Abstract
PCR-based analysis of Bacteroidales 16S rRNA genes has emerged as a promising tool to identify sources of fecal water pollution. In this study, three TaqMan real-time PCR assays (BacGeneral, BacHuman, and BacBovine) were developed and evaluated for their ability to quantitatively detect general (total), human-specific, and bovine-specific Bacteroidales 16S rRNA genetic markers. The detection sensitivity was determined to be 6.5 copies of 16S rRNA gene for the BacGeneral and BacHuman assays and 10 copies for the BacBovine assay. The assays were capable of detecting approximately one to two cells per PCR. When tested with 70 fecal samples from various sources (human, cattle, pig, deer, dog, cat, goose, gull, horse, and raccoon), the three assays positively identified the target markers in all samples without any false-negative results. The BacHuman and BacBovine assays exhibited false-positive reactions with non-target samples in a few cases. However, the level of the false-positive reactions was about 50 times smaller than that of the true-positive ones, and therefore, these cross-reactions were unlikely to cause misidentifications of the fecal pollution sources. Microbial source-tracking capability was tested at two freshwater streams of which water quality was influenced by human and cattle feces, respectively. The assays accurately detected the presence of the corresponding host-specific markers upon fecal pollution and the persistence of the markers in downstream areas. The assays are expected to reliably determine human and bovine fecal pollution sources in environmental water samples.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Waterborne pathogens cause human diseases by contaminating public water supplies, recreational water, and aquifers (Centers for Disease Control and Prevention 2008; Simpson et al. 2002). Their presence in environmental waters can also lead to economic losses through beach closure to aquaculture, fishing, and recreational water use (Dorfman et al. 2004; Rabinovici et al. 2004). Most of waterborne pathogens are known to reside in human and warm-blooded animal feces (Craun et al. 2004; Till et al. 2004) that are introduced to water bodies from various sources (Field et al. 2003; Seurinck et al. 2005). Management and remediation of fecal water pollution require the assessment of fecal pollutant loading from specific sources (Elshorbagy et al. 2005; Kildare et al. 2007; Seurinck et al. 2005). Consequently, the current waterborne disease problem stems in part from the absence of tools to reliably identify these fecal pollution sources (Bernhard and Field 2000b).
PCR-based detection of Bacteroidales 16S rRNA genes is gaining popularity as a fecal source identification tool because of its advantages over other microbial source-tracking (MST) methods. Firstly, the method directly detects diagnostic sequences (i.e., markers) and does not require culturing or maintenance of reference databases (Dick and Field 2004). Secondly, Bacteroidales markers can be relatively easily detected in environmental water samples because of their abundance in feces (e.g., Bacteroides spp. comprise 26% to 36% of human fecal flora) (Hold et al. 2002; Sghir et al. 2000). Thirdly, these markers are highly specific to the host species (Bernhard and Field 2000b; Dick et al. 2005). Fourthly, Bacteroidales detection can be associated with recent fecal pollution because they are not likely to survive for a long time in the environment due to their strict anaerobic physiology without endospore-forming capacity (Kreader 1998). Lastly, these markers are geographically conserved and found in fecal samples from various locations (Dick et al. 2005; Field et al. 2003; Layton et al. 2006; Mieszkin et al. 2009).
However, source-tracking capability of the Bacteroidales assays has not been successfully demonstrated in previous studies probably because host specificity was validated only qualitatively (i.e., based on presence or absence of the target markers in the reference fecal samples), despite the use of quantitative PCR for analysis (Kildare et al. 2007; Lamendella et al. 2009; Reischer et al. 2006). The source-tracking assays should be validated for their capability to determine fecal pollution contribution by sources over a wide range of Bacteroidales levels, which is a basis for establishing water quality management strategies (Elshorbagy et al. 2005; Simpson et al. 2002). In addition, the assays’ MST capability was rarely evaluated under clearly defined field conditions. For example, the field water samples used in previous studies appeared to contain fecal contaminants from multiple sources (i.e., mixed-source fecal pollution) (Mieszkin et al. 2009; Okabe and Shimazu 2007; Savichtcheva et al. 2007), which could have confounded the interpretation of assay outcomes and thus interfered with the validation of the assays’ source-tracking capability.
