Abstract
Light-mediated control of gene expression and thus of any protein function and metabolic process in living microbes is a rapidly developing field of research in the areas of functional genomics, systems biology, and biotechnology. The unique physical properties of the environmental factor light allow for an independent photocontrol of various microbial processes in a noninvasive and spatiotemporal fashion. This mini review describes recently developed strategies to generate photo-sensitive expression systems in bacteria and yeast. Naturally occurring and artificial photoswitches consisting of light-sensitive input domains derived from different photoreceptors and regulatory output domains are presented and individual properties of light-controlled expression systems are discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Precise and spatiotemporal control of gene expression by an external stimulus is required to analyze and control complex biological processes at the molecular level within a living cell. Light represents such an environmental stimulus as it offers a high level of flexibility without changing metabolic conditions and additionally allows studying biological systems in vivo with spatial and temporal resolution. Thus, light-mediated modulation of gene expression is a rapidly advancing research field in the areas of functional genomics, systems biology, and biotechnology.
Biological reactions can be induced by light using either photocaged molecules and enzymes or photoreceptors which possess a light-sensitive domain harboring a small light-absorbing ligand called chromophore. Due to their photophysical properties, both photocaged compounds and naturally occurring chromophoric molecules absorb light of a defined wavelength and subsequently initiate a light-triggered reaction. Such a photo-induced reaction leads either to an irreversible release of the caging group or to a reversible formation of a light-activated photoreceptor state. Therefore, both mechanisms can basically be applied to gain specific control over gene expression in a light-dependent manner.
The concept of caged molecules (Kaplan et al. 1978) carrying a protecting group which can be removed by irradiation with ultraviolet (UV) light to restore the respective biological function (Fig. 1a) has recently been reviewed, in detail, (Riggsbee and Deiters 2010; Heckel and Mayer 2010; Deiters 2010; Lee et al. 2009; Young and Deiters 2007b; Mayer and Heckel 2006; Shao and Xing 2010). Hence, this concept is only briefly described here with 2-nitrobenzyl derivatives as a well-studied example (see, for instance, Schaper et al. 2009).
Chemical caging to control gene expression. a Concept of caged effectors. Caged effector molecules are released upon UV light irradiation. b Caged small molecule effectors. Caged IPTG and doxycycline enable the control of gene expression. c Caged protein effectors. Caged inactive T7 RNA polymerase is activated by UV irradiation to allow for the photochemical regulation of orthogonal gene expression. The active effector molecules and proteins are highlighted in green
Many small effector molecules exist within cells which can bind to proteins and thus modulate their biological activity. These molecules or the respective proteins can be caged and subsequently activated by irradiation allowing for the direct or indirect control of gene expression. Examples include caged estradiol (Cruz et al. 2000), β-ecdysone (Lin et al. 2002), hydroxytamoxifen (Link et al. 2005), doxycycline (Cambridge et al. 2006), isopropyl-β-d-galactoside (IPTG; Young and Deiters 2007a), and toyocamycin (Young et al. 2009). For instance, it has been demonstrated that caged IPTG 1 (Fig. 1b, left) can be used to control the expression of target genes under the control of a lac operator in bacterial cells in vivo (Young and Deiters 2007a). While caged IPTG is unable to bind to the repressor LacI, UV light irradiation releases IPTG, which induces gene expression by binding to LacI which removes the lac repressor from its cognate promoter. It should be noted that the primary product of the light-induced reaction, the corresponding ester formed from acetal 1, needs to be hydrolyzed in the cytoplasm before IPTG is released. Despite the fact that a more defined spatial control over gene expression was achieved with caged doxycycline 2 (Fig. 1b, right) in mammalian cells (Sauers et al. 2010), it is still difficult to target a light signal to specific cells in vivo due to free diffusion of the caged and photoreleased compounds. Although these molecules were completely inactive in the caged state and full biological activity was restored upon decaging, it is reasonable to assume that several caged compounds will still exhibit residual activity and their activity might as well not completely be restored upon irradiation (for a detailed discussion, see Deiters 2010).
As an alternative to the utilization of small inducer molecules, a caging group can be incorporated directly into a protein. The most convenient approach here is the introduction of caged amino acids at specific sites in the protein. Bacteria and yeast cells with an expanded genetic code are used carrying orthogonal transfer ribonucleic acid (tRNA) synthase–tRNA pairs to produce caged enzymes (for a review, see Wang et al. 2006). In a recent example, a caged tyrosine was introduced at position 639 of the T7 RNA polymerase (Chou et al. 2010), thus rendering the polymerase inactive (Fig. 1c). Upon decaging, the polymerase activity was fully restored and gene expression enabled.
The described genetic tools that combine photochemical and biological properties to achieve light-mediated control over gene expression offer various applications in complex settings. However, in all cases, the use of caged compounds exhibits some limitations that may hamper the targeting of the light signal to specific cells in vivo as well as its application in a broad range of different organisms. Thus, the development of genetically encoded artificial photoreceptors can help to circumvent these system-inherent limitations. In microorganisms, photocontrol of gene expression is naturally implemented by different photoreceptor protein families each carrying chromophore-binding input domains and light-triggered output domains in a modular fashion. Only recently, the natural toolbox of photo-sensitive regulatory systems is exploited for the construction of recombinant photoswitches with novel regulatory properties (Deiters 2010; Möglich and Moffat 2010).
This review article will focus on the construction of genetically encoded light-triggered switches that can be used to achieve photocontrol over gene expression both in vitro and in vivo. Special emphasis is placed on the description of naturally occurring and recombinant photoreceptors to functionally link light perception to gene expression control in pro- and eukaryotic microorganisms.
Microbial photoreceptors for light-mediated control of gene expression
Photoreceptor proteins constitute highly sophisticated regulatory systems which enable the fine-tuned control of diverse biological processes. Different chromophores bound within the respective photoreceptor proteins capture the incoming radiation and thus enable different photoreceptor proteins to respond to different wavelengths of the visible spectrum (van der Horst and Hellingwerf 2004). As initial photochemical event, light absorption triggers the rapid formation of the excited state of the chromophore resulting in a change of the electron distribution which enables subsequent reactions of the chromophore usually causing a conformational change of the surrounding protein (van der Horst and Hellingwerf 2004). Since photoreceptors are often modularly built multidomain proteins, a structural perturbation in the photoreceptor domain is relayed to coupled effector modules which display or mediate different light-dependent biological activities (Losi and Gärtner 2008). In nature, light-mediated control of gene expression represents an ubiquitous mechanism by which organisms of all three kingdoms of life are able to respond to changing light conditions (Ávila-Pérez et al. 2006; Tschowri et al. 2009; Neiss et al. 2008; Takahashi et al. 2007; Braatsch et al. 2002). The chromophore photochemistry and some of the respective structural pertubations of isolated photosensory domains are well studied. However, the photo-induced processes within the full-length photoreceptor proteins, and hence the molecular mechanism of light-dependent control of gene expression is far from being resolved.
A synthetic biology approach (Khalil and Collins 2010) to establish light-mediated gene expression within a heterologous host requires detailed knowledge of the sensory event and accompanying signal transduction mechanism of a given photoreceptor. In case of light-controlled regulatory cascades, all essential components as well as the respective target deoxyribonucleic acid (DNA) sequences need to be known. In this respect, blue-light-absorbing light–oxygen–voltage (LOV) photoreceptors (Christie et al. 1999; Losi 2004), the sensor of blue light using flavin adenine dinucleotide (FAD) or BLUF domain containing photoreceptors (Losi 2007; Gomelsky and Klug 2002), and the red/far-red-light-sensitive phytochromes (Rockwell and Lagarias 2010) represent the best studied naturally occurring photoreceptor systems in prokaryotes (Losi 2007). In the following, we will provide an overview about different naturally occurring microbial photoreceptor systems, their mechanisms of signal transduction, and the involved regulatory cascades (Table 1) that offer the potential to serve as novel light-controlled expression systems.
Complex regulatory cascades with signal transduction via protein–protein interactions
Photocontrol of a complex downstream signaling cascade can be achieved by changed protein–protein interactions between the photoreceptor protein and various input and output components of the cellular signaling machinery. This eventually results in the photocontrolled expression of several target genes at a time.
Blue-light-sensing systems
In the Gram-positive soil bacterium Bacillus subtilis YtvA acts as a blue-light photoreceptor in the environmental branch of the general stress response pathway (Ávila-Pérez et al. 2006, 2009; Gaidenko et al. 2006). YtvA carries a blue-light-sensitive flavin-binding LOV domain, which is directly coupled to a sulfate-transporter anti-sigma factor antagonist (STAS) domain (Losi et al. 2002). YtvA forms together with a set of homologous STAS-containing but light-insensitive sensor proteins, a macromolecular assembly, the so-called stressosome (Gaidenko et al. 2006). The signals perceived by the stressosome are integrated in a complex phosphorylation/dephosphorylation cascade, activating the general stress response transcription factor σ B which drives the expression of about 150 genes from the σ B-RNA polymerase dependent promoters (Hecker et al. 2007). This complex system allows the bacterium to respond to a diverse array of different environmental conditions including, among others, blue light (Hecker et al. 2007).
The well-characterized circadian transcription factor white collar complex (WCC) plays the dominant role in the photobiology of Neurospora crassa and other fungi. WCC is constituted by two LOV domain containing blue-light-sensitive photoreceptor proteins, namely, VIVID (VVD) and white-collar1 (WC1) protein. Both act together with the homologous (but light-insensitive) white-collar2 (WC2) protein, allowing the organism a tight transcriptional control over 300 genes (Chen et al. 2009) in response to changing conditions during night and day cycles. Light-regulated protein–protein interactions between the three core constituents of the WCC complex enable a fine-tuned transcriptional response (Chen et al. 2010; Hunt et al. 2010; Malzahn et al. 2010). In both of the above cases, the respective target promoters which are activated or repressed in response to blue light are known (Jaubert et al. 2004; Chen et al. 2009).
