Abstract
Pseudomonas putida KT2440 strain was investigated for biosynthesis of the valuable xanthophyll zeaxanthin. A new plasmid was constructed harboring five carotenogenic genes from Pantoea ananatis and three genes from Escherichia coli under control of an l-rhamnose-inducible promoter. Pseudomonas putida KT2440 wild type hardly tolerated the plasmids for carotenoid production. Mating experiments with E. coli S17-1 strains revealed that the carotenoid products are toxic to the Pseudomonas putida cells. Several carotenoid-tolerant transposon mutants could be isolated, and different gene targets for relief of carotenoid toxicity were identified. After optimization of cultivation conditions and product processing, 51 mg/l zeaxanthin could be produced, corresponding to a product yield of 7 mg zeaxanthin per gram cell dry weight. The effect of various additives on production of hydrophobic zeaxanthin was investigated as well. Particularly, the addition of lecithin during cell cultivation increased volumetric productivity of Pseudomonas putida by a factor of 4.7 (51 mg/l vs. 239 mg/l).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Carotenoids are a class of natural fat-soluble pigments found mostly in plants, but also in some types of algae, fungi, and bacteria (Britton et al. 1995). An overall structure of carotenoids is based on a 40-carbon polyene chain which is sometimes terminated by rings. Unsubstituted hydrocarbons like alpha-carotene, beta-carotene, and lycopene are known as carotenes. Carotenoids containing oxygen, such as lutein and zeaxanthin, are known as xanthophylls.
Zeaxanthin (β,β-carotene-3,3′-diol) is a xanthophyll that occurs naturally in many vegetables like red pepper (Capsicum annuum var. grossum) and corn (Zea mays) (Meyer 2002) and also in some bacteria like Pantoea ananatis (former Eriwinia uredovora), where it can be mainly found as mono- and diglycoside (Sajilata et al. 2008). The major part of zeaxanthin in plants is esterified with fatty acids, for example, zeaxanthin dipalmitate (physalien) is the main carotenoid of Chinese wolfberry (Lycium barbarum L.).
Generally, xanthophylls are important food and feed additives or precursors for pharmaceuticals. In the European Union, zeaxanthin (E 161h) is used mainly as food additive for poultry pigmentation (BVL 2007). Furthermore, zeaxanthin and its stereoisomer lutein are reported to prevent age-related macular degeneration (Gerster 1993). The first commercially available dietary product containing zeaxanthin and lutein in a ratio 2:1 (as it can be found in a healthy macular) is on the market (Sajilata et al. 2008). Anti-carcinogenic activity of zeaxanthin has been demonstrated (Sun and Yao 2007; Tsushima et al. 1995; Nishino et al. 2002), and its use for cancer prevention is under discussion (Snodderly 1995; Nishino et al. 2002).
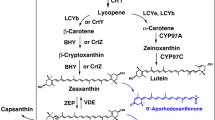
Generic scheme for zeaxanthin synthesis is presented in Fig. 1. The glucose metabolites pyruvate and d-glyceraldehyde-3-phosphate are the building blocks for the two isoprenoid precursors, dimethylallyl pyrophosphate, and isopentenyl pyrophosphate, which are built within several steps. These two substances serve as precursors for farnesyl pyrophosphate, which is transformed further in three steps into phytoene. Phytoene is then desaturated to lycopene in four steps via phytofluene, ζ-carotene, and neurosporene. Lycopene represents a skeletal structure for all carotenoids, and after neurosporene is the second colored substance in this pathway. Different modifications like cyclization and hydroxylation of lycopene lead to various carotenoids, for example, cyclization of lycopene to β-carotene and the following 3- and 3′-hydroxylation lead to formation of zeaxanthin. In this step, three stereoisomers, (3R,3′R)-, (3S,3′S)-, and (3R,3′S; meso)-zeaxanthin can be formed depending on the hydroxylase used.
Already in 1950, Karrer and Eugster published the first chemical synthesis of β-carotene (Karrer and Eugster 1950). However, the chemical synthesis of zeaxanthin starting from easily accessible inexpensive chemicals like acetone, butadiene, and isoprene requires 20 stages in 11 steps (Vaz et al. 2002) which results finally in high production costs (Meyer 2002).
With respect to renewable resources, microbial biosynthesis using recombinant microbial strains represents an environmentally friendly alternative to chemical synthesis. In several published studies, Escherichia coli was used for production of carotenoids, with focus on β-carotene and lycopene. For β-carotene, the yields between 6 mg/g cell dry weight (CDW) (Yuan et al. 2006) and 49.3 mg/g CDW (Yoon et al. 2007b) have been reported. Lycopene could be produced in amounts between 4.7 mg/g CDW (Kang et al. 2005) and 22 mg/g CDW (Yoon et al. 2007a). In all mentioned cases, the improved productivity was achieved through metabolic engineering either in the prenyl diphosphate pathway, the methylerythritol phosphate (MEP) pathway, or in the mevalonate pathway (Yuan et al. 2006). Only one study on metabolic engineering of E. coli was dedicated to production of zeaxanthin, where 0.82 mg product per 1 g CDW could be achieved (Nishizaki et al. 2007).
