hot weather in summer (Inglis et al. 1997). Indeed, outdoor temperatures near or above 40°C are major factors that decrease the persistence and performance of fungal formulations in the field, rendering them essentially ineffective. Thus, a major current research goal is to formulate conidia with greater tolerance to higher temperatures. However, the mechanisms involved in the thermotolerance of B. bassiana and other fungal biocontrol agents are poorly understood, hindering improvement of conidial thermotolerance by means of modern biotechnology.
Enzymatic or non-enzymatic defense is known to be associated with the thermotolerance of some eukaryotic species. For instance, heat shock is known to induce the expression of a suite of heat shock proteins, of which some act as chaperones in protecting proteins against denauration and others may act directly as or induce higher catalase activities in Aspergillus nidulans (Lindquist and Kim 1996; Noventa-Jordao et al. 1999). In Aspergillus niger and other fungi, a particularly effective mechanism of thermal resistance involves changes in the lipid composition of cell membrane (Morozova et al. 2001). Alternatively, the production of small thermo- and osmo-protectants such as trehalose may help protect fungal spores from thermal damage due to the effect of these compounds on increasing the stability of intracellular enzymes (Thevelein 1984; Jagdale and Grewal 2003). Conidial cell wall proteins are also likely to participate in stress resistance although the exact role(s) for many such proteins remains unknown. Conidial proteins accumulated onto or inside cell walls (Aguado et al. 1998) are often tightly associated with these structures and can be dissociated as soluble form only with formic acid, trifluoroacetic acid, or acetic acid (Wösten 2001). In filamentous fungi, such formic-acid-extractable (FAE) proteins are generally small hydrophobin-like proteins of less than 20 kDa to be implicated in fungal growth and development (Wessels et al. 1991; Wessels 1994; Wösten 2001), conidial dispersal (Whiteford and Spanu 2001), and spore protection (Temple et al. 1997) in part due to the action of cross-linking wall components (de Vocht et al. 1998). The FAE protein profiles identified from the conidia of different B. bassiana strains span in molecular weight from 6.5 to 14.0 kDa and are often related to conidial hydrophobicity (Biodochka et al. 1995a, b; Jeffs et al. 1999) and attachment to the insect cuticle (Boucias et al. 1988). Previous work has shown that overall FAE protein contents from the conidia of six B. bassiana strains were significantly correlated to conidial tolerance to the thermal stress of 48°C (Ying and Feng 2004), indicating the likelihood that at least some of the FAE proteins may be involved in conidial tolerance to thermal stress.
Among a few major FAE proteins previously identified from B. bassiana conidia, a 15-kDa conidial protein (denoted CP15 herein) was found to be constitutively expressed (Ying and Feng 2004). The present study sought to probe the role of CP15 in mediating conidial (thermal) stress tolerance using the gene knockdown method of antisense RNA-mediated silencing (de Backer et al. 2002; Nakayashiki 2005). Our results indicate that CP15 may play a role in conidial thermotolerance, but does not appear to contribute to conidial tolerance to UV-B irradiation or osmotic stress.
Materials and methods
Microbial strains and media
The fungal strain B. bassiana 2860 (ARSEF accession number, RW Holley Center for Agriculture and Health, Ithaca, NY, USA) was preserved as dry conidia mixed with sterile sand at −76°C. Escherichia coli DH5α (Invitrogen, Carlsbad, CA, USA) was cultured in Luria–Bertani medium supplemented with 100 μg ml−1 ampicillin or 50 μg ml−1 kanamycin depending upon the plasmid resistance marker used. Fungal transformants were selected on Czapek’s medium containing 200 μg ml−1 phosphinothricin (PPT) (Ying and Feng 2006).
Extraction and purification of conidial proteins
B. bassiana was cultured for 7 days on modified Sabouraud dextrose agar plates (w/v: 0.5% glucose, 1% peptone, 1% yeast extract, and 1.5% agar; SDAY) at 25 °C and 12:12 h (light:dark cycle). Aerial conidia harvested from the agar plates were used for protein extraction with formic acid, and the extracts were dissolved in 2% (w/v) sodium dodecyl sulfate (SDS) as described previously (Ying and Feng 2004). After extraction, the protein solution was mixed 3:1 (v/v) with 75% (v/v) isopropanol, and allowed to precipitate overnight at 4°C. Proteins were collected by centrifugation (10 min, 7370× g). The pellets were rinsed once with 75% isopropanol and then dissolved in 30% (v/v) acetonitrile. The proteins in the acetonitrile solution were precipitated and rinsed again as above and then dissolved in 5 mM K2HPO4–KH2PO4 buffer (pH 7.0). Protein extracts and purified CP15 were analyzed by SDS-PAGE. The gel was stained with 0.12% Coomassie Blue R-250 (0.12 g R-250, 25 ml ethanol, 8 ml acetic acid, and 67 ml dd-H2O) and visualized in a reagent containing 25% (v/v) ethanol, and 8% (v/v) glacial acetic acid.
Sequencing the N-terminus of CP15
Proteins resolved on SDS-PAGE gels were transferred onto polyvinylidene difluoride (PVDF) membrane (Schleicher & Schuell Biosience, Dassel, Germany) in a semi-dry electrophoretic transfer cell (Bio-Rad, Hercules, CA, USA) following the manufacturer’s protocols. Protein bands were visualized with 1% Ponceau S (dissolved in 5% acetic acid) and excised from the membrane for further analysis. The N-terminus of the purified CP15 was determined on the ABI491A protein sequencer (Applied Biosystems, Carlsbad, CA, USA) using Edman method at the Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, Shanghai, China.