Blue-light-dependent gene expression in Rhodobacter sphaeroides is mediated by the AppA protein which controls the expression of photosynthesis genes by antagonizing PpsR, the global repressor of all photosynthesis genes of this organism (Gomelsky and Kaplan 1997; Masuda and Bauer 2002). The light signal is perceived here by a so-called BLUF sensor domain (Gomelsky and Klug 2002). The Escherichia coli YcgF protein represents another blue-light-sensitive system also relying on a BLUF sensory domain and a so-called EAL domain as potential effector (Rajagopal et al. 2004). Usually, EAL domains are enzymatically active modules degrading the general secondary messenger molecule bis-(3′-5′)-cyclic dimeric guanosine monophosphate (c-di-GMP; see below; Schirmer and Jenal 2009). However, when studied in vitro, the EAL domain of YcgF was shown to be enzymatically inactive, but instead serves as a light-regulated interaction module for the downstream MerR-like helix-turn-helix (HTH) DNA-binding transcriptional regulator YcgE (Tschowri et al. 2009). This transcriptional regulator, when bound to YcgF, is released from its operator DNA in response to blue light, thereby affecting the expression of eight genes, among them ymgA and ymgB (Tschowri et al. 2009). The respective YmgA and YmgB proteins activate the production of the biofilm matrix substance colanic acid as well as other processes related to biofilm formation via the RcsC/RcsD/RcsB phosphorelay system (Hagiwara et al. 2003; Tschowri et al. 2009). For all of the above mentioned cases, details of regulatory mechanisms as well as the respective target promoter structures are known (Petersohn et al. 1999; Gomelsky and Kaplan 1995; Tschowri et al. 2009; Elsen et al. 2005).
Red-light sensing systems
A red/far-red-light-sensitive gene expression system is constituted by the bacteriophytochromes of Bradhyrhizobium (BrBphP) and of the closely related phototrophic bacterium Rhodopseudomonas palustris (Giraud et al. 2002). Both phytochromes stringently control the synthesis of the complete photosynthetic apparatus (Giraud et al. 2002). As the carboxy-terminal domain of both phytochromes lack histidine kinase-like features, their molecular mode of action is apparently not based on a phosphorylation/dephosphorylation cascade. However, direct antagonistic protein–protein interactions have been suggested with a homologue of PpsR, the global repressor of photosynthesis genes (Giraud et al. 2002; Jaubert et al. 2004).
In conclusion, photosensory systems that base on protein–protein interactions represent an efficient means to control cellular processes in a variety of species. The complexity of these regulatory cascades, for example, the B. subtilis stressosome pathway, apparently hampers a functional transfer from one host organism to another thus preventing their immediate application for synthetic biology.
Light-sensitive two-component systems
Blue-light-sensing two-component systems
Sensory two-component systems are utilized to achieve light-mediated control of gene expression in various microbes (Mitrophanov and Groisman 2008). Those systems consist in their minimal configuration of one light-responsive sensory histidine kinase and a corresponding response regulator (Mitrophanov and Groisman 2008) which can control the transcription of a set of genes in response to illumination resulting in changed physiological behavior (Purcell et al. 2007; Swartz et al. 2007). Examples with known biological function include the LOV blue-light photoreceptor of the notorious mammalian pathogen Brucella abortus (Swartz et al. 2007). Moreover, the LOV histidine kinase proteins of several Brucella strains undergo a light-mediated autophosphorylation in vitro (Swartz et al. 2007). Probably, the phosphate is subsequently transferred to an as yet unidentified response regulator which enables the organism to respond to light stimuli with an increase in virulence (Swartz et al. 2007). In Caulobacter crescentus, an analogous system, the light-dependent histidine kinase LovK, is used to control cell-to-cell attachment (Purcell et al. 2007, 2010). Also in this case, both the target response regulator and the respective promoter structures are unknown. A related protein which in contrast to the above systems contains a fused response regulator C-terminally linked to the kinase sub-domain has been described in the proteobacterial plant pathogen Pseudomonas syringae pv. tomato (Cao et al. 2008). Here, neither its physiological role nor the DNA target sequences are known.
Red-light-sensing two-component systems
The role of red/far-red-light-sensitive phytochromes is well studied in plants where they were initially identified. However, their role in the control of gene expression in bacteria is less clear. Currently, the physiological role of bacterial phytochrome-based two-component systems was studied only in photosynthetic prokaryotes. The structurally and functionally best characterized two-component phytochrome systems are represented by the cyanobacterial phytochromes Cph1 of Synechocystis sp. PCC6803 (Yeh et al. 1997), RcaE of Fremyella diplosiphon (Kehoe and Grossman 1996; Terauchi et al. 2004) and CcaS of Synechocystis sp. PCC 6803 (Kehoe and Grossman 1997; Hirose et al. 2008). All three proteins probably act as light-dependent sensor histidine kinases in a light-dependent two-component system. For cph1, rcaE, and ccaS, genes encoding corresponding response regulators are located immediately downstream of the respective phytochrome genes in the genome (Yeh et al. 1997; Kehoe and Grossman 1997; Hirose et al. 2008). Light-controlled target genes including promoters are known or have been suggested only for RcaE and CcaS, both controlling the green-red photoreversible process of complementary chromatic adaptation (CCA; Kehoe and Gutu 2006; Hirose et al. 2008). However, the molecular basis of the involved regulatory cascade is not established in much detail. Moreover, for RcaE biochemical evidence for the bound chromophore species and an in vitro test of its green/red light photochromicity is missing.
Two-component systems are usually much less complex with respect to the number of regulatory proteins involved in a specific response. With respect to promoter recognition, a single response regulator targeted by a specific histidine kinase can regulate the expression of a number of target genes. Nevertheless, cross-talk between sensory histidine kinases and non-cognate or orphan response regulators has been reported (Wang et al. 2009) which might reduce the in vivo target specificity of these systems. Unfortunately, for most of the light-mediated control processes outlined above either the target response regulator and/or the controlled promoter sequence is yet unknown or has not been experimentally validated.
Light-dependent control over gene expression via secondary messengers
Besides the direct control of transcription factor activities, light absorption in a photosensory domain can also trigger a change in an enzymatically active effector module which may catalyze the synthesis or degradation of secondary messengers involved in the control of gene expression.
In bacteria, 3′,5′-cyclic adenosine monophosphate (cAMP) and cyclic-di-guanosine monophosphate (c-di-GMP) act as secondary messenger molecules which can induce or repress transcription at several target promoters through binding to their respective receptors (Schirmer and Jenal 2009; Won et al. 2009). In the following, we briefly describe how light as an environmental signal can affect gene expression via controlling intracellular levels of such secondary messengers.
c-di-GMP and light
The c-di-GMP molecule is an important bacterial secondary messenger involved in complex behavioral responses such as biofilm formation, virulence and many other surface-associated traits in bacteria (Hengge 2009). In a number of sensor systems, the level of c-di-GMP in the cell is controlled by GGDEF (diguanylate cyclases) and EAL (phosphodiesterases) domains (D’Argenio and Miller 2004) with GGDEF domains catalyzing the synthesis and EAL domains the hydrolysis of these molecules (Hengge 2009; Schirmer and Jenal 2009). The wide range of cellular processes which are controlled by c-di-GMP implies the presence of multiple signal transduction mechanisms for this secondary messenger molecule (Hengge 2009). In that context, proteins carrying a PilZ domain are the best studied c-di-GMP receptors identified so far (Schirmer and Jenal 2009). Recently, degenerated GGDEF- and EAL containing proteins have been identified that have lost their enzymatic activity and instead have adopted new roles as allosteric modules comparable to the YcgF protein of E. coli (Tschowri et al. 2009; Christen et al. 2005). Other degenerated GGDEF or EAL modules act as c-di-GMP receptor proteins (Newell et al. 2009).
Recently, sequences of various GGDEF and EAL domains have been identified, that are naturally fused to light-sensitive receptor domains (Losi and Gärtner 2008). For example, one photoreceptor protein from Rhodobacter sphaeroides bearing both of the respective enzymatic effector domains has been characterized. The protein BphG1 consists of a GGDEF and an EAL domain that are attached to a red/far-red-light-controllable phytochrome sensory module as well as to other auxiliary domains (Tarutina et al. 2006). However, when studied in vitro, full-length constructs only showed light-independent phosphodiesterase activity but no cyclase activity (Tarutina et al. 2006). Remarkably, deletion of the EAL domain led to a light-mediated control of c-di-GMP synthesis (Tarutina et al. 2006). Similar photoreceptor architectures exhibiting either a GGDEF, an EAL, or both of those enzymatic effector modules coupled to a blue-light-sensitive BLUF domain instead of the phytochrome sensory module have been described recently. However, for both the BLUF-photoreceptor BlrP1 of Klebsiella pneumonia (Barends et al. 2009) and the LOV-GGDEF-EAL protein SL2 of Synechococcus elongatus (Cao et al. 2010), no functional roles or downstream targets have been identified.
cAMP and light
The ubiquitous cAMP secondary messenger molecule not only can act as a small regulatory molecule in bacteria (Schünke et al. 2009) but is also involved in complex processes in higher eukaryotes including mammals and humans (Kaupp and Seifert 2002). A photoreceptor bearing an adenylyl cyclase effector domain which catalyzes the formation of cAMP from adenosine triphosphate has been described in the unicellular euglenoid Euglena gracilis (Iseki et al. 2002). Here, the heterotetrameric photoactivated adenylyl cyclase (PAC) is probably involved in controlling the phototactic behavior of the organism (Iseki et al. 2002; Ntefidou et al. 2003). Very recently, a bacterial BLUF domain containing bacterial PAC (bPAC) from the marine filamentous proteobacterium Beggiatoa sp. was characterized in vitro and used in E. coli as heterologous host to light-dependently control intracellular cAMP (Ryu et al. 2010; Stierl et al. 2011) and, after site-directed mutagenesis of the cyclase domain, cGMP levels (Ryu et al. 2010). Therefore, using a standard E. coli lac promoter-based expression system, light-dependent bPAC-mediated control of cAMP synthesis basically allows controlling the expression level of target genes by modulating the activity of the cAMP-binding catabolite activator protein (CAP; Wilson et al. 2007; Kolb et al. 1993). However, the response of an organism to the intracellular concentration of the individual secondary messenger takes place on a global regulatory level and is thus rather complex, possibly introducing unwanted cross-talk and off-target effects into the rewired circuitry.