In the present study, Pseudomonas putida KT2440 was investigated regarding its applicability for carotenoid production. Pseudomonads are a ubiquitous group of gram-negative bacteria, which belong to the gamma subclass of the Proteobacteria. Pseudomonas putida KT2440 is a plasmid-free derivative of Pseudomonas putida mt-2 (Vilchez et al. 2000) and known as a root-colonizer and considered as a rhizosphere microorganism (Molina et al. 2000). Because of their ability to utilize xenobiotics, Pseudomonas strains have a great potential for bioremediation (Dejonghe et al. 2001). They are also applied for biocatalysis (Schmid et al. 2001) and plant protection (Walsh et al. 2001).
A new plasmid harboring several carotenogenic genes from Pantoea ananatis and E. coli was constructed and inserted in Pseudomonas putida. The Pseudomonas putida recombinant system was adopted for production of zeaxanthin and compared to the established recombinant E. coli system.
Material and methods
Material
All chemicals were purchased from Sigma-Aldrich Labor Chemie GmbH (Steinheim, Germany) or VWR International GmbH (Darmstadt, Germany) in analytical quality. β-Carotene was purchased from Fluka (Seelze, Germany). β-Cryptoxanthin, Chinese wolfberries (Lycium barbarum L.) and (3R,3′R)-zeaxanthin were gifts from the Institute of Food Chemistry, Universität Hohenheim (Stuttgart, Germany). Water for high-performance liquid chromatography (HPLC) was filtered through a Modulab Bioscience (Liquipure Europe Ltd., Bicester, United Kingdom).
Media
Luria–Bertani (LB) and terrific broth (TB) media were prepared according to standard protocols (Sambrook et al. 2001). For preparation of minimal salt medium MEK medium, 50 ml phosphate buffer (20 g/l K2HPO4, 70 g/l Na2HPO4 2 H2O, pH 7.1), 10 ml mineral salt solution (100 g/l (NH4)2SO4, 20 g/l MgSO4 7H2O, 1 g/l ammonium ferric citrate, 100 ml/l trace element solution), 1 ml calcium solution (50 g/l Ca(NO3)2 7H2O), and 10 ml sodium succinate solution (18% w/v) were mixed and filled up with demineralized water to 1 L. The trace element solution was composed of 100 mg/l ZnSO4 7H2O, 30 mg/l MnCl2 4H2O, 300 mg/l H3BO3, 200 mg/l CoCl2 6H2O, 10 mg/l CuCl2 2H2O, 20 mg/l NiCl2 6H2O, and 30 mg/l Na2MoO4 2H2O.
Construction of plasmids for carotenoid production
The broad host range and mobilizable expression vector pJeM1 (Jeske and Altenbuchner 2010) based on the Bordetella bronchiseptica pBBR1MCS-2 (Kovach et al. 1995) and the E. coli l-rhamnose-inducible promoter system (Stumpp et al. 2000) was used for cloning. Besides the genes for carotenoid biosynthesis from Pantoea ananatis (DSM 30080) (Misawa et al. 1990), three genes from E. coli encoding enzymes for bottleneck reactions in isoprenoid synthesis (Matthews and Wurtzel 2000; Wang et al. 1999) were cloned accessorily. Genes were amplified with the primers shown in Table 1. All plasmids constructed and used in this study are shown in Table 2. For construction of pJH13 and pJH14, the In-Fusion Dry-Down PCR Cloning Kit (Taka Bio Europe/Clontech, France) was used. Further details on cloning strategy applied for the construction of all other plasmids are included in the “Results” section.
Mating experiments
For all mating experiments, plasmid-carrying E. coli S17-1 strains (Simon et al. 1983) were used as donors. The experiments were done under standard conditions. Donor and recipient strains were cultured overnight in LB broth. For every experiment, 5 × 108 donor cells were mixed with an equal number of recipient cells in 200 μl 0.9% NaCl, dripped on LB agar and incubated overnight at 30 °C. The cells were then floated off the plate with 5 ml 0.9% NaCl. The particular number of donor, recipient, and transconjugant cells in the obtained suspension was determined by dilution and subsequent cultivation on agar containing an appropriate antibiotic for the separate selection of each cell type. Random samples were taken among the transconjugants; plasmids were extracted and checked by restriction digest. The experiments were carried out with Pseudomonas putida KT2440, Pseudomonas putida D7L3 and, as a control, with E. coli JM109. Experiments were repeated independently three to five times.
Transposon mutagenesis of Pseudomonas putida KT2440
The mutant library of Pseudomonas putida KT2440 (DSM 6125) was constructed by transferring the mini-Tn5 containing vector pJOE5777.3 from E. coli S17-1 into Pseudomonas putida. The plasmid pJOE5777.3 is a derivative of the pUT-mini-Tn5-CmR which had the chloramphenicol resistance gene deleted and replaced by a BsaI fragment from pACYC184 carrying a tetracycline resistance gene and the plasmid origin of replication. The plasmid cannot replicate in Pseudomonas putida and so the tetracycline resistance is only rescued by integration of the mini-Tn5 into the chromosome. After mating, the obtained cell suspension was diluted and spread on LB agar containing 20 μg/ml tetracycline and 10 μg/ml nalidixic acid. About 5,000 Pseudomonas putida mutant colonies were obtained, and 100–200 of them at time were replica plated on LB agar covered with a lawn of E. coli S17-1 pJOE5573.3. The replica plates were incubated at 30 °C overnight and afterwards again replica plated on LB agar containing 50 μg/ml kanamycin and 10 μg/ml nalidixic acid. The obtained kanamycin resistant colonies were identified on the master plates and used for further investigations.