Cloning and sequencing of the CP15 gene
Based on the N-terminal amino acid sequence of CP15, the degenerate primer B15p was designed as 5′-CTCTTCTCNGGNCARTT-3′ (N = A, T, C or G; R = A or G) by referring to the B. bassiana codon bias table found in the Codon Usage Database at the Kazusa DNA Research Institute (Chiba, Japan; URL: http://www.kazusa.or.jp/codon/).
Total RNA was extracted from mycelia from 3 to 4-day colonies grown on the SDAY plates using TRIzol® Reagent (Invitrogen) as described elsewhere (Zou et al. 2006). The cDNA was synthesized via reverse transcription PCR (RT-PCR) of the total RNA using oligo(T) primer [5′-GACCTGAGCTGACACTGA(T)17-3′] and M-MLV reverse transcriptase (Promega, Shanghai, China). The synthesis reaction was performed in a GeneAmp® PCR System 9700 (Applied Biosystems, Foster City, CA, USA) at 25°C for 5 min, 42°C for 60 min, and 70°C for 15 min, respectively, generating the cDNA template for amplification of the target gene CP15. The CP15 cDNA clone was amplified using the primers B15p and oligo(T) in a 50-μl reaction system (2 μl cDNA solution, 0.2 μM each primer, 0.2 mM each dNTP, 2.5 mM MgCl2, 1× Taq Polymerase Buffer, and 2.5 U Taq Polymerase) by denaturation at 94°C for 2 min, followed by 35 cycles of 30 s at 95°C, 30 s at 55°C, and 60 s at 72°C, and terminating with a final extension at 72°C for 7 min. The amplified fragments were resolved in 1.5% agarose gel and cloned into pGEM-T Easy (Promega, Madison, WI, USA) using E. coli DH5α as the host strain. PCR verified clones were sequenced at Invitrogen (Shanghai, China). The plasmid carrying the CP15 cDNA clone was designated as pGEM-T-CP15.
The full-length genomic clone encoding CP15 and its 5′ upstream sequence was cloned by Y-shaped adaptor-dependent extension (YADE) (Fang et al. 2005). B. bassiana genomic DNA was isolated using the method of Raeder and Broda (1985). Restriction enzymes HincII, ScaI, EcoRV, SmaI, DraI, HindIII and XbaI (New England Biolabs, Beijing, China) were used for digesting the genomic DNA. The cDNA sequence was used to design CP15-specific primers as B15pR1 (5′-cgcttagaaatacgctggatgt-3′) and B15pR2 (5′-cggtaggactccgcagaac-3′) for linear and exponential amplification. The amplified products were cloned and sequenced as previously indicated.
The primers CP15F (5′-atgctcttctctggggggcagtctacta-3′) and CP15R (5′-aatacgctggatgct ccagcggcgg-3′) were designed to amplify the genomic sequence of the CP15 ORF via PCR using the genomic DNA as template under the following conditions: 2-min denaturation at 94°C, followed by 35 cycles of 30 s at 95°C, 30 s at 55°C and 40 s at 72°C, and a final extension at 72°C for 7 min. The amplified fragment was also cloned and sequenced as described above.
Sequence similarity of the deduced amino acid sequence of the target protein CP15 was compared by online BLAST analysis (http://www.ncbi.nlm.nih.gov/BLAST/).
Detection of CP15 in genomic DNA
Southern blotting was performed using genomic DNA (10 μg) digested with either HindIII, EcoRI, or BglII (New England Biolabs). The digested DNA was resolved on 0.7% agarose gel and was transferred onto a Biodyne B nylon membrane (Gelman Laboratory, Shelton, WA, USA). The PCR product of the CP15 gene was used as a probe. Probe preparation, membrane hybridization, and visualization were performed using DIG High Prime DNA Labeling and Detection Starter Kit II with chemiluminescence detection method (Roche, Penzberg, Germany). The hybridization and stringent washing procedures were performed overnight at 42°C and then at 68°C for 2 × 15 min, respectively.
Construction of CP15-silenced transformants
The antisense fragment of the cloned gene CP15 was amplified from genomic DNA using the primers antiB15pF (5′-ctgccatggagacctaaatacgctggatg-3′) and antiB15pR (5′-gctggactcataacctcgtaacaatgctctt-3′). The primers were designed to introduce NcoI and BamHI downstream and upstream of CP15, respectively. The fragment was digested with NcoI/BamHI and cloned into pAN52-1N (Punt et al. 1990) cut with NcoI/BamHI, resulting in an inverted insertion of CP15 between the promoter PgpdA and terminator TtrpC of A. nidulans in the pAN52-1N, yielding the antisense plasmid pAN52-antiCP15. To insert the PPT marker (bar gene) into the antisense plasmid, the bar gene was isolated from the BglII/HindIII restriction sites of pAN52-bar (Ying and Feng 2006) and then inserted into the BamHI/HindIII sites of pET29b(+) (Novagen, San Diego, CA, USA), yielding pET29b-bar. An XbaI–XbaI fragment containing the bar element was cut from the pET29b-bar and then inserted into the XbaI site of pAN52-antiCP15, forming the binary plasmid pAN52-antiCP15-bar. The integrity of the final construct was confirmed by enzyme digestion.
The binary plasmid was linearized with HindIII and then transformed into the wild-type strain B. bassiana 2860 using the methods of restriction enzyme mediated integration (REMI) and blastospore-based transformation (Ying and Feng 2006). The presence of the bar gene in putative transformants was examined by PCR and Southern blotting.
Characterization of antisense transformants
Five randomly selected B. bassiana antisense transformants (T1–T5), the wild-type parent (WT), and a control transformant (CT) bearing the empty vector pAN52-bar were grown on the plates of 1% glucose, 4% NH4NO3, 1% yeast extract, and 2% agar (w/v) at 25°C for 10–14 days. Conidia harvested from the plates were vacuum-dried and used in FAE protein analysis and stress assays. Extraction, quantification and SDS-PAGE analysis of the FAE proteins from the conidial preparations of the fungal strains were performed following a previous protocol (Ying and Feng 2004).