Light-sensitive transcription factors
The most elegant way to control gene expression by light comprises a direct transcriptional control exerted by a DNA-binding protein fused to a photosensory domain. Light absorption in the sensory module would thus trigger a conformational change in the DNA-binding domain which results in light-mediated change of its DNA-binding properties. However, although putative light-sensitive transcription factors harboring a LOV domain can readily be identified in sequence databases (Krauss et al. 2009; Briggs 2007; Losi and Gärtner 2008), almost no functional information is available for this class of photoreceptors. The Aureochrome protein found in a number of stramenopile algae, such as Vaucheria frigida, represents the only example of a naturally occurring light-dependent DNA-binding photoreceptor that has been functionally characterized (Takahashi et al. 2007). In general, Aureochromes contain a LOV domain as the photosensory module fused with a DNA-binding basic region leucine zipper (bZIP) motif at the carboxy-terminus of the photoreceptor. Photoexcitation results in a change in DNA-binding affinity of the protein and thus controls photomorphogenesis of the organism in response to blue light. Target consensus sequences to which the Aureochrome bZIP domain binds are known (Takahashi et al. 2007). Besides stramenopile algae, Aureochrome-like sequences have also been identified in different microbes as the unicellular marine diatoms Phaeodactylum tricornutum and Thalassiosira pseudonana (Ishikawa et al. 2009; Takahashi et al. 2007).
Recombinant photoreceptors for light-mediated control of gene expression
Naturally occurring photoreceptor proteins exhibit an intrinsic sensitivity to light-signals which can be used as genetically encoded photoswitches especially for in vivo applications. However, the complexity of these signal transduction cascades generally limits their use since most of them are not completely characterized or the respective target genes are unknown.
Alternatively, artificial light-sensitive regulatory systems comprising a well-characterized light perception domain and a regulatory output domain can be used to build up novel light-inducible expression systems. This strategy makes use of the observation that, in nature, those regulatory proteins are modularly composed of sensory modules (input domains) and signal transduction modules (output domains; see, e.g., Grünberg and Serrano 2010; Pawson and Nash 2003 for comprehensive reviews). Natural photoreceptors in all kingdoms of life only utilize a limited number of conserved input and output domain families in various combinations rather than a diverse array of unique proteins to convert the environmental light stimulus into a specific intracellular response (Losi 2004; Losi and Gärtner 2008; Purcell and Crosson 2008). In the following, we summarize how natural light-sensing domains have been exploited as input modules for constructing novel light-triggered gene expression systems in bacteria and yeast.
Photocontrolled bacterial two-component systems
Phytochrome-based systems
The synthetic phytochrome sensor kinase Cph8 represents the first recombinant bacterial light regulator used for gene expression (Levskaya et al. 2005). This chimeric red-light receptor consists of the photoreceptor domain from cyanobacterial phytochrome Cph1 and the histidine kinase domain from E. coli EnvZ.
The phytochrome Cph1 from the cyanobacterium Synechocystis sp. PCC6803 is the first member of the plant photoreceptor family that has been identified in bacteria (Kehoe and Grossman 1996; Yeh et al. 1997). Together with the response regulator Rcp1, the light-responsive histidine kinase forms a phytochrome two-component system (Yeh et al. 1997). Mutational analysis and genomewide expression profiling in Synechocystis revealed that, besides other photoreceptors, Cph1 is involved in red and far-red-light-dependent regulation of up to 20 genes (Hübschmann et al. 2005). Cph1 is a dimeric receptor protein that binds phycocyanobilin (PCB) as a red-light-absorbing chromophore. Similar to plant phytochromes Cph1 is able to reversibly switch between a red-light-absorbing ground state (Pr) and a far-red-light-absorbing photoactivated state (Pfr). As a light-sensitive, two-component histidine kinase, it basically consists of a sensor module [including the chromophore-binding GAF domain as well as the phytochrome (PHY)-specific domain] and a transmitter module that exhibits the mentioned kinase activity (Hughes 2010).
EnvZ, a dimeric osmosensor, is a multidomain transmembrane protein and one of the best characterized two-component histidine kinases from E. coli. Besides the periplasmic receptor domain that is flanked by two transmembrane helices, it possesses a C-terminal 228-residue histidine kinase domain that is located in the cytoplasm (Dutta et al. 1999). Upon changes of extracellular osmolarity, EnvZ specifically phosphorylates its cognate response regulator OmpR, which, in turn, regulates the expression of ompF and ompC genes encoding two outer membrane porines (Forst and Roberts 1994).
The fusion protein Cph8 was generated to combine the light-sensing function of Cph1 with the output function of EnvZ, which is the phosphorylation of the response regulator OmpR, followed by the activation of the ompC promoter (Fig. 2a). The chimera Cph8 consisted of the C-terminal cytoplasmic histidine kinase domain (229 amino acids of EnvZ), whereas the natural EnvZ sensor domain has been completely removed as described previously (Utsumi et al. 1989). Instead, the 517 N-terminal residues of Cph1 encompassing the Per-ARNT-Sim (PAS), GAF, and PHY domains without the transmitter module were fused to the EnvZ histidine kinase domain as light-sensitive input domain. In vivo expression experiments in E. coli could demonstrate that Cph8 specifically activated the response regulator OmpR in the dark, whereas illumination inhibited the phosphotransfer activity of the recombinant sensor kinase and thus repressed transcription from the P ompC promoter. Expression of the β-galactosidase-encoding reporter gene lacZ, which was under control of the ompC promoter, further showed that the phytochrome-based light-controlled expression system gradually responded to decreasing light intensities with a moderate induction factor of 2.4 (Tabor et al. 2009).
Chimeric photoswitches for light-mediated control of gene expression in bacteria. a Phytochrome-based two-component system with the chimeric photoreceptor Cph8. b LOV-based two-component system with recombinant histidine kinase YF1 (YtvA LOV−FixL HK). c LOV-based repressor LovTAP. d Recombinant transcriptional activator GCN∆25-PYP-v2 with light-sensitive PYP domain. The light-activated chromophores and photoreceptor domains are shown in red (Phy) and blue (LOV and PYP). The active regulatory domains are highlighted in green. The respective target promoters are shown as green arrows. Phosphorylated domains are labeled with a P in a red circle. HK Histidine kinase, LOV light–oxygen–voltage domain, P promoter, Phy phytochrome domain, PYP photoactive yellow protein, (+) activation of transcription, (-) repression of transcription
Light-control of lacZ expression was also used to generate bacterial populations exhibiting photographic properties (Levskaya et al. 2005). Here, a confluent lawn of E. coli cells was able to convert a light-projected image into a high-definition chemical image via β-galactosidase-catalyzed synthesis of a black pigment. Furthermore, based on predictive mathematic models, the artificial light sensor and output modules Cph8/OmpR could be genetically connected to a second regulatory cascade to design a higher-order genetic circuit. The novel genetic logic gate enabled E. coli communities of genetically identical clones to detect and to process edges of a light pattern (Tabor et al. 2009). In detail, the Cph8-mediated light response triggers the expression of the quorum sensing gene luxI from Vibrio fischeri. The acyl-homoserine lactone synthase LuxI then catalyzes the synthesis of the autoinducer-1 (3-oxohexanoyl-homoserine lactone, AHL), which finally activates the constitutively expressed regulator LuxR to enable lacZ expression. As a result, neighboring E. coli cells are able to communicate to each other to accurately identify the light–dark edges and consequently “print” the contour of different light-projected images.
More recently, Tabor et al. (2011) generated a bichromatically controlled transcription system. Here, the red-light-sensing Cph8 was combined with the cyanobacterial phytochrome two-component system CcaS/CcaR (Hirose et al. 2008). Although both phytochrome domains share the same chromophore, sensor kinase CcaS needs green light (535 nm) for its photoactivation, whereas red light (672 nm) inactivates the kinase activity. For green light induction of target gene expression, the photoactivated CcaS kinase phosphorylates its cognate response regulator CcaR. Coexpression of both photoswitch systems (Cph8/EnvZ and CcaS/CcaR) allowed to create a higher-order regulatory circuit exhibiting sensitivity to two-dimensional light gradients. The red/green-light sensor system could be successfully used to gain two-colour control over gene expression in a single cell.
The synthetic light regulator Cph8 as well as the cyanobacteriochrome CcaS offer a tremendous potential for various applications; however, they also share some system-inherent disadvantages. First of all, the functional expression of a phytochrome domain in E. coli requires the biosynthesis of the respective bilin chromophore PCB. It has been shown that expression of ho1 and pcyA from Synechocystis encoding a heme oxygenase and bilin reductase, respectively, is essential to convert heme into PCB (Gambetta and Lagarias 2001). Thus, in order to efficiently utilize Cph8- and CcaS-dependent light-mediated gene expression control in E. coli, the respective PCB biosynthesis genes need to be co-expressed along with the regulatory genes.
Furthermore, in case of Cph8, the use of a recombinant light-sensitive two-component system that predominantly consists of homologous components might lead to unwanted cellular responses since two-component systems generally form regulatory networks via cross-regulation. In the case of EnvZ/OmpR, for example, the two-component system was shown to be involved directly and indirectly in the expression of 125 genes (Oshima et al. 2002). Thus, functional interaction of synthetic light regulators as Cph8 with other regulators might affect the light-controlled expression system as well as the expression of other genes, and hence, the physiology of E. coli or other host organisms would prevent a strict and controllable gene regulation and also aggravate data interpretation in systems biology approaches.
LOV-based systems
Recently, an alternative design concept has been proposed to construct an artificial light-sensing two-component system suitable for gene expression control in E. coli. Based on the natural diversity of sensor proteins that contain PAS domains (Möglich et al. 2009b) as the central input module, Möglich et al. (2009a) designed the light-sensitive sensor kinase YF1 (Fig. 2b) . The heme-binding PAS domain from the Bradyrhizobium japonicum oxygen sensor kinase FixL (Gilles-Gonzalez et al. 1991; Gong et al. 1998) was replaced by the blue-light sensor module derived from the B. subtilis photoreceptor YtvA (Losi et al. 2002; Losi 2004). YtvA consists of a blue-light receptor domain called LOV and a STAS output domain that are linked by a short flexible α-helix named Jα. Unlike phytochromes, YtvA as a member of the LOV family non-covalently binds flavin mononucleotide (FMN) as chromophore and undergoes a reversible photocycle upon blue-light excitation (Losi 2004; Losi et al. 2002). Similar to EnvZ, the sensor kinase FixL basically comprises two modules. The N-terminal input module consist of two different PAS domains (PAS A and B), of which one, PAS B, binds heme as oxygen-sensitive cofactor, whereas the C-terminal output module harbors the histidine kinase domain. As described for YtvA, both FixL modules are interconnected through a helical linker. The response regulator FixJ forms a two-component system together with FixL that activates the expression of nitrogen fixation genes under anaerobic conditions (Fischer 1994).