Carotenoid biosynthesis in bacteria
Pseudomonas putida D7L3 and E. coli JM109 harboring pBBR1MCS-2 derived plasmids were grown in 50–200 ml medium containing 30 μg/ml kanamycin in baffled flasks at 30 °C (Pseudomonas putida) or 37 °C (E. coli), respectively, and were shaken at 160 min−1. For induction of gene expression, l-rhamnose solution (final concentration in medium 0.2% w/v) was added to the cultures at an optical density at 600 nm (OD600) between 0.3 and 0.5. After induction, the cultures were cultivated at 25 °C for further 48–72 h. Every 24 h, one tenth of the initial volume of fivefold-concentrated corresponding medium was added. Finally cells were harvested by centrifugation (4,000×g, 4 °C, 15 min) and stored at −20 °C until further investigations. The effect of the hydrophobic additives oleic acid, choline, lecithin, or Amberlite® XAD-7 on productivity of E. coli JM109 and Pseudomonas putida D7L3 was investigated in 50 ml TB medium, supplemented with 1%, 5%, or 10% of a corresponding additive. Cell cultures without additives were used as negative controls. Each experiment was repeated at least three times in duplicate.
Extraction and analysis of carotenoids
Cell samples were taken during cultivation, pelletized by centrifugation and extracted with acetone. The first sample was taken 3.5 h after inoculation (2.5 h after induction) and further every 10 h. Cell pellets were extracted with small portions of acetone until the cells became colorless. Due to the difficulties in separation of the additives choline, lecithin, and oleic acid by centrifugation, 1 ml culture aliquots were added to 2 ml of a mixture of methanol, ethyl acetate, and n-hexane (1:1:1) and stirred. This procedure was repeated until the organic phase remained colorless. The colored organic extracts were combined and dried over sodium sulfate. They were evaporated to dryness after adding 20 μg of trans-8′-β-apocarotenal as internal standard. The residue was resolved in 1 ml mixture of methyl-tert-butyl ether (MTBE) and methanol (1:1) with 1% of 2,6-di-tert-butyl-4-methylphenol and analyzed by HPLC. For HPLC, a Shimadzu device (Shimadzu Deutschland GmbH, Duisburg, Germany) equipped with a controller SCL-10Avp and a diode array detector SPD-M10Avp was used. For gradient elution, two solvents were used: solvent A containing methanol and water (9:1) and solvent B containing MTBE and methanol (9:1). The time program was as follows: 0 min 6.5% solvent B, 38 min 100% solvent B; 42 min 6.5% solvent B, 47 min 6.5% solvent B. Phytoene was detected at 286 nm, lycopene, β-carotene, β-cryptoxanthin, and zeaxanthin at 450 nm. For product separation, YMC-Pack YMC30 column (250 × 4.6 mm, 3 μm, YMC Europe GmbH, Dinslaken, Germany) equipped with a pre-column YMC30 (10 × 4 mm, 3 μm) was used. The column was kept at room temperature.
Substances were identified by comparison of their retention times and UV/Vis spectra with authentic standards. Ratios of the investigated substances were calculated only for the corresponding all-E-isomers. Quantification was done using calibration curves of zeaxanthin, β-carotene, and β-cryptoxanthin. For the curves, at least five different concentrations of the authentic substance were measured. Peak areas were plotted against the concentration as described elsewhere (Lindsay 1996).
Extraction of zeaxanthin from Chinese wolfberries
For the extraction of zeaxanthin, as a reference substance, Chinese wolfberries were used. For these fruits it is known that they contain a high amount of zeaxanthin dipalmitate (physalien). Some dried fruits were crushed in a mortar and extracted several times with small portions (approx 30 ml in total) of a mixture of methanol, ethyl acetate, and n-hexane (1:1:1). The extract was evaporated to dryness and resolved in 5 ml of diethyl ether. To the resulting yellow solution, 10 ml ethanol and 5 ml potash lye (30%) were added, and the mixture was stirred at room temperature overnight in the dark for saponification. The organic phase was separated from the aqueous phase. The aqueous phase was extracted three times with an equal amount of diethyl ether. The combined organic phases were then washed twice with water, dried over anhydrous sodium sulfate, and evaporated to dryness. The obtained zeaxanthin was used as a reference for the calibration curve.
Derivatization of zeaxanthin
For investigation of the stereochemistry of the produced zeaxanthin, its enantiomers where derivatized with (+)-(S)-1-1(1-naphthyl)-ethyl isocyanate as described elsewhere (Eugster 1995; Schiedt et al. 1988). Briefly, the extract was evaporated to dryness and resolved in 100–200 μl methylene chloride. To the solution, two drops of triethylamine, one drop of the derivatizing agent, and one tip of a spatula of 4-(dimethylamino)pyridine were added, and the mixture was stirred at room temperature overnight. It was dissolved in a mixture of ethyl acetate and hexane and investigated by HPLC.