Comparison of CP15 expression levels in fungal colonies
All strains (T1–T5, CT and WT) were grown on SDAY plates for 3 days at 25°C. RNA was extracted from the colony culture (mycelia) of each strain using TRIzol® Reagent (Invitrogen) as described above. Quantitative real-time PCR (qRT-PCR) for CP15 was performed in 20-μl reactions containing 1 μl synthesized first strand cDNA, 10 μl 2× SYBR Green Master mix (TaKaRa), 0.4 μl 10 μM forward primer C15sf (5′-GGAGGGCTACTGTGGCGATGG-3′) and reverse primer C15sr (5′-CGGCGGGCAGGATCACCAG-3′), and 8.2 μl H2O by 10 s denaturing at 95°C, followed by 40 cycles of 5 s at 95°C and 34 s at 62°C. The 18S rRNA of B. bassiana was used as an internal standard and amplified with the paired primers Bb18Sf (5′-TGGTTTCTAGGACCGCCGTAA-3′) and Bb18Sr (5′-CCTTGGCAAATGCTTTCGC-3′). The relative expression level of CP15 was calculated as the ratio of its expression level (normalized to the 18S rRNA signal) in each transformant over that in the WT strain using the \( {2^{ - \Delta \Delta {\text{Ct}}}} \) method (Livak and Schmittgen 2001). All reactions were repeated three times, and the results were subjected to one-way variance of analysis (ANOVA).
Detection of expressed CP15 in fungal conidia
CP15 protein levels and distribution in the wild-type and transformed strains were examined by Western blotting and immunogold localization (Li et al. 2010). For antibody preparation, a His-tag version of CP15 was constructed in which the CP15 ORF was PCR-amplified using the sense primer CP15EF (5′-AGATATACATATGGCCATGGCGTCTCTTGGGGGGCA-3′) and the antisense primer CP15ER (5′-CGCAAGCTTAATACGCTGGATGCCTCAGCGGCG-3′) and then inserted into pET-29b(+) cut with NdeI/HindIII, yielding pET29-CP15. This plasmid was transformed into E. coli BL21 for expression of the (His)6-tagged recombinant protein. The expressed protein was purified from 50-ml cell culture via Ni-affinity chromatography, and the purified protein was suspended in normal saline (1 mg ml−1). The protein solution was then mixed with Freund’s complete adjuvant at a volume ratio of 1:1 and injected into 4-month-old New Zealand white rabbits three times at 14-day interval (2 ml per injection). Rabbit blood was taken 14 days after the final injection. The serum separated by 3,000× g centrifugation at 4°C was used as the polyclonal antibody (anti-CP15).
To detect the presence of CP15 in the conidia of all concerned strains, FAE protein extracts resolved on SDS-PAGE gels were electrophoretically transferred onto PVDF membranes for Western blotting with the anti-CP15. All blots were probed with 500-fold dilution of the antibody and visualized with goat anti-rabbit IgG-alkaline phosphatase conjugate (Novagen).
For immunogold localization, the conidia of T1, CT, and WT were separately fixed in 1% paraformaldehyde and 2.5% glutaraldehyde in 0.1 M phosphate buffer saline (PBS, pH 7.2) at 4°C for 2 h, followed by washing three times in water. After 1 min centrifugation at 16,100× g, the pellets were dehydrated at −20°C using a gradient ethanol series and finally embedded in fresh pre-cooled resin of Lowicryl K4M (Plano, Wetzlar, Germany). The resins were photopolymerized in capsules under 360-nm ultraviolet illumination at −35°C for 72 h and then at room temperature for 48 h. The final resin pyramids were cut into 50 to 70-nm sections. These ultrathin sections were then mounted on 200-mesh Bioden Meshcement (Oken Shoji, Tokyo, Japan) coated with nickel grids (TAAB Laboratories, Berkshire, UK). The obverse side of the grids was treated for 30 min with a solution of 50 mM PBS, 1% bovine serum albumin, 0.02% PEG-20000, 100 mM NaCl and 0.1% NaNO3, followed by incubation with 100× anti-CP15 dilution for 2 h and then with 100× dilution of 10-nm colloidal gold goat anti-rabbit IgG (Sigma) for another 2 h. The sections were finally contrasted with 1% uranyl acetate for 12 min and observed under a transmission electron microscope.
Assays for stress tolerance of antisense transformants
Thermotolerance assay
For each of the seven strains (T1–T5, CT and WT), conidia were suspended in 1 ml of 0.02% Tween 80 in 1.5 ml glass vials and exposed to thermal stress by immersing the vials in a water bath at 48°C for up to 75 min as described previously (Ying and Feng 2004; Li and Feng 2009). Conidial viability (24-h germination rate on SDAY plates at 25°C) of a stressed sample relative to that of the unstressed control was defined as the survival index (I s). Such estimates were made by taking 100-μl samples during a time course of thermal stress. All assays were repeated three times (three vials per strain), with at least two independent batches of cells.
The inverted sigmoid I s trends of various strains over the stress time (t) were subjected to two-way ANOVA and then fitted to the logistic equation \( {I_{\text{s}}} = K/\left[ {1 + \exp \left( {a + rt} \right)} \right] \) where K = 1 due to I s ≤ 1, a was the intercept for the fitted curve, and r the rate of decline in conidial viability under the thermal stress (relative to the unstressed conidia). When I s = 0.5, the fitted equation gave a solution (−a/b) to median lethal time (LT50). This value was used to quantify the conidial thermotolerance of each strain.