The chimeric light sensor YF1 was created by fusing the light perception domain of YtvA to the FixL kinase domain using the α-helix as a natural linker. In vivo studies in E. coli expressing the redesigned YF1 as well as the response regulator FixJ revealed that transcription from the target promoter P fixK2 is induced approximately 70-fold by maximal induction (dark) as compared to repressing conditions under saturating photoexcitation (Möglich et al. 2009a).
Remarkably, analysis of variants of both recombinant light-sensitive sensor kinases, Cph8 and YF1, revealed that the length and composition of the linker, located between the input and output module, tremendously influenced the intra-molecular signal transduction affecting the overall activity of the kinase as well as the mode of light regulation.
The LOV photoswitch YF1 could further be used successfully to expand the regulatory functionality of the artificial gene circuit for the perception of an additional signal. The insertion of the O2-detecting heme-binding domain PAS B from FixL into YF1 resulted in YHF, a sensor kinase which is sensitive toward two environmental stimuli (Möglich et al. 2010). The authors could demonstrate that light and oxygen acted cooperatively on the activity of the output module: upon blue-light irradiation and in the presence of oxygen, YHF kinase activity was downregulated by a factor of 280, whereas the presence of one of both signals just led to a twofold to threefold reduction of activity.
Photocontrolled DNA-binding proteins
Besides two-component systems, in vivo gene expression can principally also be directly controlled using DNA-binding proteins. In contrast to two-component systems, DNA-binding proteins sense a signal by an input domain and directly control gene transcription by binding to their target promoters.
LOV-based systems
The first light-sensitive repressor was engineered by fusing the blue-light-sensitive LOV2 domain derived from Arvena sativa phototropin 1 (AsLOV2) with the E. coli l-tryptophan-responsive TrpR repressor (Strickland et al. 2008). As already described for the YtvA LOV domain, AsLOV2 non-covalently binds FMN and exhibits a characteristic photocycle including the reversible formation of a covalent bond between the chromophore and a conserved photoactive cysteine residue upon excitation with blue light (Crosson and Moffat 2002; Swartz et al. 2002). The dimeric E. coli Trp repressor is one of the smallest proteins that shows sequence-specific DNA binding and is also allosterically controlled by l-tryptophan (Gunsalus and Yanofsky 1980; Marmorstein and Sigler 1989). The TrpR aporepressor is naturally inactive; it is activated in the presence of the effector molecule tryptophan and subsequently binds to at least five different promoters in the E. coli genome including the P trp of the corresponding trp operon essential for tryptophan synthesis (Jeeves et al. 1999).
The TrpR-based LOV photoswitch was constructed by joining the two regulator modules through the LOV Jα-helix as a shared helical linker. The chimeric repressor designated as LovTAP (Fig. 2c) selectively binds to its cognate operator in saturating light (Strickland et al. 2008). Comparable to YHF, the regulatory modules of the LOV and Trp proteins worked cooperatively since light-induced DNA binding of LovTAP requires free tryptophan as an effector. Initial light-mediated increase of LovTAP DNA-binding capacity was comparatively low, but could be optimized by rational improvement of the Jα-linker characteristics. Upon photoexcitation, the best variant exhibited a DNA-binding affinity enhanced by a factor of 70 in comparison to the original LovTAP, where the binding affinity increased only sixfold upon blue-light illumination (Strickland et al. 2008, 2010).
PYP-based systems
The bZIP DNA-binding domain of the activator protein GCN4 that is required for the general amino acid control de-repression response in yeast has also been used to create a photo-switchable transcription factor. The well-characterized bZIP domain consists of a leucine zipper peptide and a basic region that preferentially binds to the pseudo-palindromic DNA site AP1 (Arndt and Fink 1986; Koldin et al. 1995). In the presence of its cognate DNA, two GCN4-bZIP monomers form a stable dimer via the interaction between the α-helical zipper domains, whereas the basic regions are responsible for the interaction with the target DNA (Ellenberger et al. 1992).
In the past, different strategies have been pursued to utilize bZIP-type DNA-binding proteins for light-dependent regulation of DNA binding. For example, the incorporation of a photoisomerizable azobenzene cross-linking reagent introduced into the GCN4 bZIP domain led to a reversible photomodulation of its DNA-binding characteristics (Caamano et al. 2000; Woolley et al. 2006). Here, illumination results in conformational changes within the zipper domain of the regulator which affects its DNA-binding affinity. Recently, the photoactive yellow protein (PYP) from Halorhodospira halophila was applied to create an alternative light-sensitive bZIP activator (Morgan et al. 2010). PYP is a small soluble blue-light photoreceptor that belongs to the family of xanthopsins (Kort et al. 1996; van der Horst and Hellingwerf 2004). The α/β-fold of PYP is regarded as structural prototype of PAS domains (Borgstahl et al. 1995; Pellequer et al. 1998) and thus shares overall structural properties with LOV domains. PYP is proposed to be involved in blue-light-mediated negative phototaxis (Sprenger et al. 1993; Hoff et al. 2009), although the signal transduction pathway of the photoreceptor is still not unravelled. PYP covalently binds p-coumaric acid (also designated in literature as p-hydroxycinnamic acid) as chromophore that undergoes a light-induced photocycle triggered by its reversible photoisomerization. The formation of the PYP signaling state includes a partial unfolding of its N-terminal region (van der Horst and Hellingwerf 2004; Imamoto and Kataoka 2007).
For designing a GCN4-PYP chimera capable of controlling its DNA-binding capacity in response to blue-light irradiation, the N-terminally truncated PYP lacking 25 amino acid residues was fused to the 58-residue GCN4 fragment encompassing the zipper domain as well as the basic DNA-binding region (Morgan et al. 2010). A structure-based strategy allowed generating a light-dependent regulator simultaneously harboring structural characteristics of GCN4 in the light and of PYP in the dark. The resulting chimera GCN∆25PYP-v2 (Fig. 2d) specifically binds to its AP1 recognition site exhibiting a twofold higher binding affinity of the irradiated light regulator in comparison to its dark-adapted state as shown by electrophoretic mobility shift assays. Mechanistically, the GCN4 effector domain is shielded by the PYP input domain in the absence of light, thereby preventing the dimerization of the leucine zipper motif. Under illumination, PYP reversibly unfolds which, in turn, allows dimerization of bZIP on the target DNA and thereby increases its binding affinity. Remarkably, deletion of the eight C-terminal amino acid residues of the leucine zipper region within the GCN4-PYP fusion protein significantly affected the photo-controlled DNA-binding properties. In detail, the resulting derivative GCN4(S)∆25PYP exhibited an inverted photoresponse, leading to a threefold increased DNA-binding capacity in the dark in comparison to its light-adapted state (Morgan and Woolley 2010).
PYP derivatives used as photoswitches in heterologous organisms also require the supply of a unique chromophore in sufficient amounts as do light-sensing phytochrome domains. To this end, heterologous production of holo-PYP for in vivo applications can be achieved either by the supplementation of the activated chromophore to the medium and its subsequent uptake into the cell or by co-expression of two p-coumaric acid biosynthesis genes from Rhodobacter capsulatus or H. halophila encoding the tyrosine ammonia lyase and p-hydroxycinnamic acid ligase (Kyndt et al. 2003; Hori et al. 2009; Morgan et al. 2010).
Y2H systems
Allosteric coupling of sensor and effector domains via light-mediated conformational changes is regarded as the basic principle underlying the construction of photoswitches, where signal transduction is usually mediated by an α-helical linker region connecting these two modules. Engineered light-modulated protein–protein interactions represent an alternative approach to generate light-sensitive expression systems. This method was pioneered in yeast (Shimizu-Sato et al. 2002), and it is based on the yeast two-hybrid (Y2H) system (Fields and Song 1989). Here, the light-triggered interaction of two proteins called bait and prey fused to different domains of a yeast-derived transcription factor allows for a rapid and reversible induction of a target promoter by light. Plant photoreceptors seem to be particularly suited for designing Y2H-based photoswitches.
Phytochrome-based systems
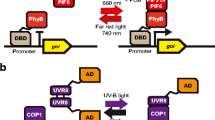
The first artificial photoswitch developed for light-mediated gene expression in yeast combines the reversible binding of A. thaliana phytochrome PhyB to its signaling partner phytochrome interacting factor 3 (PIF3) employing the classical Y2H system (Shimizu-Sato et al. 2002). The transcription factor GAL4 separated into its DNA-binding domain [GAL-binding domain (GBD)], and the GAL4 promoter activation domain (GAD) is activated when the chimeric proteins PhyB–GBD and PIF3–GAD form a stable complex. Plant phytochromes contain an N-terminal chromophore-binding domain covalently linked to the chromophore phytochromobilin and a C-terminal kinase-related output domain responsible for dimerization (see, e.g., Sharrock 2008). The photoreceptor reversibly changes its conformation upon absorption of red (λ max = 660 nm) or far-red light (λ max = 730 nm), in which only the activated, far-red-light-absorbing conformer Pfr conveys numerous transcriptional responses by direct interaction with some nuclear, basic helix-loop-helix (bHLH) transcription factors, the so-called PIFs (see Leivar and Quail 2010 for a recent review). In A. thaliana, all members of the PIF family comprise the C-terminal bHLH domain responsible for dimerization and DNA binding and a conserved N-terminal PAS-related domain for PhyB(Pfr) interaction.
The Y2H-based expression system was established with the N-terminal chromophore containing domain derived from PhyB (PhyB(NT)) and the full-length PIF3 protein (Fig. 3a; Shimizu-Sato et al. 2002). Red-light-dependent interaction of PhyB(NT)–GBD and PIF3–GAD was verified in vivo by GAL1 promoter elements mediated lacZ gene expression under various light conditions. The active photosensing domain was reconstituted in yeast by exogenous supply of the heterologous chromophore PCB to the growth medium. Since the Pr-to-Pfr photo-interconversion process only needs milliseconds to complete, this system reacts very fast, dose-dependently and simultaneously in all illuminated cells of an isogenic cell population. Under standard expression conditions, a 50-fold induction of LacZ activity was reached within 30 min after a red light pulse. However, the need for exogenously supplied or biosynthetically produced chromophore constitutes the same drawback as for the bacterial Cph8 regulator.