Results
Construction of an expression vector for carotenoid production
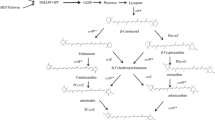
Instead of cloning the whole operon for carotenoid biosynthesis from Pantoea ananatis at once, the biosynthesis genes were amplified separately by PCR and inserted step by step into pJeM1. In the future this will allow to add new genes at any step of the carotenoid biosynthesis and to redirect the synthesis to new products. To enhance the synthesis of the carotenoid precursors, first of all three genes from E. coli were inserted downstream the rhaP BAD promoter, encoding enzymes which catalyze bottleneck reactions in the biosynthesis. The first one added was the idi gene encoding the isopentenyl pyrophosphate isomerase converting isopentenyl pyrophosphate to dimethylallyl pyrophosphate and vice versa. The gene was inserted in pJeM1 as an NdeI/BsrGI fragment, hereby replacing the eGFP gene of the vector. With the primer used for the amplification of the idi gene, a PmeI site was added at the 3′-end which allowed the insertion of the second fragment. All the fragments added next had a SmaI or DraI site at the 5′-end and a PmeI site at the 3′-end. When these blunt ending fragments were inserted in the right orientation, the PmeI site at the 3′-end was regenerated and on the 5′-end destroyed which allows theoretically an endless insertion of new genes into this synthetic operon. Behind the idi gene, the E. coli genes ispA encoding the geranyl/farnesyl pyrophosphate synthase and dxs encoding the 1-deoxy-xylulose-5-phosphate synthase were added on one fragment. This was followed by the crtE, crtB, crtI, crtY, and finally crtZ genes from Pantoea ananatis. The crtB gene was added alone to give plasmid pJH13 and together with crtI to give plasmid pJOE5573.3 (see Table 2). All the fragments were amplified with the primers shown in Table 1, cloned into the positive selection vector pJOE4786.1, and sequenced before they were cut out from pJOE4786.1 and inserted into the expression vector. The cloning strategy is outlined in Fig. 2. The production of carotenoids by E. coli carrying the various vectors could be easily seen by the red (lycopene), orange (β-carotene), and yellowish (zeaxanthin) color of the colonies, even without induction of the rhaP BAD promoter.
Effect of carotenoid production on Pseudomonas putida KT2440
The plasmids containing the genes for production of lycopene, β-carotene, and zeaxanthin were brought into E. coli S17-1, and the transformants mated with Pseudomonas putida KT2440. With all three plasmids pJOE5573.3, pJOE5607.5, and pJH14, the transfer efficiency dropped about 105-fold compared to the control vector pJeM1. The few obtained transconjugants exhibited heterogeneous phenotypes in terms of color and colony size. All of them lost the plasmids within a few generations when their maintenance was not selected with kanamycin. Similar results were obtained in mating experiments with other pseudomonads, like Pseudomonas stutzeri, Pseudomonas fluorescens, and Pseudomonas oleovorans (data not shown). The plasmids of transconjugants that were obtained by mating experiments or by electroporation were isolated and subjected to restriction digest analysis. Among the tested plasmids, several were found to be smaller or bigger than the original plasmid. The decrease in size was caused by gene deletions which were all located in the carotenoid biosynthesis gene cluster. Plasmids with increased size were found to contain IS elements (e. g., ISPpu15). The IS elements had either integrated into the carotenoid biosynthesis gene cluster or even into the rep gene of the plasmid which mediates plasmid replication and copy number control. Judging from the extracted DNA amount, the plasmid with the IS element in its rep gene had a dramatically reduced copy number.
In principle, there were two explanations for these findings: (1) the carotenoids are toxic to Pseudomonas putida and (2) one of the enzymes encoded by the plasmids produces a toxic compound from a Pseudomonas putida specific metabolite or depletes an essential metabolite by converting it into another useless substance. To identify the gene or intermediate which causes this problem, the mating experiments were repeated with various plasmids carrying just the idi gene (pJOE5473.2) up to the plasmid pJOE5573.3 with all genes necessary for lycopene production. The experiments were carried out under standard conditions, and as a control, mating was also done into E. coli JM109 for comparison. The results are shown in Table 3. The transfer efficiency of a particular plasmid from E. coli S17-1 into E. coli JM109, Pseudomonas putida wild type, or mutant D7L3 is given as the percentage of recovered transconjugants in proportion to the total number of recipient cells.
Whereas the plasmid transfer between E. coli strains was unaffected by the genes inserted into pJeM1, there was strong decrease of transconjugants in the mating between E. coli and Pseudomonas putida with the increasing number of genes in the vector. The most dramatic reduction of plasmid mobilization (about 50,000-fold) was seen from plasmid pJH13 for the synthesis of phytoene to pJOE5573.3 for synthesis of lycopene. The difference between the two plasmids is the additional gene crtI for desaturase in pJOE5573.3. To see if the lycopene or may be the desaturase activity causes the problem in Pseudomonas putida, the plasmid pJOE5901.2 was constructed which carries the crtI gene like pJOE5573.3 but lacks the crtB gene. The plasmid was transferred to Pseudomonas putida at about the same frequency as pJH13 for phytoene production which clearly demonstrates that lycopene is the carotenoid which is toxic to the cells.