Oxidative stress assay
The seven strains were grown on SDAY plates containing 0.1 mM menadione for their tolerance to the oxidative stress of the superoxide-generating compound (Xie et al. 2010). Briefly, 10-μl aliquots of spore suspension (106 conidia ml−1) were spread evenly on the plates, followed by 24-h incubation at 25°C. Percent germinations were determined using the microscopic counts of germinated and non-germinated conidia. The menadione survival index was calculated as the ratio of the percent germination of a given strain in the presence of menadione over that in the absence of menadione.
Hyperosmotic stress assay
For each strain, 10-μl aliquots of spore suspension (106 conidia ml−1) were spread evenly on the plates of Czapek’s agar supplemented with 0–1.5 M NaCl for different degrees of osmotic stress (Zhang et al. 2009). After 24-h incubation at 25°C, percent germinations on the plates were determined as above, and the osmotic survival index was estimated as the ratio of the percent germination on the salt-inclusive plate over that on the salt-free plate.
UV-B stress assay
All transformed and wild-type strains were assayed for their tolerance to UV-B irradiation as described elsewhere (Huang and Feng 2009). Briefly, 10-μl aliquots of spore suspension (107 conidia ml−1) in 0.02% Tween 80 containing 0.5% peptone and 2% sucrose were spotted centrally on glass slides. Dried in air at room temperature, the slides were placed on the sample tray of Bio-Sun++ UV irradiation chamber (Vilber Lourmat, Marne-la-Vallée, France) and then exposed to a UV-B irradiation of 312 nm weighted wavelength at a controlled dosage of 0.4 J cm−2. After exposure, the slides were incubated for 24 h at 25°C under saturated humidity, and the percent germination of the irradiated conidia was determined using microscopic counts. The UV-B survival index of each strain was assessed as the ratio of the percent germination of the irradiated conidia over that in the non-irradiated control.
All survival indices of the tested strains in the oxidative, hyperosmotic, or UV-B stress assay were subjected to one-way ANOVA and then compared by Fisher’s least significant difference (Fisher’s LSD).
Results
Purification of CP15 and its N-terminal sequence
The FAE proteins from the conidia of the wild-type strain B. bassiana 2860 (WT) were extracted with formic acid, precipitated by isopropanol and dissolved in 2% SDS solution as detailed in the Materials and methods section. After the final step of isopropanol precipitation, the cell wall extract was resuspended in 30% acetonitrile and analyzed by SDS-PAGE (Fig. 1a). These data showed a major protein band of ∼15 kDa. The first 11N-terminal amino acids of the 15 kDa protein (i.e., CP15) were determined to be SFPDGQFIIRN after transfer to a PVDF membrane.
Gene cloning of the conidial protein CP15 of B. bassiana 2860. a SDS-PAGE profile for the purified CP15 (lane 1) from the crude extract (lane C) after selective precipitation (M mol. marker). b Agarose gel electrophoresis of the 3′-RACE CP15 products (∼500 bp) amplified with RT-PCR (M DNA marker). c Full-length sequence of the cloned CP15 with the uppercased 393-bp ORF encoding a 131 amino acid protein. Partial up- and downstream sequences are lowercased (underlined putative polyadenylation signal). d Southern blotting of the CP15 gene in the genomic DNA of B. bassiana using linearized pGEM-T-CP15 (3.5 kbp) and pAN52-antiCP15-bar (9.6 kbp) as markers (M). Lanes 1–3 loaded with genomic DNA digested with EcoRI, HindIII and BglII, respectively
Molecular cloning of the CP15 gene
A 500-bp product (Fig. 1b) was amplified by RT-PCR from a B. bassiana cDNA library using a degenerate primer based on the first seven amino acids of the deduced N-terminus as the forward primer and an oligo-dT as the reverse primer. The resultant band was cloned and sequenced, and an analysis of the deduced OFR revealed that the subsequent 4 amino acids (after the first seven used to design the primer) were identical to those of the sequenced N-terminus of CP15, indicating that the amplified fragment was the cDNA clone of CP15 which included a 3′ untranslated region and a putative polyadenylation signal of AAAATT (Fig. 1c).
The full-length gene CP15 (GenBank accession number: EF177489) was then isolated by cloning its upstream sequence (Fig. 1c) as detailed in the Materials and methods section. The upstream sequence was A/T rich (69%), and sequence analysis showed that CP15 consisted of a 393-bp ORF and no introns, yielding a protein product of 131 amino acids. Intriguingly, two apparent copies of the cloned gene were found in the B. bassiana genome, as shown in Southern blotting using the entire ORF as a probe (Fig. 1d). The deduced protein sequence of CP15 was rich in Asp, Asn (both 20%), and Arg (12.5%) residues. No similarity was found between the deduced amino acid sequence and any known proteins in the GenBank database.
Characterization of CP15-silenced transformants
Based upon the isolated cDNA sequence of CP15, an antisense construct was made and transformed into the REMI-mediated competent blastospores of B. bassiana 2860 using the bar gene as selective marker. PCR and Southern blot analysis of five transformants (T1–T5), plus one transformed with the bar marker plasmid with no insert of the CP15 antisense construct (designated as CT), indicated that the inserted bar gene was present in these strains but absent in the wild-type parent (Fig. 2a, b). Three parameters were examined in order to confirm CP15 silencing. (1) The qRT-PCR analysis showed ∼80 to 95% decrease in CP15 transcripts in the various T1–T5 strains as compared to the WT or CT strain (Fig. 2e). Transformant T2 appeared to have the highest level of silencing, whereas T5 still expressed significant levels of the CP15 transcript. (2) The overall FAE protein contents of T1–T5 conidia ranged from 20.4 to 22.4 μg–mg−1 (Fig. 2f), which were significantly lower than the measurements from the WT (24.1 μg mg−1) and CT (23.2 μg mg−1) strains (F 6,12 = 105.9, p < 0.01). The measured FAE protein contents were linearly correlated to the levels of the CP15 transcripts in the seven strains (r 2 = 0.71, F 1,5 = 12.4, p = 0.017). (3) SDS-PAGE analysis indicated loss of the CP15 protein in T1–T5 extracts (Fig. 2c) while Western blots confirmed loss or reduction of the CP15 protein in the T1–T5 cells (Fig. 2d), although faint amounts of CP15 could be detected in the T1 and T4 cells. In contrast, two other major FAE proteins (12.0 and 17.5 kDa) were expressed in T1–T5, but showed no cross reactivity with the anti-CP15 antibody.