Eukaryotic photoswitches based on the system. Two fusion proteins interact in response to light and bring the fused GBD and the activator domain (AD) in close proximity, thus forming an active transcription factor for the expression of a target gene. a Phytochrome-based red-light regulation. b Cryptochrome-based blue-light regulation. c ZTL family-based blue-light regulation. Photoreceptor moieties are depicted by squares. Colors other than grey represent the active status of photoreceptors (red or blue) and interacting partner proteins (green). P Gal Promoter with UAS of GAL1 or GAL4 promoter, GBD GAL-binding domain, GAD GAL activator domain, VP16 AD viral protein activator domain, Phy phytochrome domain, (+) activation of transcription
Another Phy-based light-regulated yeast expression system employed phytochrome PhyA and its interaction partners FHY1 (far-red elongated hypocotyl 1) and FHL (FHY1 like protein) analogously to the PhyB(NT)–GBD/PIF3–GAD system (Sorokina et al. 2009). These small (20 and 23 kDa) plant-specific proteins contain functional nuclear localization and nuclear export signals and have been shown to control the nuclear import of PhyA after Pfr-specific interaction within the N-terminus of PhyA (Hiltbrunner et al. 2006). The three artificial yeast Phy-based photoswitches were tested in vivo using the firefly luciferase gene (luc) as a reporter fused to the GAL1 promoter. Comparative expression studies revealed that all Phy–Y2H systems show a fast in vivo response after irradiation with red light. In contrast to the PhyB(NT)+PIF3 systems, where LUC expression was only 40-fold increased, the novel PhyA+FHY1 and PhyA+FHL system led to a significantly increased induction rate of 300 and 150, respectively, upon irradiation (Sorokina et al. 2009). Surprisingly, the PhyA–GBD/FHY1–GAD system was able to act as a photoswitch responsive to red light even in the absence of the supplemented chromophore PCB. Since the reduced induction capacity (110-fold) was still higher than with PhyB(NT)+PIF3, the supplementation of PCB to functionally synthesize a Phy–Y2H system can, in principle, be omitted. Possibly, an as yet unidentified compound naturally present in yeast can serve as a substitute for the natural chromophore.
In principle, Phy-based photoswitches can be fine-tuned by using different phytochrome-interacting protein combinations as the “light-sensing” unit for expression of any gene of interest fused to GAL4-responsive promoter elements. Likewise, cytoplasmic phytochrome-interacting ROP guanine-nucleotide exchange factor (PIRF1), a small GTPase activator protein involved in light-regulated root development of A. thaliana, might be also employed since Pr conformer-dependent interaction with the N-terminal PhyA or PhyB domain in Y2H screens has already been demonstrated (Shin et al. 2010).
Cryptochrome-based systems
In contrast to the red-light-responsive phytochromes, Arabidopsis cryptochromes (CRY) mediate blue-light regulation of cell growth, entrainment of circadian expression, and photoperiodic flowering (Cashmore et al. 1999). Since yeasts harbor cryptochrome-related DNA photolyases that contain identical chromophores — methyltetrahydrofolate (MTHF) and FAD (Jorns 1990) — the formation of active plant cryptochrome and thus its light-dependent protein–protein interactions in yeast should function independent of chromophore supply (Liu et al. 2008). CIB1 was identified via Y2H screens as a nuclear cryptochrome-interacting bHLH protein, regulating gene expression in plant cells (Lin et al. 2002). In yeast, interaction of CRY2–GBD and CIB1–GAD (Fig. 3b) was not detectable in darkness or red-light-irradiated cells and required relatively high intensities of blue light as well as long irradiation times.
LOV-based systems
Apart from the phototropin genes PHOT1 and PHOT2, A. thaliana contains a set of additional LOV domain containing blue-light receptor genes such as the Zeitlupe (ZTL) gene family (Somers 2001). ZTL, LOV Kelch Protein 2 (LKP2), and Flavin-binding Kelch F-Box 1 (FKF1) possess an N-terminal LOV domain binding the FMN chromophore and a C-terminal output domain with an F-box motif and six Kelch repeats (Somers 2001). The three proteins are either involved in the regulation of the circadian clock (ZTL, LKP2) or the induction of flowering (FKF1) by controlling proteasome-dependent protein degradation (Somers 2001; Yasuhara et al. 2004). In addition, a fourth LOV protein, PAS/LOV protein (PLP), exhibits a different structural organization with an N-terminal PAS domain and a C-terminal LOV domain (Ogura et al. 2008). Three splicing variants of the PLP gene lead to PLPA, PLPB, and PLPC protein of which PLPC contains a truncated LOV domain.
As already described for cryptochrome, Y2H screens enabled the isolation of PLP-interacting proteins that might be employed in a Y2H-based photoswitch. The interaction partners Vitamin C defective 2 (VTC2), VTC2-like (VTC2L) and BEL1-like homeodomain 10 proteins (BEL10A and BEL10B) were fused to the GAL4 activator domain GAD (prey) to interact with PLPA–GDB or PLPB–GDB (bait; Ogura et al. 2008). Protein–protein interaction was tested under different light conditions (dark, blue, red, far-red, and green lights). Only blue-light irradiation led to a specific photoresponse of some interaction partners. However, in contrast to light-induced interaction of the other described systems, increasing blue-light intensity led to a significant reduction of GBD–GAD interaction, in which PLPA+BLH10A showed highest sensitivity followed by PLPB+VTC2L, PLPA+BLH10B, and PLPA+VTC2L. Although a detailed characterization of the PLP-based Y2H systems is still missing, the identified combinations of interaction partners could be useful for the design of new blue-light photoswitches.
The members of the ZTL photoreceptor family, FKF1, LKP2, and ZTL, also showed interesting blue-light responsive protein–protein interactions, especially with the flowering time regulator Gigantea (GI). The interaction of the LOV domain of FKF1, LKP2, and ZTL with the GI N-terminus, as suggested from preliminary Y2H studies, was shown to be enhanced by blue light in planta (Kim et al. 2007; Sawa et al. 2007).
The combined results of both expression systems were used to construct a Y2H-like light-activated dimerization (LAD) system with GI−GAL4-binding domain (GI−GBD) and FKF1-VP16 viral-derived transactivation domain (FKF1-VP16 AD) forming a potent transcription factor in mammalian HEK cells (Fig. 3c; Yazawa et al. 2009). Here, the artificial light-responsive transcription factor acts on a 5 × GAL4−upstream activation sequence (UAS) promoter fused to a luciferase reporter gene. Likewise, employment of ZTL for blue light Y2H-like photoswitches is conceivable, either based on ZTL+GI interaction analyzed in Y2H and in planta (Kim et al. 2007) or by using ZTL interactions found with the C-termini of PhyB and CRY1 in Y2H screens (Jarillo et al. 2001).
In summary, different Arabidopsis photosensors and their respective interaction partners have already been used to regulate GAL-derived promoters via protein–protein complexes responsive to red-/far-red or blue light. One benefit of the classical Y2H system is its almost unlimited modularity allowing to optimize each photoswitch for any application by just modifying it via the integration of other interaction partners with various strength and light reactivity. Furthermore, the yeast systems can be easily transferred to other eukaryotic expression systems as shown for the FKF1+GI system that was successfully used also in mammalian cells (Yazawa et al. 2009).
Conclusions
An experimental strategy allowing for the regulation and dynamic modulation of gene expression in living cells is of utmost importance for systems biology as well as for many bioprocesses and their industrial applications. Regulatory networks can be designed and used to simultaneously achieve control over expression of multiple genes in vivo. We have described here novel light-sensing regulatory modules with defined photochemical properties that allow light-modulated control of a defined target promoter. However, to set up complex interoperable genetic circuits with broad functionality, such regulatory cascades have to be further developed to respond simultaneously and independently to more than one environmental stimulus. Basically, two different approaches can be used to set up genetically encoded higher-order regulatory circuits. First, the spectrum of sensed signals can be expanded by adding additional sensor domains. The modular combination of a LOV light input domain with a second sensor domain responding to a redox signal (PAS) and cellular metabolites (TrpR), respectively, resulted in recombinant transcription factors that were able to simultaneously integrate two environmental signals (Möglich et al. 2010; Strickland et al. 2008). Second, light-sensitive regulators can be further used to add a light switch to a user-defined regulatory circuit that responds to multiple signals. Hence, sensor properties of two or more differently triggered expression systems can be combined within one joint higher-order regulatory cascade. Here, the expression of one of the regulator-encoding genes is controlled by the activity of another transcription factor as realized, for instance, for the synthetic bacterial edge detection system (Tabor et al. 2009). Such a genetic circuit basically consists of a two-level regulatory cascade: (1) the light–dark sensor Cph8 exerts control over the expression of genes; (2) their products trigger the autoinducer-dependent expression of the target gene.
In nature, several distinct photosensory systems can reside within the same organism and respond to identical or different wavelengths of light (Losi and Gärtner 2008; van der Horst et al. 2007). For synthetic biology approaches, it would be highly desirable to employ distinct light qualities, such as red, green, and blue light, for the differential control of various target genes within the same host with spatiotemporal resolution. Nature uses this strategy to fine-tune the response of organism to changing light conditions. In synthetic biology, the same strategy would enable the generation of complex regulatory networks, logic gates, and circuits (Greber and Fussenegger 2007).
Light input modules are currently regarded as the most promising tools for tuning the expression of multiple target genes independently and simultaneously in response to noninvasive stimuli and with high spatiotemporal resolution. Nevertheless, they suffer from a major drawback. The majority of the artificially generated recombinant photoreceptors display a modest photoswitching dynamic range. The LovTAP (Strickland et al. 2008) and the PYP-GCN4 chimera (Morgan et al. 2010; Morgan and Woolley 2010) only showed a difference in activity of twofold to fivefold between the light- and dark-adapted states. In contrast to these artificial chimeras, naturally occurring photoreceptors represent evolutionary optimized systems, in which interactions between sensor and effector modules along with the inherent signal transduction mechanisms evolved over millions of years and hence, by far, exceeds the efficiency reached so far with artificially constructed systems. Several factors can influence the photoswitching efficiency:
First, to generate a functional photoswitch, the chimeric fusion of a sensor domain to the effector module of choice needs to effectively inhibit the activity of the effector module in one of the two sensor states (dark or light). However, at least in case of the AsLOV2 sensor system, which has been widely employed for the generation of artificial photoswitches, it was shown that dark- and light-state do not represent “kinetically trapped” stable structural conformations, but co-exist in a dynamic equilibrium (Yao et al. 2008). This means that, even in the dark, the sensor domain exists in two conformational states: an energetically favored dark-state (helix-docked conformation in case of AsLOV2) and a disfavored light-state (helix-undocked conformation in case of AsLOV2; Yao et al. 2008). Light excitation provides the energy for a shift of this equilibrium. Fusion of such a dynamic sensor module to an effector protein may result in a change of this equilibrium by lowering the energy barrier for transition which would create a photoswitch that is constitutively active in both states (Strickland et al. 2008; Lee et al. 2008). Recently improved variants of LovTAP with an increased dynamic range were engineered by introducing mutations into the LOV core domain and the C-terminal Jα-helix. These mutations obviously stabilized the helix-docked conformation in the dark-state thereby increasing the dynamic range of the system significantly. Second, the intrinsic photochemical properties of a photosensory domain might hamper its application as an artificial photoswitch. Here, the lifetime of the activated state plays an important role for cycling photoreceptors such as LOV domains and the PYP. After illumination, changes in activity can only be observed as long as the sensory domain remains in the light (activated) state. This problem particularly pertains to fast cycling LOV domains which display a dark recovery time constant of only a few tens to hundreds of seconds (Kasahara et al. 2002; Jentzsch et al. 2009) resulting in a reduced active state population and consequently a reduced dynamic range. Several recent mutagenesis studies have provided a structural and functional explanation for the lifetime of the light (activated) state and thus identified several amino acid positions in LOV domains that strongly influence the dark recovery process (Christie et al. 2007; Jentzsch et al. 2009; Zoltowski et al. 2009).