Isolation of Pseudomonas putida transposon mutants which tolerate carotenoid production
A transposon mutant library of Pseudomonas putida KT2440 was screened for high transfer efficiency of pJOE5573.3 from E. coli S17-1 into the mutants. Four colonies were obtained which produced lycopene in a stable way. The strains were named C7L, D7L3, D8L1, and E3L2. All of them were tested again for transfer frequencies of the plasmid pJOE5573.3 from E. coli S17-1 to mutant, and all of them had an at least 100-fold higher transfer efficiency compared to Pseudomonas putida wild type. A detailed analysis was done for the mutant D7L3, and the data are given in Table 3. From all four mutants, chromosomal DNA was isolated, cut with various enzymes, and the DNA ligated under conditions which favored religation of the fragments to circles. By transformation of E. coli JM109 and selection of tetracycline resistance, the mini-transposons were recovered as autonomously replication plasmids which contained the flanking regions of the Pseudomonas putida integration sites. DNA sequencing showed that the transposons were inserted in four different genes, in the mutant D7L3 in a major facilitator family transporter gene, GeneID 1044254, in the mutant C7L in pepA, encoding an aminopeptidase, GeneID 1044903, in the mutant D8L1 in dsbD encoding a thiol disulphide interchange protein, GeneID 1042590, and in the mutant E3L2 in a gene for sensory box protein, GeneID 1046331. Obviously, there are a series of targets allowing tolerance of lycopene production in Pseudomonas putida. In the following experiments, Pseudomonas putida D7L3 mutant as well as E. coli JM109 were investigated for zeaxanthin biosynthesis.
Exclusion of the presence of a β-carotene hydroxylase in Pseudomonas putida KT2440
There is a β-carotene hydroxylase gene annotated in the genome of Pseudomonas putida KT2440 (crtZ, GeneID 1046895) but no genes leading to zeaxanthin precursors, so the expression of the β-carotene hydroxylase should be induced by β-carotene. In that case, β-cryptoxanthin and/or zeaxanthin should be found besides β-carotene. To investigate this, a culture of Pseudomonas putida KT2440 with the plasmid pJOE5607.5 leading to β-carotene was cultivated in TB medium. Gene expression was induced with l-rhamnose, and the culture was then incubated for 24 h at 25 °C and 160 min−1. The final HPLC analysis revealed only β-carotene but none of its hydroxylated products in the cell culture, which suggests that no natural β-carotene hydroxylase activity is present in Pseudomonas putida KT2440.
Time course of zeaxanthin production
In this work, both purity and yield of zeaxanthin produced in the mutant strain Pseudomonas putida D7L3 pJH14 in a mineral MEK medium and in two commonly used rich media (TB and LB) were investigated. For this purpose, 100 ml medium was inoculated from an overnight culture and cultivated at 30 °C before and at 25 °C after induction with l-rhamnose. For comparison, E. coli JM109 pJH14 was cultivated at 37 °C before and at 30 °C after induction. E. coli and Pseudomonas putida D7L3 cultures grew comparably (data not shown). The highest cell dry weight (CDW) of 7.3 g/l (OD600 of 15) was reached with Pseudomonas putida D7L3 in TB medium and the lowest one at about 0.7 g/l (OD600 of 2.5) in mineral medium. E. coli reached a maximum CDW of 9.2 g/l (OD600 of 23) in TB medium. The lowest CDW with this strain was observed in MEK medium (0.4 g/l, OD600 about 1). CDWs of about 3.2 g/l (OD600 of about 8) were observed in LB medium for both E. coli and Pseudomonas putida.
In our experiments, the concentrations of both carotene precursors phytoene and lycopene were below the detection limit, which means less than 0.5 μg/ml. In contrast, β-carotene, β-cryptoxanthin, and zeaxanthin could easily be detected in all Pseudomonas putida D7L3 samples that were analyzed. Less than 0.1 mg of β-carotene per gram CDW was found in TB medium; in LB and MEK medium, only traces of the substance could be detected. β-Cryptoxanthin was detected in all samples at yields of up to 0.6 mg/g CDW. The amount of zeaxanthin increased continuously upon cultivation of Pseudomonas putida D7L3 within 48 h. The highest yield of zeaxanthin (7.0 mg/g CDW) was obtained in TB medium (Fig. 3), followed by mineral medium MEK supplemented with glucose (2.5 mg/g CDW) and LB medium (1.3 mg/g CDW). The volumetric yield was highest in TB medium (51.3 mg/l) followed by LB medium (2.2 mg/l) and MEK medium (1.7 mg/l). The difference between the yield per CDW and volumetric productivities in LB and MEK medium is easily explained by differences in cell masses. Higher cell productivity in TB compared to LB media can be explained by higher concentration of yeast extract and other nutritions in TB medium, which are important for Pseudomonas putida metabolism.
For recombinant E. coli, a maximum zeaxanthin yield of 2.4 mg/g CDW in TB medium, 1 mg/g CDW in LB medium, and 1.6 mg/g CDW in MEK medium was found (Table 4).
The ratio of zeaxanthin (reflecting its purity) in the mixture of the carotenoids produced during cultivation was different at different cultivation stages (Fig. 4). Both (Z)-isomers of zeaxanthin were considered as impurities as well. While in TB medium nearly 80% product purity could be achieved after 48 h, in LB medium a purity of approximately 55% and in MEK medium of 32% was obtained.
Optimization of zeaxanthin production
In order to increase productivity of microbial cells, in situ product extraction into a second non-polar layer was applied. The tested organic solvents were either harmful to the cells (like n-octanol, log p = 2.9) or did not extract the carotenoids (like heptane, log p = 4.4), and therefore did not improve production rate (data not shown). In the next set, the effect of hydrophobic substances such as choline, lecithin, Amberlite XAD-7, and oleic acid was tested. All substances were added at the beginning of cell cultivation. The cells taken from all cultures after 24 h and spread on agar plates after respective dilution were able to grow. Nevertheless, addition of 1% choline inhibited cell growth and so resulted in lower cell densities as compared to the control culture without any additives. Addition of 1% oleic acid, in contrast, significantly increased cell density. Lecithin and Amberlite XAD-7 had no effect on cell growth. With respect to zeaxanthin production, the best results were achieved with lecithin and oleic acid. Judging for the yellow color, Amberlite XAD-7 was able to bind zeaxanthin; however, the carotenoid could not be extracted from the matrix afterwards.