Silenced expression of CP15 in five antisense transformants (T1–T5) of B. bassiana. a PCR detection of the bar gene in the genomic DNAs of T1–T5, WT, (wild-type strain) and CT (a control transformant bearing the bar gene marker only). b Southern blotting of the bar in the genomic DNAs of T1–T5, WT, and CT strains using linearized pABeG (10.0 kbp; Ying and Feng 2006) and pAN52-bar (6.28 kbp)] as markers. c SDS-PAGE profiles of CP15 (arrowed) in the formic-acid conidial extracts of T1–T5, WT, and CT. d Western blotting of CP15 (arrowed) in the extracts of T1–T5 (negative), WT (positive), and CT (positive). Lane E loaded with the recombinant CP15 purified from the cell culture of transgenic E. coli. e The relative expression levels of the CP15 gene in the 3-day SDAY colonies of various strains, determined by qRT-PCR. f Overall contents of the FAE proteins extracted from the conidia of various strains. The different letters on bars denote significant differences (Fisher’s LSD, p < 0.05; error bars SD)
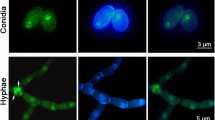
The same antibody was also used in immunogold localization experiments. These data indicated more abundant labeling in the conidial cytoplasm of WT and CT than T1 (Fig. 3).
Electron micrographs of CP15 in B. bassiana conidia. The CP15 protein molecules in the conidia of the wild-type strain (a), the CP15-silenced strain T1 (b), and the control transformant (c) were sequentially labeled with the polyclonal antibody (anti-CP15) and goat anti-rabbit IgG antibody coupled to 10-nm colloidal gold particles (enlarged in framed squares). Scale bars 0.5 μm in conidial sections or 0.25 μm in enlarged squares
Stress tolerance changes in CP15-silenced transformants
Several phenotypic features were examined in the CP15-silenced transformants. The survival indices (I s) of the T1–T5 conidia after 30-min heat stress at 48°C ranged from 0.388 (T2) to 0.546 (T3) and were significantly reduced compared to those of WT (0.664) and CT (0.660) (F 6,12 = 19.7, p < 0.01) strains. These I s values were also linearly correlated to the overall FAE protein contents of the seven strains (r 2 = 0.928, F 1,5 = 64.4, p < 0.0001). During the 75-min stress assays at 48°C, the declining I s trends varied significantly with the tested strains (F 6,70 = 65, p < 0.01) and stress time (F 4,70 = 3082, p < 0.01) based on two-way ANOVA. However, no such significant difference was found between the I s trends of WT and CT strains (Fisher’s LSD, p > 0.05). These data indicated that the conidial survival indices of all the strains decreased with incubation time at 48°C. The observed I s trends over stress time (t) fit well the equation I s = 1 / [1 + exp(a + rt)] (r 2 > 0.98, p < 0.01). The fitted I s–t relationships resulted in the LT50 estimates (mean ± SD) of 30.0 ± 1.23, 27.4 ± 0.32, 33.6 ± 0.44, 31.8 ± 0.26, 31.4 ± 1.66, 38.9 ± 0.94, and 41.2 ± 1.02 min for T1, T2, T3, T4, T5, WT and CT strains (Fig. 4a), respectively. Thus, silencing of the CP15 gene appeared to be responsible for a reduction of 5.3–11.5 min in conidial LT50 under the thermal stress of 48°C, contributing 13.6–29.5% of the conidial thermotolerance to the wild-type strain.
Phenotypic comparisons of the CP15-silenced strains (T1–T5), wild-type strain (WT), and control transformant (CT). a The conidial LT50s estimated from the fitted I s–t relationships for the tested strains under the thermal stress of 48°C. b–d The conidial survival indices of the tested strains under the stresses of 0.1 mM menadione (oxidative), UV-B irradiation (0.4 J cm−2), and 0.5–1.5 M NaCl (hyperosmotic), respectively. The grouped bars from left to right in d represent WT, T1–T5, and CT, respectively. The different letters on the bars of each graph denote significant differences (Fisher’s LSD, p < 0.05). Error bars SD
Moreover, the survival indices of the T1–T5 strains (Fig. 4b) on menadione-inclusive agar plates ranged from 0.469 (T1) to 0.828 (T3), which were significantly lower than those of WT (0.982) and CT (0.964) strains (F 6,12 = 74.3, p < 0.01). However, the conidial survival indices (0.54–0.57) of the seven strains (Fig. 4c) irradiated at the UV-B dosage of 0.4 J cm−2 did not differ from one to another (F 6,12 = 0.20, p = 0.97). Nor were the survival indices significantly different between the T1–T5, WT, and CT strains with respect to the increasing concentration of 0.5–1.5 M NaCl, because the indices were equally reduced from ∼0.91 to ∼0.05 for all the tested strains (Fig. 4d).