In conclusion, recent novel insights into structure and function of photoreceptors, in particular, of the LOV and PYP type, now allow their application as artificial photoswitches and, at the same time, enable to tune their photochemical and structural properties. The design of recombinant photoswitches which may mimic naturally occurring photoreceptor system in terms of sensitivity and dynamic range comes within reach. As a next stage, “light-driven artificial switchboards” will be constructed to independently and simultaneously tune multiple biological functions in vivo in response to variable environmental stimuli.
References
Arndt K, Fink GR (1986) GCN4 protein, a positive transcription factor in yeast, binds general control promoters at all 5′ TGACTC 3′ sequences. Proc Natl Acad Sci USA 83(22):8516–8520
Ávila-Pérez M, Hellingwerf KJ, Kort R (2006) Blue light activates the sigmaB-dependent stress response of Bacillus subtilis via YtvA. J Bacteriol 188(17):6411–6414
Ávila-Pérez M, Vreede J, Tang Y, Bende O, Losi A, Gärtner W, Hellingwerf K (2009) In vivo mutational analysis of YtvA from Bacillus subtilis: mechanism of light activation of the general stress response. J Biol Chem 284(37):24958–24964
Barends TR, Hartmann E, Griese JJ, Beitlich T, Kirienko NV, Ryjenkov DA, Reinstein J, Shoeman RL, Gomelsky M, Schlichting I (2009) Structure and mechanism of a bacterial light-regulated cyclic nucleotide phosphodiesterase. Nature 459(7249):1015–1018
Borgstahl GE, Williams DR, Getzoff ED (1995) 1.4 ºA structure of photoactive yellow protein, a cytosolic photoreceptor: unusual fold, active site, and chromophore. Biochemistry 34(19):6278–6287
Braatsch S, Gomelsky M, Kuphal S, Klug G (2002) A single flavoprotein, AppA, integrates both redox and light signals in Rhodobacter sphaeroides. Mol Microbiol 45(3):827–836
Briggs WR (2007) The LOV domain: a chromophore module servicing multiple photoreceptors. J Biomed Sci 14(4):499–504
Caamano AM, Vázquez ME, Martínez-Costas J, Castedo L, Mascarenas JL (2000) A light-modulated sequence-specific DNA-binding peptide. Angew Chem Int Ed 39(17):3104–3107
Cambridge SB, Geissler D, Keller S, Cürten B (2006) A caged doxycycline analogue for photoactivated gene expression. Angew Chem Int Ed 45:2229–2231
Cao Z, Buttani V, Losi A, Gärtner W (2008) A blue light inducible two-component signal transduction system in the plant pathogen Pseudomonas syringae pv. tomato. Biophys J 94(3):897–905
Cao Z, Livoti E, Losi A, Gärtner W (2010) A blue light-inducible phosphodiesterase activity in the cyanobacterium Synechococcus elongatus. Photochem Photobiol 86(3):606–611
Cashmore AR, Jarillo JA, Wu YJ, Liu D (1999) Cryptochromes: blue light receptors for plants and animals. Science 284(5415):760–765
Chen CH, Ringelberg CS, Gross RH, Dunlap JC, Loros JJ (2009) Genome-wide analysis of light-inducible responses reveals hierarchical light signalling in Neurospora. EMBO J 28(8):1029–1042
Chen CH, Demay BS, Gladfelter AS, Dunlap JC, Loros JJ (2010) Physical interaction between VIVID and white collar complex regulates photoadaptation in Neurospora. Proc Natl Acad Sci USA 107(38):16715–16720
Chou C, Young DD, Deiters A (2010) Photocaged T7 RNA polymerase for the light activation of transcription and gene function in pro- and eukaryontic cells. Chembiochem 11:972–977
Christen M, Christen B, Folcher M, Schauerte A, Jenal U (2005) Identification and characterization of a cyclic di-GMP-specific phosphodiesterase and its allosteric control by GTP. J Biol Chem 280(35):30829–30837
Christie JM, Salomon M, Nozue K, Wada M, Briggs WR (1999) LOV (light, oxygen, or voltage) domains of the blue-light photoreceptor phototropin (nph1): binding sites for the chromophore flavin mononucleotide. Proc Natl Acad Sci USA 96(15):8779–8783
Christie JM, Corchnoy SB, Swartz TE, Hokenson M, Han IS, Briggs WR, Bogomolni RA (2007) Steric interactions stabilize the signaling state of the LOV2 domain of phototropin 1. Biochemistry 46(32):9310–9319
Crosson S, Moffat K (2002) Photoexcited structure of a plant photoreceptor domain reveals a light-driven molecular switch. Plant Cell 14(5):1067–1075
Cruz FG, Koh JT, Link KH (2000) Light-activated gene expression. J Am Chem Soc 122:8777–8778
D’Argenio DA, Miller SI (2004) Cyclic di-GMP as a bacterial second messenger. Microbiology 150:2497–2502
Deiters A (2010) Principles and applications of the photochemical control of cellular processes. Chembiochem 11:47–53
Dutta R, Qin L, Inouye M (1999) Histidine kinases: diversity of domain organization. Mol Microbiol 34(4):633–640
Ellenberger TE, Brandl CJ, Struhl K, Harrison SC (1992) The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted α helices: crystal structure of the protein–DNA complex. Cell 71(7):1223–1237
Elsen S, Jaubert M, Pignol D, Giraud E (2005) PpsR: a multifaceted regulator of photosynthesis gene expression in purple bacteria. Mol Microbiol 57(1):17–26
Fields S, Song O (1989) A novel genetic system to detect protein–protein interactions. Nature 340(6230):245–246
Fischer HM (1994) Genetic regulation of nitrogen fixation in Rhizobia. Microbiol Rev 58(3):352–386
Forst SA, Roberts DL (1994) Signal transduction by the EnvZ–OmpR phosphotransfer system in bacteria. Res Microbiol 145(5–6):363–373
Gaidenko TA, Kim TJ, Weigel AL, Brody MS, Price CW (2006) The blue-light receptor YtvA acts in the environmental stress signaling pathway of Bacillus subtilis. J Bacteriol 188(17):6387–6395
Gambetta GA, Lagarias JC (2001) Genetic engineering of phytochrome biosynthesis in bacteria. Proc Natl Acad Sci USA 98(19):10566–10571
Gilles-Gonzalez MA, Ditta GS, Helinski DR (1991) A haemoprotein with kinase activity encoded by the oxygen sensor of Rhizobium meliloti. Nature 350(6314):170–172
Giraud E, Fardoux J, Fourrier N, Hannibal L, Genty B, Bouyer P, Dreyfus B, Vermeglio A (2002) Bacteriophytochrome controls photosystem synthesis in anoxygenic bacteria. Nature 417(6885):202–205
Gomelsky M, Kaplan S (1995) Genetic evidence that PpsR from Rhodobacter sphaeroides 2.4.1 functions as a repressor of puc and bchF expression. J Bacteriol 177(6):1634–1637
Gomelsky M, Kaplan S (1997) Molecular genetic analysis suggesting interactions between AppA and PpsR in regulation of photosynthesis gene expression in Rhodobacter sphaeroides 2.4.1. J Bacteriol 179(1):128–134
Gomelsky M, Klug G (2002) BLUF: a novel FAD-binding domain involved in sensory transduction in microorganisms. Trends Biochem Sci 27(10):497–500
Gong W, Hao B, Mansy SS, Gonzalez G, Gilles-Gonzalez MA, Chan MK (1998) Structure of a biological oxygen sensor: a new mechanism for heme-driven signal transduction. Proc Natl Acad Sci USA 95(26):15177–15182
Greber D, Fussenegger M (2007) Mammalian synthetic biology: engineering of sophisticated gene networks. J Biotechnol 130(4):329–345
Grünberg R, Serrano L (2010) Strategies for protein synthetic biology. Nucleic Acids Res 38(8):2663–2675
Gunsalus RP, Yanofsky C (1980) Nucleotide sequence and expression of Escherichia coli trpR, the structural gene for the trp aporepressor. Proc Natl Acad Sci USA 77(12):7117–7121
Hagiwara D, Sugiura M, Oshima T, Mori H, Aiba H, Yamashino T, Mizuno T (2003) Genome-wide analyses revealing a signaling network of the RcsC–YojN–RcsB phosphorelay system in Escherichia coli. J Bacteriol 185(19):5735–5746
Heckel A, Mayer G (2010) Light-responsive nucleic acids for the spatiotemporal control of biological processes. In: Mayer G (ed) The chemical biology of nucleic acids. Wiley, pp 279–306
Hecker M, Pane-Farre J, Volker U (2007) SigB-dependent general stress response in Bacillus subtilis and related gram-positive bacteria. Annu Rev Microbiol 61:215–236
Hengge R (2009) Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol 7(4):263–273
Hiltbrunner A, Tscheuschler A, Viczian A, Kunkel T, Kircher S, Schäfer E (2006) FHY1 and FHL act together to mediate nuclear accumulation of the phytochrome A photoreceptor. Plant Cell Physiol 47(8):1023–1034
Hirose Y, Shimada T, Narikawa R, Katayama M, Ikeuchi M (2008) Cyanobacteriochrome CcaS is the green light receptor that induces the expression of phycobilisome linker protein. Proc Natl Acad Sci USA 105(28):9528–9533
Hoff WD, van der Horst MA, Nudel CB, Hellingwerf KJ (2009) Prokaryotic phototaxis. Methods Mol Biol 571:25–49
Hori Y, Ueno H, Mizukami S, Kikuchi K (2009) Photoactive yellow protein-based protein labeling system with turn-on fluorescence intensity. J Am Chem Soc 131(46):16610–16611
Hübschmann T, Yamamoto H, Gieler T, Murata N, Börner T (2005) Red and far-red light alter the transcript profile in the cyanobacterium Synechocystis sp. PCC 6803: impact of cyanobacterial phytochromes. FEBS Lett 579(7):1613–1618