In the next experiment, 1%, 5%, and 10% of the most promising additives lecithin and oleic acid were tested. The samples were taken after 1 and 2 days of cultivation and investigated by HPLC. Concentration of zeaxanthin in recombinant cells increased with increasing lecithin concentrations. In the presence of 1% lecithin, a 3.3-fold higher volumetric yield could be reached, with 5% the volumetric yield was enhanced by a factor of 4.2 and with 10% by a factor of 10.4; however, in the latter case, the standard deviation was quite high. After 48 h the effect is less pronounced with the lower concentrations but was still manifested in the presence of 10% lecithin (4.7 times compared to the control on day 1). With oleic acid, the best result could be observed with 1%. The low product recovery from cell suspensions containing 5% and 10% oleic acid was caused by the formation of an insoluble fatty layer from which extraction was very difficult. In the presence of 1% oleic acid, the yield of zeaxanthin was increased by a factor of 2.3 on the first and 1.2 on the second day, respectively (Fig. 5). Since exact CDWs could not be estimated after adding the hydrophobic compounds, only volumetric yields are provided here.
Stereochemistry of zeaxanthin produced
The last step in the present study was to determine stereochemistry of the target product. Zeaxanthin can occur as three optical isomers, namely (3R,3′R)-zeaxanthin, (3 S,3′S)-zeaxanthin, and (3R,3′S; meso)-zeaxanthin. These enantiomers cannot be separated on a non-chiral column without derivatization. After derivatization with (+)-(S)-1-1(1-naphthyl)-ethyl isocyanate, the zeaxanthin carbamate ester could be separated. The HPLC analysis revealed three signals in an area ratio of 33:4:1. This observation suggests that the β-carotene hydroxylase from Pantoea ananatis has a preference for one stereoisomer. Pure commercially available (3R,3′R)-zeaxanthin carbamate was used as a reference substance and appeared at the same retention time as the main signal. Further investigations are required to identify the absolute configuration of the isomers.
Discussion
In this work, we cloned five zeaxanthin-producing genes from Pantoea ananatis (crtE, crtI, crtB, crtY, and crtZ) and three genes from the MEP pathway from E. coli (idi, ispA, and dxs) in the shuttle vector pJH14 (derivate of pBBR1MCS-2) under control of the l-rhamnose-inducible promoter and expressed them both in E. coli and Pseudomonas putida.
We showed that Pseudomonas putida KT2440 does not exhibit its own β-carotene hydroxylase activity under the conditions tested, although a putative β-carotene hydroxylase gene is annotated within the genome. This is an advantage for the stereoselective synthesis of zeaxanthin using a heterologous β-carotene hydroxylase with the desired specificity without limitation to the isomer formed by the organism’s hydroxylase. Furthermore, the synthesis of β-carotene and hydroxylated products with hydroxyl groups at other positions is also possible.
Pseudomonas putida wild type hardly tolerated plasmids for lycopene, β-carotene, and zeaxanthin production. Transfer efficiencies of plasmids containing different gene combinations for isoprenoid precursor, phytoene, or lycopene biosynthesis were investigated in mating experiments with E. coli S17-1. Dramatically reduced transfer efficiency could be observed exclusively when all genes for the production of lycopene were present on the plasmid. Therefore, we conclude that the product lycopene itself and not the activity of any enzyme is toxic to the cells. Four transposon mutants of Pseudomonas putida with higher tolerance of carotenoid synthesis could be isolated. Mating experiments were also done for the mutant D7L3, and the latter exhibited a strong increase in transfer efficiency for the lycopene plasmid (about 15,000-fold compared to the wild type). Analysis of the transposon integration sites of the four mutants proposed that there are various gene targets for the relief of lycopene toxicity.
Media composition had a strong effect on both product yield and purity. TB medium was found to be more appropriate for zeaxanthin production with E. coli and Pseudomonas putida. Using Pseudomonas putida D7L3, 7 mg zeaxanthin/g CDW with 80% purity was produced in TB medium. This amount is 8.5 times higher than published for E. coli (Nishizaki et al. 2007). The yield of zeaxanthin in our experiments is in the same range which was published for both lycopene and β-carotene in E. coli (Das et al. 2007). The high yields of 49.3 mg/g CDW for β-carotene (Yoon et al. 2007b) and 22 mg/g CDW for lycopene (Yoon et al. 2007a) were achieved in the strains where the mevalonate pathway was changed via metabolic engineering. Further, only a small pronounced accumulation of β-cryptoxanthin was observed. This fits with the results obtained by Choi et al. (Choi et al. 2006). They reported that the hydroxylase CrtZ did not demonstrate any substrate preference either for β-carotene or for one time hydroxylated β-cryptoxanthin during astaxanthin production in E. coli (Choi et al. 2006).