Discussion
B. bassiana mediated virulence towards insects represents a unique model system with which to address host–pathogen interactions (Lewis et al. 2009; Wanchoo et al. 2009; Bidochka et al. 2010). Recently, the use of molecular methods has expanded research in this field to examining the genes and molecular mechanisms involved in pathogenesis (Fang et al. 2008, 2009; Jin et al. 2010; Zhang et al. 2010). To date, however, gene silencing methods have not been used in B. bassiana. The new CP15 gene cloned from B. bassiana in this study corresponded to a 393-bp ORF, with no introns in its genomic sequence, encoding for a 131 amino acid protein product of 15.0 kDa deduced molecular weight. Immunogold labeling experiments indicated that CP15 was present mainly in conidial cytoplasm despite its extractability with formic acid. Phenotypic analysis of CP15 antisense bearing transformants indicated that CP15 contributes to conidial thermotolerance and resistance to the reactive oxygen species (ROS) generated by menadione, but is not involved in conidial resistance to UV-B irradiation and hyperosmotic stress. These findings support our hypothesis that FAE conidial proteins may play important role(s) in conidial thermotolerance in B bassiana and possibly other entomogenous fungi (Ying and Feng 2004).
FAE proteins are abundant in the conidia of fungal biocontrol agents (Bidochka et al. 1995a, b; Jeffs et al. 1999; Shan et al. 2010), and CP15 is one of three major proteins that can be dissociated from the B. bassiana conidia using formic acid in the previous study (Ying and Feng 2004). In this study, CP15 was well resolved from other FAE proteins by means of selective dissolution in 30% acetonitrile. FAE proteins with low water solubility can be blotted onto PVDF membrane for N-terminal sequencing after being separated by SDS-PAGE (Mankel et al. 2002; Gribun et al. 2004), and our data indicate that this method is effective for the purification of CP15. N-terminal amino acid sequencing of CP15 indicated that the protein CP15 matured after cleaving the first residue (Met), since this amino acid was present in the determined amino acid sequence. Many FAE conidial proteins from filamentous fungi including B. bassiana are known as hydrophobins or hydrophobin-like proteins (Bidochka et al. 1995a, b; Jeffs et al. 1999). These proteins may favor conidial attachment and infection to insect pests (Boucias et al. 1988; Holder and Keyhani 2005; Cho et al. 2007). Our data indicate that CP15 is a hitherto uncharacterized protein and unique to B. bassiana, since no significant matches were found in the GenBank database. Recently, a non-hydrophobic FAE protein (CWP10) of Metarhizium anisopliae was found enhancing conidial hydrophobicity when expressed in transgenic B. bassiana (Li et al. 2010). Also, the reduction of FAE protein contents in the conidia of chemically induced B. bassiana mutants was associated with an increase in benzimidazole resistance but a decrease in conidial thermotolerance (Zou et al. 2006). These data indicate that non-hydrophobin FAE conidial proteins are likely to have important functions, potentially in relation to stress resistance. As a cytoplasmic FAE protein, the protein CP15 is likely associated with certain intracellular component and may act as a cytoplasm structural component to contribute to conidial thermotolerance. This is similar to some cytoplasm proteins (e.g. small heat protein) that protect cells from stress damages (Klis et al. 2002; Imazu and Sakurai 2005; Sakthivel et al. 2009). The suppressed expression of CP15 in the antisense transformants is likely to make conidial cytoplasm looser or unstable, as clued from the reduced contents of their FAE proteins. Thus, their conidia became more susceptible to the thermal and oxidative stresses.
Interestingly, the B. bassiana CP15 gene appeared to possess two genomic copies and an atypical (for fungi) polyadenylation signal, which is more similar to mammalian poly(A) signals (Tabaska and Zhang 1999). The upstream A/T rich sequence, however, was similar to that found for fungal genes (Saito et al. 1998). The function of CP15 was probed via silencing in B. bassiana using an antisense-RNA derived method. This method can be used to suppress target gene expression essentially irrespective of the genomic inserted position of the antisense plasmid, i.e., no need to target the gene of interest via homologous recombination which would not have been possible due to the presence of two copies of CP15 in the B. bassiana genome (Nakayashiki 2005). Our data indicated that CP15 silencing in the five antisense transformants apparently via an RNAi mediated pathway did not interfere with the expression of the genes coding for other FAE proteins, e.g., 12.0 and 17.0 kDa. The suppression of CP15 expression in the transformants is likely due to the high efficiency of the PgpdA promoter (Punt et al. 1990), which drove the expression of the antisense transcript, and to our knowledge this is the first example of antisense silencing of a gene in B. bassiana. The phenotypes of these strains included reduced FAE protein contents, decreased conidial tolerance to thermal and oxidative stresses, and presumably showed the effectiveness of using an antisense approach method in functional studies of genes with two or more copies.
In conclusion, B. bassiana CP15 appears to contribute to conidial thermotolerance and resistance to oxidative stress but is not involved in either UV-B or osmotic stress, indicating that individual FAE proteins may be involved in discrete stress responses. Thus, future experiments examining other FAE proteins for their possible functions in fungal resistance or tolerance to environmental stresses are warranted. From a practical point of view, our results will help to explore means to manipulating conidial environmental fitness of the fungal biocontrol agents for more persistent control of insect pests in summer.