Hughes J (2010) Phytochrome three-dimensional structures and functions. Biochem Soc Trans 38(2):710–716
Hunt SM, Thompson S, Elvin M, Heintzen C (2010) VIVID interacts with the WHITE COLLAR complex and FREQUENCY-interacting RNA helicase to alter light and clock responses in Neurospora. Proc Natl Acad Sci USA 107(38):16709–16714
Imamoto Y, Kataoka M (2007) Structure and photoreaction of photoactive yellow protein, a structural prototype of the PAS domain superfamily. Photochem Photobiol 83(1):40–49
Iseki M, Matsunaga S, Murakami A, Ohno K, Shiga K, Yoshida K, Sugai M, Takahashi T, Hori T, Watanabe M (2002) A blue-light-activated adenylyl cyclase mediates photoavoidance in Euglena gracilis. Nature 415(6875):1047–1051
Ishikawa M, Takahashi F, Nozaki H, Nagasato C, Motomura T, Kataoka H (2009) Distribution and phylogeny of the blue light receptors aureochromes in eukaryotes. Planta 230(3):543–552
Jarillo JA, Capel J, Tang RH, Yang HQ, Alonso JM, Ecker JR, Cashmore AR (2001) An Arabidopsis circadian clock component interacts with both CRY1 and phyB. Nature 410(6827):487–490
Jaubert M, Zappa S, Fardoux J, Adriano JM, Hannibal L, Elsen S, Lavergne J, Vermeglio A, Giraud E, Pignol D (2004) Light and redox control of photosynthesis gene expression in Bradyrhizobium: dual roles of two PpsR. J Biol Chem 279(43):44407–44416
Jeeves M, Evans PD, Parslow RA, Jaseja M, Hyde EI (1999) Studies of the Escherichia coli Trp repressor binding to its five operators and to variant operator sequences. Eur J Biochem 265(3):919–928
Jentzsch K, Wirtz A, Circolone F, Drepper T, Losi A, Gärtner W, Jaeger KE, Krauss U (2009) Mutual exchange of kinetic properties by extended mutagenesis in two short LOV domain proteins from Pseudomonas putida. Biochemistry 48(43):10321–10333
Jorns MS (1990) DNA photorepair: chromophore composition and function in two classes of DNA photolyases. Biofactors 2(4):207–211
Kaplan JH, Forbush B III, Hoffmann JF (1978) Rapid photolytic release of adenosine 5′-triphosphate from a protected analog: utilization by the sodium:potassium pump of human red blood cell ghosts. Biochemistry 17:1929–1935
Kasahara M, Swartz TE, Olney MA, Onodera A, Mochizuki N, Fukuzawa H, Asamizu E, Tabata S, Kanegae H, Takano M, Christie JM, Nagatani A, Briggs WR (2002) Photochemical properties of the flavin mononucleotide-binding domains of the phototropins from Arabidopsis, rice, and Chlamydomonas reinhardtii. Plant Physiol 129(2):762–773
Kaupp UB, Seifert R (2002) Cyclic nucleotide-gated ion channels. Physiol Rev 82(3):769–824
Kehoe DM, Grossman AR (1996) Similarity of a chromatic adaptation sensor to phytochrome and ethylene receptors. Science 273(5280):1409–1412
Kehoe DM, Grossman AR (1997) New classes of mutants in complementary chromatic adaptation provide evidence for a novel four-step phosphorelay system. J Bacteriol 179(12):3914–3921
Kehoe DM, Gutu A (2006) Responding to color: the regulation of complementary chromatic adaptation. Annu Rev Plant Biol 57:127–150
Khalil AS, Collins JJ (2010) Synthetic biology: applications come of age. Nat Rev Genet 11(5):367–379
Kim WY, Fujiwara S, Suh SS, Kim J, Kim Y, Han L, David K, Putterill J, Nam HG, Somers DE (2007) ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 449(7160):356–360
Kolb A, Busby S, Buc H, Garges S, Adhya S (1993) Transcriptional regulation by cAMP and its receptor protein. Annu Rev Biochem 62:749–795
Koldin B, Suckow M, Seydel A, von Wilcken-Bergmann B, Müller-Hill B (1995) A comparison of the different DNA binding specificities of the bZip proteins C/EBP and GCN4. Nucleic Acids Res 23(20):4162–4169
Kort R, Hoff WD, Van West M, Kroon AR, Hoffer SM, Vlieg KH, Crielaand W, Van Beeumen JJ, Hellingwerf KJ (1996) The xanthopsins: a new family of eubacterial blue-light photoreceptors. EMBO J 15(13):3209–3218
Krauss U, Minh BQ, Losi A, Gärtner W, Eggert T, von Haeseler A, Jaeger K-E (2009) Distribution and phylogeny of light–oxygen–voltage-blue-light-signaling proteins in the three kingdoms of life. J Bacteriol 191(23):7234–7242
Kyndt JA, Vanrobaeys F, Fitch JC, Devreese BV, Meyer TE, Cusanovich MA, Van Beeumen JJ (2003) Heterologous production of Halorhodospira halophila holo-photoactive yellow protein through tandem expression of the postulated biosynthetic genes. Biochemistry 42(4):965–970
Lee J, Natarajan M, Nashine VC, Socolich M, Vo T, Russ WP, Benkovic SJ, Ranganathan R (2008) Surface sites for engineering allosteric control in proteins. Science 322(5900):438–442
Lee MM, Larson DR, Lawrence DS (2009) Illuminating the chemistry of life: design, synthesis, and applications of ‘caged’ and related photoresponsive compounds. ACS Chem Biol 4:409–427
Leivar P, Quail PH (2010) PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci 16(1):19–28
Levskaya A, Chevalier AA, Tabor JJ, Simpson ZB, Lavery LA, Levy M, Davidson EA, Scouras A, Ellington AD, Marcotte EM, Voigt CA (2005) Synthetic biology: engineering Escherichia coli to see light. Nature 438(7067):441–442
Lin W, Albanese C, Pestell RG, Lawrence DS (2002) Spatially discrete, light-driven protein expression. Chem Biol 9:1347–1353
Link KH, Shi Y, Koh JT (2005) Light activated recombination. J Am Chem Soc 127:13088–13089
Liu H, Yu X, Li K, Klejnot J, Yang H, Lisiero D, Lin C (2008) Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 322(5907):1535–1539
Losi A (2004) The bacterial counterparts of plant phototropins. Photochem Photobiol Sci 3(6):566–574
Losi A (2007) Flavin-based blue-light photosensors: a photobiophysics update. Photochem Photobiol 83(6):1283–1300
Losi A, Gärtner W (2008) Bacterial bilin- and flavin-binding photoreceptors. Photochem Photobiol Sci 7(10):1168–1178
Losi A, Polverini E, Quest B, Gärtner W (2002) First evidence for phototropin-related blue-light receptors in prokaryotes. Biophys J 82(5):2627–2634
Malzahn E, Ciprianidis S, Káldi K, Schafmeier T, Brunner M (2010) Photoadaptation in Neurospora by competitive interaction of activating and inhibitory LOV domains. Cell 142(5):762–772
Marmorstein RQ, Sigler PB (1989) Stereochemical effects of l-tryptophan and its analogues on trp repressor’s affinity for operator-DNA. J Biol Chem 264(16):9149–9154
Masuda S, Bauer CE (2002) AppA is a blue light photoreceptor that antirepresses photosynthesis gene expression in Rhodobacter sphaeroides. Cell 110(5):613–623
Mayer G, Heckel A (2006) Biologically active molecules with a ‘light switch’. Angew Chem Int Ed 45:4900–4921
Mitrophanov AY, Groisman EA (2008) Signal integration in bacterial two-component regulatory systems. Genes Dev 22(19):2601–2611
Möglich A, Ayers RA, Moffat K (2009a) Design and signaling mechanism of light-regulated histidine kinases. J Mol Biol 385(5):1433–1444
Möglich A, Ayers RA, Moffat K (2009b) Structure and signaling mechanism of Per-ARNT-Sim domains. Structure 17(10):1282–1294
Möglich A, Ayers RA, Moffat K (2010) Addition at the molecular level: signal integration in designed Per-ARNT-Sim receptor proteins. J Mol Biol 400(3):477–486
Möglich A, Moffat K (2010) Engineered photoreceptors as novel optogenetic tools. Photochem Photobiol Sci 9(10):1286–1300
Morgan SA, Woolley GA (2010) A photoswitchable DNA-binding protein based on a truncated GCN4-photoactive yellow protein chimera. Photochem Photobiol Sci 9(10):1320–1326
Morgan SA, Al-Abdul-Wahid S, Woolley GA (2010) Structure-based design of a photocontrolled DNA binding protein. J Mol Biol 399(1):94–112
Neiss A, Schafmeier T, Brunner M (2008) Transcriptional regulation and function of the Neurospora clock gene white collar 2 and its isoforms. EMBO Rep 9(8):788–794
Newell PD, Monds RD, O’Toole GA (2009) LapD is a bis-(3′, 5′)-cyclic dimeric GMP-binding protein that regulates surface attachment by Pseudomonas fluorescens Pf0–1. Proc Natl Acad Sci USA 106(9):3461–3466
Ntefidou M, Iseki M, Watanabe M, Lebert M, Hader DP (2003) Photoactivated adenylyl cyclase controls phototaxis in the flagellate Euglena gracilis. Plant Physiol 133(4):1517–1521
Ogura Y, Komatsu A, Zikihara K, Nanjo T, Tokutomi S, Wada M, Kiyosue T (2008) Blue light diminishes interaction of PAS/LOV proteins, putative blue light receptors in Arabidopsis thaliana, with their interacting partners. J Plant Res 121(1):97–105
Oshima T, Aiba H, Masuda Y, Kanaya S, Sugiura M, Wanner BL, Mori H, Mizuno T (2002) Transcriptome analysis of all two-component regulatory system mutants of Escherichia coli K-12. Mol Microbiol 46(1):281–291
Pawson T, Nash P (2003) Assembly of cell regulatory systems through protein interaction domains. Science 300(5618):445–452