We observed a difference in the yields between E. coli and Pseudomonas putida. Especially in TB medium, Pseudomonas putida seems to be more suitable for carotenoid production than E. coli (Table 4). The difference of yields between E. coli and Pseudomonas putida may arise from the differences between their metabolisms in conjunction with the nutrient content of the TB medium. E. coli is facultative anaerobe, while Pseudomonas putida is obligate aerobe. TB is a very rich medium, and the strong nutrient excess might exceed the capacity of the oxidative metabolism. E. coli is known to produce acetic acid when this occurs (Han et al. 1992). Due to its inability to grow under anaerobic conditions, Pseudomonas putida does not produce acids in the presence of nutrient excess. For this reason, Pseudomonas putida might be able to convert more of the supplied nutrients into the product zeaxanthin, while E. coli converts a certain amount of them into acetic acid.
Further, we demonstrated that the addition of lecithin can increase volumetric yield of zeaxanthin by a factor of 10.4. In contrast to organic solvents, this additive is not harmful to cells.
A preliminary test showed that the β-carotene hydroxylase from Pantoea ananatis has a preference or is even selective for one isomer. HPLC analysis carried out after derivatization of formed zeaxanthin revealed three signals: one major and two minor peaks. A comparison with authentic (3R,3′R)-zeaxanthin carbamate showed that the major peak had the same retention time as the reference. For the exact identification of the minor signals, further investigations are required. The results from the literature (Aasen et al. 1972; Bartlett et al. 1969; De Ville et al. 1969; Maoka et al. 1986) also let assume that the (3R,3′R)-form is a preferred form. Maoka et al. found that (3R,3′R)-zeaxanthin is either the main or even the only isomer in 30 of 36 investigated zeaxanthin-producing species (Maoka et al. 1986).
In summary, we demonstrated that the production of zeaxanthin is possible in recombinant Pseudomonas putida D7L3 and that solid additives can increase the product yield.
References
Aasen AJ, Borch G, Liaaen-Jensen S (1972) Carotenoids of flexibacteria V. chirality of zeaxanthin from different natural sources. Acta Chem Scand 26:404–405
Bartlett L, Klyne W, Mose WP, Scopes PM, Galasko G, Mallams AK, Weedon BCL, Szabolcs J, Toth G (1969) Optical rotatory dispersion of carotenoids. J Chem Soc Perkin 1:2527–2544
Britton G, Liaaen-Jensen S, Pfander H (1995) Carotenoids today and challenges for the future. In: Britton G, Liaaen-Jensen S, Pfander H (eds) Carotenoids 1A: isolation and analysis, 1st edn. Birkhäuser Verlag Basel, Berlin, pp 13–26
BVL (2007) Zusatzstoffe zugelassen nach Richtlinie 70/524/EWG und Übergangsregelung Verordnung EG 1831/2003. Bundesamt für Verbraucherschutz und Lebensmittelsicherheit.
Choi SK, Matsuda S, Hoshino T, Peng X, Misawa N (2006) Characterization of bacterial beta-carotene 3, 3′-hydroxylases, CrtZ, and P450 in astaxanthin biosynthetic pathway and adonirubin production by gene combination in Escherichia coli. Appl Microbiol Biotechnol 72:1238–1246
Das A, Yoon SH, Lee SH, Kim JY, Oh DK, Kim SW (2007) An update on microbial carotenoid production: application of recent metabolic engineering tools. Appl Microbiol Biotechnol 77:505–512
De Ville TE, Hursthouse MB, Russell SW, Weedon BCL (1969) Absolute configuration of carotenoids. J Chem Soc Chem Commun 1311–1312
Dejonghe W, Boon N, Seghers D, Top EM, Verstraete W (2001) Bioaugmentation of soils by increasing microbial richness: missing links. Environ Microbiol 3:649–657
Eugster CH (1995) Chemical derivatization: microscale tests for the presence of common functional groups in carotenoids. In: Britton G, Liaaen-Jensen S, Pfander H (eds) Carotenoids 1A: isolation and analysis. Birkhäuser Verlag Basel, Berlin, pp 71–80
Gerster H (1993) Anticarcinogenic effect of common carotenoids. Int J Vitam Nutr Res 63:93–121
Han K, Lim HC, Hong J (1992) Acetic acid formation in Escherichia coli fermentation. Biotechnol Bioeng 39:663–671
Jeske M, Altenbuchner J (2010) The Escherichia coli rhamnose promoter rhaP (BAD) is in pseudomonas putida KT2440 independent of Crp-cAMP activation. Appl Microbiol Biotechnol 85:1923–1933
Kang MJ, Lee YM, Yoon SH, Kim JH, Ock SW, Jung KH, Shin YC, Keasling JD, Kim SW (2005) Identification of genes affecting lycopene accumulation in Escherichia coli using a shot-gun method. Biotechnol Bioeng 91:636–642
Karrer P, Eugster CH (1950) Synthese von carotinoiden II. Totalsynthese des beta-carotins I. Helv Chim Acta 33:1172–1174
Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, Peterson KM (1995) Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176
Lindsay S (1996) Einführung in die HPLC, 1st edn. Friedr. Viehweg & Sohn Braunschweig, Wiesbaden, pp 187–191