References
Aguado C, Rufz-Herrera J, Iranzo M, Sentandreu R, Mormeneo S (1998) Reaggregation and binding of cell protein from Candida albican to structural polysaccharides. Res Microbiol 149:327–338
Bidochka MJ, St Leger RJ, Joshi L, Robert DW (1995a) The rodlet layer from aerial and submerged conidia of entomopathogenic fungus Beauveria bassiana contains hydrophobin. Mycol Res 99:403–406
Bidochka MJ, St Leger RJ, Joshi L, Robert DW (1995b) An inner cell wall protein (cwp1) from conidia of the entomopathogenic fungus Beauveria bassiana. Microbiology 141:1075–1080
Bidochka MJ, Clark DC, Lewis MW, Keyhani NO (2010) Could insect phagocytic avoidance by entomogenous fungi have evolved via selection against soil amoeboid predators? Microbiology 156:2164–2171
Boucias DG, Pendland JC, Latge JP (1988) Nonspecific factors involved in attachment of entonomopathogenic Deuterromycetes to host insect cuticle. Appl Environ Microbiol 54:1795–1805
Cho EM, Kirkland BH, Holder DJ, Keyhani NO (2007) Phage display cDNA cloning and expression analysis of hydrophobins from the entomopathogenic fungus Beauveria (Cordyceps) bassiana. Microbiology 153:3438–3447
de Backer MD, Raponi M, Arndt GM (2002) RNA-mediated gene silencing in non-pathogenic and pathogenic fungi. Curr Opin Microbiol 5:323–329
de Vocht ML, Scholtmeijer K, van der Vegte EW, de Vries OMH, Sonveaux N, Wösten HAB, Ruysschaert JM, Hadziioannou G, Wessels JGH, Robillard GT (1998) Structural characterization of the hydrophobin SC3, as a monomer and after self-assembly at hydrophobic/hydrophilic interfaces. Biophys J 74:2059–2068
Fang WG, Leng B, Xiao YH, Jin K, Ma JC, Fan YH, Feng J, Yang XY, Zhang YJ, Pei Y (2005) Cloning of Beauveria bassiana chitinase gene Bbchit1 and its application to improve fungal strain virulence. Appl Environ Microbiol 71:363–370
Fang WG, Scully LR, Zhang L, Pei Y, Bidochka MJ (2008) Implication of a regulator of G protein signalling (BbRGS1) in conidiation and conidial thermotolerance of the insect pathogenic fungus Beauveria bassiana. FEMS Microbiol Lett 279:146–156
Fang WG, Feng J, Fan YH, Zhang YJ, Bidochka MJ, St Leger RJS, Pei Y (2009) Expressing a fusion protein with protease and chitinase activities increases the virulence of the insect pathogen Beauveria bassiana. J Invertebr Pathol 102:155–159
Feng MG, Poprawski TJ, Khachatourians GG (1994) Production, formulation and application of the entomopathogenic fungus Beauveria bassiana for insect control: current status. Biocontrol Sci Technol 4:3–34
Gribun A, Kactoff DJ, Hershkovits G, Pechatnikov I, Nitzan Y (2004) Cloning and characterization of the gene encoding for OMP-PD porin: the major Photobacterium damsela outer membrane protein. Curr Microbiol 48:167–174
Holder DJ, Keyhani NO (2005) Adhesion of the entomopathogenic fungus Beauveria (Cordyceps) bassiana to substrata. Appl Environ Microbiol 71:5260–5266
Huang BF, Feng MG (2009) Comparative tolerances of various Beauveria bassiana isolates to UV-B irradiation with a description of a modeling method to assess lethal dose. Mycopathologia 168:145–152
Imazu H, Sakurai H (2005) Saccharomyces cerevisiae heat shock transcription factor regulates cell wall remodeling in response to heat shock. Eukaryot Cell 4:1050–1056
Inglis GD, Johnson DL, Goettel MS (1997) Efftects of temperature and sunlight on mycosis (Beauveria bassiana) (Hyphomycetes: Sympodulosporae) of grasshoppers under field conditions. Environ Entomol 26:400–409
Jagdale GB, Grewal PS (2003) Acclimation of entomopathogenic nematodes to novel temperatures: trehalose accumulation and the acquisition of thermotolerance. Int J Parasitol 33:145–152
Jeffs LB, Xavier HJ, Matai RE, Khachatourians GG (1999) Relationships between fungal spore morphologies and surface properties for entomopathogenic members of the genus Beauveria, Metarhizium, Paecilomyces, Tolypocladium, and Verticillium. Can J Microbiol 45:936–948
Jin K, Zhang YJ, Fang WG, Luo ZB, Zhou YH, Pei Y (2010) Carboxylate transporter gene JEN1 from the entomopathogenic fungus Beauveria bassiana is involved in conidiation and virulence. Appl Environ Microbiol 76:254–263
Klis FM, Mol P, Hellingwerf K, Brul S (2002) Dynamics of cell wall structure in Saccharomyces cerevisiae. FEMS Microbiol Rev 26:239–256
Lewis MW, Robalino IV, Keyhani NO (2009) Uptake of the fluorescent probe FM4-64 by hyphae and haemolymph-derived in vivo hyphal bodies of the entomopathogenic fungus Beauveria bassiana. Microbiology 155:3110–3120
Li J, Feng MG (2009) Intraspecific tolerance of Metarhizium anisopliae conidia to the upper thermal limits of summer with a description of a quantitative assay system. Mycol Res 113:93–99
Li J, Ying SH, Shan LT, Feng MG (2010) A new non-hydrophobic cell wall protein (CWP10) of Metarhizium anisopliae enhances conidial hydrophobicity when expressed in Beauveria bassiana. Appl Microbiol Biotechnol 85:975–984
Lindquist S, Kim G (1996) Heat-shock protein 104 expression is sufficient for thermotolerance in yeast. Proc Natl Acad Sci USA 93:5301–5306
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the \( {2^{ - \Delta \Delta {\text{Ct}}}} \) method. Methods 25:402–408