Pellequer JL, Wager-Smith KA, Kay SA, Getzoff ED (1998) Photoactive yellow protein: a structural prototype for the three-dimensional fold of the PAS domain superfamily. Proc Natl Acad Sci USA 95(11):5884–5890
Petersohn A, Bernhardt J, Gerth U, Hoper D, Koburger T, Volker U, Hecker M (1999) Identification of sigma(B)-dependent genes in Bacillus subtilis using a promoter consensus-directed search and oligonucleotide hybridization. J Bacteriol 181(18):5718–5724
Purcell EB, Crosson S (2008) Photoregulation in prokaryotes. Curr Opin Microbiol 11(2):168–178
Purcell EB, Siegal-Gaskins D, Rawling DC, Fiebig A, Crosson S (2007) A photosensory two-component system regulates bacterial cell attachment. Proc Natl Acad Sci USA 104(46):18241–18246
Purcell EB, McDonald CA, Palfey BA, Crosson S (2010) An analysis of the solution structure and signaling mechanism of LovK, a sensor histidine kinase integrating light and redox signals. Biochemistry 49(31):6761–6770
Rajagopal S, Key JM, Purcell EB, Boerema DJ, Moffat K (2004) Purification and initial characterization of a putative blue light-regulated phosphodiesterase from Escherichia coli. Photochem Photobiol 80(3):542–547
Riggsbee CW, Deiters A (2010) Recent advances in the photochemical control of protein function. Trends Biotech 28:468–475
Rockwell NC, Lagarias JC (2010) A brief history of phytochromes. Chemphyschem 11(6):1172–1180
Ryu MH, Moskvin OV, Siltberg-Liberles J, Gomelsky M (2010) Natural and engineered photoactivated nucleotidyl cyclases for optogenetic applications. J Biol Chem 285(53):41501–41508
Sauers DJ, Temburni MK, Biggins JB, Ceo LM, Galileo DS, Loh JT (2010) Light-activated gene expression directs segregation of co-cultured cells in vitro. ACS Chem Biol 5(3):313–320
Sawa M, Nusinow DA, Kay SA, Imaizumi T (2007) FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318(5848):261–265
Schaper K, Etinski M, Fleig T (2009) Theoretical investigation on the excited states of 2-nitrobenzyl and 4, 5-methylendioxy-2-nitrobenzyl caging groups. Photochem Photobiol 85:1075–1081
Schirmer T, Jenal U (2009) Structural and mechanistic determinants of c-di-GMP signalling. Nat Rev Microbiol 7(10):724–735
Schünke S, Stoldt M, Novak K, Kaupp UB, Willbold D (2009) Solution structure of the Mesorhizobium loti K1 channel cyclic nucleotide-binding domain in complex with cAMP. EMBO Rep 10(7):729–735
Shao Q, Xing B (2010) Photoactive molecules for applications in molecular imaging and cell biology. Chem Soc Rev 39:2835–2846
Sharrock RA (2008) The phytochrome red/far-red photoreceptor superfamily. Genome Biol 9(8):230
Shimizu-Sato S, Huq E, Tepperman JM, Quail PH (2002) A light-switchable gene promoter system. Nat Biotechnol 20(10):1041–1044
Shin DH, Cho MH, Kim TL, Yoo J, Kim JI, Han YJ, Song PS, Jeon JS, Bhoo SH, Hahn TR (2010) A small GTPase activator protein interacts with cytoplasmic phytochromes in regulating root development. J Biol Chem 285(42):32151–32159
Somers DE (2001) Clock-associated genes in Arabidopsis: a family affair. Philos Trans R Soc Lond B Biol Sci 356(1415):1745–1753
Sorokina O, Kapus A, Terecskei K, Dixon LE, Kozma-Bognar L, Nagy F, Millar AJ (2009) A switchable light-input, light-output system modelled and constructed in yeast. J Biol Eng 3:15
Sprenger WW, Hoff WD, Armitage JP, Hellingwerf KJ (1993) The eubacterium Ectothiorhodospira halophila is negatively phototactic, with a wavelength dependence that fits the absorption spectrum of the photoactive yellow protein. J Bacteriol 175(10):3096–3104
Stierl M, Stumpf P, Udwari D, Gueta R, Hagedorn R, Losi A, Gärtner W, Petereit L, Efetova M, Schwarzel M, Oertner TG, Nagel G, Hegemann P (2011) Light-modulation of cellular cAMP by a small bacterial photoactivated adenylyl cyclase, bPAC, of the soil bacterium Beggiatoa. J Biol Chem 286:1181–1188
Strickland D, Moffat K, Sosnick TR (2008) Light-activated DNA binding in a designed allosteric protein. Proc Natl Acad Sci USA 105(31):10709–10714
Strickland D, Yao X, Gawlak G, Rosen MK, Gardner KH, Sosnick TR (2010) Rationally improving LOV domain-based photoswitches. Nat Methods 7(8):623–626
Swartz TE, Wenzel PJ, Corchnoy SB, Briggs WR, Bogomolni RA (2002) Vibration spectroscopy reveals light-induced chromophore and protein structural changes in the LOV2 domain of the plant blue-light receptor phototropin 1. Biochemistry 41(23):7183–7189
Swartz TE, Tseng TS, Frederickson MA, Paris G, Comerci DJ, Rajashekara G, Kim JG, Mudgett MB, Splitter GA, Ugalde RA, Goldbaum FA, Briggs WR, Bogomolni RA (2007) Blue-light-activated histidine kinases: two-component sensors in bacteria. Science 317(5841):1090–1093
Tabor JJ, Salis HM, Simpson ZB, Chevalier AA, Levskaya A, Marcotte EM, Voigt CA, Ellington AD (2009) A synthetic genetic edge detection program. Cell 137(7):1272–1281
Tabor JJ, Levskaya A, Voigt CA (2011) Multichromatic control of gene expression in Escherichia coli. J Mol Biol 405:315–324
Takahashi F, Yamagata D, Ishikawa M, Fukamatsu Y, Ogura Y, Kasahara M, Kiyosue T, Kikuyama M, Wada M, Kataoka H (2007) AUREOCHROME, a photoreceptor required for photomorphogenesis in stramenopiles. Proc Natl Acad Sci USA 104(49):19625–19630
Tarutina M, Ryjenkov DA, Gomelsky M (2006) An unorthodox bacteriophytochrome from Rhodobacter sphaeroides involved in turnover of the second messenger c-di-GMP. J Biol Chem 281(46):34751–34758
Terauchi K, Montgomery BL, Grossman AR, Lagarias JC, Kehoe DM (2004) RcaE is a complementary chromatic adaptation photoreceptor required for green and red light responsiveness. Mol Microbiol 51(2):567–577
Tschowri N, Busse S, Hengge R (2009) The BLUF-EAL protein YcgF acts as a direct anti-repressor in a blue-light response of Escherichia coli. Genes Dev 23(4):522–534
Utsumi R, Brissette RE, Rampersaud A, Forst SA, Oosawa K, Inouye M (1989) Activation of bacterial porin gene expression by a chimeric signal transducer in response to aspartate. Science 245(4923):1246–1249
van der Horst MA, Hellingwerf KJ (2004) Photoreceptor proteins, star actors of modern times: a review of the functional dynamics in the structure of representative members of six different photoreceptor families. Acc Chem Res 37(1):13–20
van der Horst MA, Key J, Hellingwerf KJ (2007) Photosensing in chemotrophic, non-phototrophic bacteria: let there be light sensing too. Trends Microbiol 15(12):554–562
Wang L, Xie J, Schultz PG (2006) Expanding the genetic code. Annu Rev Biophys Biomol Struct 35:225–249
Wang W, Shu D, Chen L, Jiang W, Lu Y (2009) Cross-talk between an orphan response regulator and a noncognate histidine kinase in Streptomyces coelicolor. FEMS Microbiol Lett 294(2):150–156
Wilson CJ, Zhan H, Swint-Kruse L, Matthews KS (2007) The lactose repressor system: paradigms for regulation, allosteric behavior and protein folding. Cell Mol Life Sci 64(1):3–16
Won HS, Lee YS, Lee SH, Lee BJ (2009) Structural overview on the allosteric activation of cyclic AMP receptor protein. Biochim Biophys Acta 1794(9):1299–1308
Woolley GA, Jaikaran AS, Berezovski M, Calarco JP, Krylov SN, Smart OS, Kumita JR (2006) Reversible photocontrol of DNA binding by a designed GCN4-bZIP protein. Biochemistry 45(19):6075–6084
Yao X, Rosen MK, Gardner KH (2008) Estimation of the available free energy in a LOV2-J alpha photoswitch. Nat Chem Biol 4(8):491–497
Yasuhara M, Mitsui S, Hirano H, Takanabe R, Tokioka Y, Ihara N, Komatsu A, Seki M, Shinozaki K, Kiyosue T (2004) Identification of ASK and clock-associated proteins as molecular partners of LKP2 (LOV kelch protein 2) in Arabidopsis. J Exp Bot 55(405):2015–2027
Yazawa M, Sadaghiani AM, Hsueh B, Dolmetsch RE (2009) Induction of protein–protein interactions in live cells using light. Nat Biotechnol 27(10):941–945
Yeh KC, Wu SH, Murphy JT, Lagarias JC (1997) A cyanobacterial phytochrome two-component light sensory system. Science 277(5331):1505–1508
Young DD, Deiters A (2007a) Photochemical activation of protein expression in bacterial cells. Angew Chem Int Ed 46:4290–4292
Young DD, Deiters A (2007b) Photochemical control of biological processes. Org Biomol Chem 5:999–1005
Young DD, Garner RA, Yoder JA, Deiters A (2009) Light-activation of gene function in mammalian cells via ribozymes. Chem Commun 568–570
Zoltowski BD, Vaccaro B, Crane BR (2009) Mechanism-based tuning of a LOV domain photoreceptor. Nat Chem Biol 5(11):827–834
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Drepper, T., Krauss, U., Meyer zu Berstenhorst, S. et al. Lights on and action! Controlling microbial gene expression by light. Appl Microbiol Biotechnol 90, 23–40 (2011). https://doi.org/10.1007/s00253-011-3141-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-011-3141-6