Maoka T, Arai A, Shimizu M, Matsuno T (1986) The 1st isolation of enantiomeric and meso-zeaxanthin in nature. Comp Biochem Physiol B Biochem Mol Biol 83:121–124
Matthews PD, Wurtzel ET (2000) Metabolic engineering of carotenoid accumulation in Escherichia coli by modulation of the isoprenoid precursor pool with expression of deoxyxylulose phosphate synthase. Appl Microbiol Biotechnol 53:396–400
Meyer K (2002) Farbenfrohe antioxidantien–carotinoide: bedeutung und technische synthesen. Chem unserer Zeit 36:178–192
Misawa N, Nakagawa M, Kobayashi K, Yamano S, Izawa Y, Nakamura K, Harashima K (1990) Elucidation of the Erwinia uredovora carotenoid biosynthetic-pathway by functional-analysis of gene-products expressed in Escherichia coli. J Bacteriol 172:6704–6712
Molina L, Ramos C, Duque E, Ronchel MC, Garcia JM, Wyke L, Ramos JL (2000) Survival of Pseudomonas putida KT2440 in soil and in the rhizosphere of plants under greenhouse and environmental conditions. Soil Biol Biochem 32:315–321
Nishino H, Murakoshi M, Ii T, Takemura M, Kuchide M, Kanazawa M, Mou XY, Wada S, Masuda M, Ohsaka Y, Yogosawa S, Satomi Y, Jinno K (2002) Carotenoids in cancer chemoprevention. Cancer Metastasis Rev 21:257–264
Nishizaki T, Tsuge K, Itaya M, Doi N, Yanagawa H (2007) Metabolic engineering of carotenoid biosynthesis in Escherichia coli by ordered gene assembly in Bacillus subtilis. Appl Environ Microbiol 73:1355–1361
Sajilata MG, Singhal RS, Kamat MY (2008) The carotenoid pigment zeaxanthin—a review. Compr Rev Food Sci Food Saf 7:29–49
Sambrook J, Fritsch EF, Maniatis T (2001) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Woodbury
Schiedt K, Mayer H, Vecchi M, Glinz E, Storebakken T (1988) Metabolism of carotenoids in salmonids. Part II. Distribution and absolute-configuration of idoxanthin in various organs and tissues of one Atlantic salmon (Salmo salar, L) fed with Astaxanthin. Helv Chim Acta 71:881–886
Schmid A, Dordick JS, Hauer B, Kiener A, Wubbolts M, Witholt B (2001) Industrial biocatalysis today and tomorrow. Nature 409:258–268
Simon R, Priefer U, Pühler A (1983) A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat Biotechnol 9:784–791
Snodderly DM (1995) Evidence for protection against age-related macular degeneration by carotenoids and antioxidant vitamins. Am J Clin Nutr 62:1448 S–1461 S
Stumpp T, Wilms B, Altenbuchner J (2000) Ein neues, L-Rhamnoseinduzierbares Expressionssystem für Escherichia coli. Biospektrum 1:33–36
Sun Z, Yao HY (2007) The influence of di-acetylation of the hydroxyl groups on the anti-tumor-proliferation activity of lutein and zeaxanthin. Asia Pac J Clin Nutr 16:447–452
Tsushima M, Maoka T, Katsuyama M, Kozuka M, Matsuno T, Tokuda H, Nishino H, Iwashima A (1995) Inhibitory effect of natural carotenoids on Epstein–Barr virus activation activity of a tumor promoter in Raji cells—a screening study for antitumor promoters. Biol Pharm Bull 18:227–233
Vaz B, Alvarez R, de Lera AR (2002) Synthesis of symmetrical carotenoids by a two-fold Stille reaction. J Org Chem 67:5040–5043
Vilchez S, Manzanera M, Ramos JL (2000) Control of expression of divergent Pseudomonas putida put promoters for proline catabolism. Appl Environ Microbiol 66:5221–5225
Walsh UF, Morrissey JP, O’Gara F (2001) Pseudomonas for biocontrol of phytopathogens: from functional genomics to commercial exploitation. Curr Opin Biotechnol 12:289–295
Wang CW, Oh MK, Liao JC (1999) Engineered isoprenoid pathway enhances astaxanthin production in Escherichia coli. Biotechnol Bioeng 62:235–241
Yoon SH, Kim JE, Lee SH, Park HM, Choi MS, Kim JY, Lee SH, Shin YC, Keasling JD, Kim SW (2007a) Engineering the lycopene synthetic pathway in E. coli by comparison of the carotenoid genes of Pantoea agglomerans and Pantoea ananatis. Appl Microbiol Biotechnol 74:131–139
Yoon SH, Park HM, Kim JE, Lee SH, Choi MS, Kim JY, Oh DK, Keasling JD, Kim SW (2007b) Increased beta-carotene production in recombinant Escherichia coli harboring an engineered isoprenoid precursor pathway with mevalonate addition. Biotechnol Prog 23:599–605
Yuan LZ, Rouviere PE, LaRossa RA, Suh W (2006) Chromosomal promoter replacement of the isoprenoid pathway for enhancing carotenoid production in E. coli. Metab Eng 8:79–90
Acknowledgment
The authors want to thank the Institute of Food Chemistry, Universität Hohenheim for the carotenoids and wolfberries. The authors further thank the Landesstiftung Baden-Württemberg for the financial support.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Beuttler, H., Hoffmann, J., Jeske, M. et al. Biosynthesis of zeaxanthin in recombinant Pseudomonas putida . Appl Microbiol Biotechnol 89, 1137–1147 (2011). https://doi.org/10.1007/s00253-010-2961-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-010-2961-0