Lomer CJ, Bateman RP, Johnson DL, Langewald J, Thomas M (2001) Biological control of locusts and grasshoppers. Annu Rev Entomol 46:667–702
Mankel A, Krause K, Kothe E (2002) Identification of a hydrophobin gene that is developmentally regulated in the ectomycorrhizal fungus Tricholoma terreum. Appl Environ Microbiol 68:1408–1413
Morozova EV, Baranova MV, Kozlov VP, Tereshina VM, Memorskaya AS, Feofilova EP (2001) Peculiarities of exogenous dormancy of Aspergillus niger conidia. Microbiology 70:527–534
Nakayashiki H (2005) RNA silencing in fungi: mechanisms and applications. FEBS Lett 579:5950–5957
Noventa-Jordao MA, Couto RM, Goldman MHS, Aguirre J, Lyer S, Caplan A, Terenzi H, Goldman GH (1999) Catalase activity is necessary for heat-shock recovery in Aspergillus nidulans germlings. Microbiology 145:3229–3234
Punt PJ, Dingemanse MA, Kuyvenhoven A, Soede RDM, Pouwels PH, van den Hondel CAMJJ (1990) Functional elements in the promoter region of the Aspergillus nidulans gpdA gene encoding glyceraldehyde-3-phosphate dehydrogenase. Gene 93:101–109
Raeder U, Broda P (1985) Rapid preparation of DNA from filamentous fungi. Lett Appl Microbiol 1:17–20
Saito K, Yamazaki H, Ohnishi Y, Fujimoto S, Takahashi E, Horinouchi S (1998) Production of trehalose synthase from a basidiomycete, Grifola frondosa, in Escherichia coli. Appl Microbiol Biotechnol 50:193–198
Sakthivel K, Watanabe T, Nakamoto H (2009) A small heat-shock protein confers stress tolerance and stabilizes thylakoid membrane proteins in cyanobacteria under oxidative stress. Arch Microbiol 191:319–328
Shan LT, Wang ZL, Ying SH, Feng MG (2010) Hydrophobicity-related protein contents and surface areas of aerial conidia are useful traits for formulation design of fungal biocontrol agents. Mycopathologia 169:483–494
Tabaska JE, Zhang MQ (1999) Detection of polyadenylation signals in human DNA sequences. Gene 231:77–86
Temple B, Horge PA, Bernier L, Hintz WE (1997) Cerato-ulmin, a hydrophobin secreted by the causal agents of Ducth elm disease, is a parasitic fitness factor. Fungal Genet Biol 22:39–53
Thevelein JM (1984) Regulation of trehalose mobilization in fungi. Microbiol Rev 48:42–59
Wanchoo A, Lewis MW, Keyhani NO (2009) Lectin mapping reveals stage-specific display of surface carbohydrates in vitro and haemolymph-derived cells of the entomopathogenic fungus Beauveria bassiana. Microbiology 155:3121–3133
Wessels JGH (1994) Developmental regulation of fungal cell wall formation. Annu Rev Phytopathol 32:413–437
Wessels JGH, de Vries OMH, Asgeirsdottir SA, Schuren FHJ (1991) Hydrophobin genes involved in formation of aerial hyphae and fruit bodies in Schizophyllum commune. Plant Cell 3:793–799
Whiteford JR, Spanu PD (2001) The hydrophobin HCf-1 of Cladosporium fulvum is required for efficient water-mediated dispersal of conidia. Fungal Genet Biol 32:159–168
Wösten HAB (2001) Hydrophobins: multipurpose proteins. Annu Rev Microbiol 55:625–646
Xie XQ, Wang J, Huang BF, Ying SH, Feng MG (2010) A new manganese superoxide dismutase identified from Beauveria bassiana enhances virulence and stress tolerance when overexpressed in the fungal pathogen. Appl Microbiol Biotechnol 86:1543–1553
Ye SD, Ying SH, Chen C, Feng MG (2006) New solid-state fermentation chamber fits up bulk production of aerial conidia of fungal biocontrol agents on rice. Biotechnol Lett 28:799–804
Ying SH, Feng MG (2004) Relationship between thermotolerance and hydrophobin- like proteins in aerial conidia of Beauveria bassiana and Paecilomyces fumosoroseus as fungal biocontrol agents. J Appl Microbiol 97:323–331
Ying SH, Feng MG (2006) Novel blastospore-based transformation system for integration of phosphinothricin resistance and green fluorescence protein genes into Beauveria bassiana. Appl Microbiol Biotechnol 72:206–210
Zhang YJ, Zhao JH, Fang WG, Zhang JQ, Luo ZB, Zhang M, Fang YH, Pei Y (2009) Mitogen-activated protein kinase hog1 in the entomopathogenic fungus Beauveria bassiana regulates environmental stress responses and virulence to insects. Appl Environ Microbiol 75:3787–3795
Zhang S, Fan Y, Xia YX, Keyhani NO (2010) Sulfonylurea resistance as a new selectable marker for the entomopathogenic fungus Beauveria bassiana. Appl Microbiol Biotechnol 87:1151–1156
Zou G, Ying SH, Shen ZC, Feng MG (2006) Multi-sited mutations of beta-tubulin are involved in benzimidazole resistance and thermotolerance of fungal biocontrol agent Beauveria bassiana. Environ Microbiol 8:2095–2106
Acknowledgements
The authors expressed their sincere thanks to Nemat O. Keyhani (University of Florida, USA) for valuable comments during manuscript preparation. Funding of this study was provided jointly by the Natural Science Foundation of China (30930018 and 30971960) and the Ministry of Science and Technology (2009CB118904 and 2007DFA3100).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ying, SH., Feng, MG. A conidial protein (CP15) of Beauveria bassiana contributes to the conidial tolerance of the entomopathogenic fungus to thermal and oxidative stresses. Appl Microbiol Biotechnol 90, 1711–1720 (2011). https://doi.org/10.1007/s00253-011-3205-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-011-3205-7