In the present study, we characterized the source of fecal water pollution using TaqMan real-time PCR assays based on host specificity of Bacteroidales. Specific aims included (a) the design of TaqMan real-time PCR assays for the quantitative detection of general (total), human-specific and bovine-specific Bacteroidales markers, (b) validation of the assays’ sensitivity and specificity using fecal samples from various host animals, and (c) evaluation of the assays for their MST capability using field water samples polluted by human or bovine feces.
Materials and methods
Sample collection
Genomic DNA of Bacteroides fragilis ATCC 25285D was purchased from American Type Culture Collection (Rockville, MD, USA).
Individual fecal samples were collected from farm animals (18 cows, 10 pigs, and one horse), domestic animals (four dogs and three cats), wildlife (three deer, 10 geese, four gulls, and one raccoon), and human sources (16 raw wastewater influents) at various locations in southern Ontario, Canada. The raw influent samples were obtained from municipal sewage treatment plants across Ontario and used as the source of human fecal materials. Cattle and hog feces were sampled from three cattle farms and a hog farm, respectively. Fecal samples of dogs, cats, and a horse were kindly donated by the owners. Wildlife fecal samples were collected from various locations, such as municipal parks, beaches, and wildlife farms in southern Ontario. Fecal samples were stored on ice during transport to the laboratory and immediately used for DNA extraction.
Water samples were collected from the Grand River and Duffins Creek, Ontario, Canada during October and November 2009. Water quality of the Grand River sampling sites was predominantly influenced by a municipal wastewater treatment plant, whereas the upstream water quality was affected by cattle farming. The Duffins Creek sampling sites were characterized by heavy cattle farming activities along the watershed without any significant point sources of human fecal pollutants. The Grand River water samples were collected on two different days at (a) where the wastewater effluent was discharged into the river (FI or fecal inflow), (b) about 15 m upstream of the effluent discharge point (US or upstream), and (c) about 60 m downstream of the effluent discharge point (DS or downstream). Sodium thiosulfate was added to the water samples immediately after collection to quench the activity of any chlorine residues in the effluent. The Duffins Creek water samples were collected on one day in November 2009 at (d) where cattle wastewater from adjacent farms was suspected to flow in (FI), (e) about 120 m upstream of the inflow point (US), and (f) about 150 m downstream of the inflow point (DS). Water samples were stored on ice during transport to the laboratory and immediately used for DNA extraction.
Sequence analysis and assay design
The following published 16S rRNA gene sequences of Bacteroidales strains were analyzed to determine their host specificity: AF233400, AF233401, AF233406∼AF233411 (Bernhard and Field 2000b), AY597127∼AY597131, AY597136∼AY597175, AY597198∼AY597206 (Layton et al. 2006), FJ596691∼FJ596695 (Lamendella et al. 2009), FJ221192∼FJ221194 (Jeter et al. 2009), and X83935. The Bacteroidales strains carrying these genes were originally isolated from various warm-blooded animal hosts (human, cattle, pig, dog, and gull). The DNA sequences were obtained from the NCBI GenBank Database (Benson et al. 2008) and aligned using ClustalX (Thompson et al. 1997). Consensus sequences and host-specific marker sequences were identified for the order Bacteroidales (general Bacteroidales marker), human-originated Bacteroidales (human-specific marker), and bovine-originated Bacteroidales (bovine-specific markers I and II) (Table 1). These marker sequences were used to design primers for novel real-time PCR assays on the order Bacteroidales (BacGeneral), cattle-specific Bacteroidales (BacBovine I and II), and human-specific Bacteroidales (BacHuman).
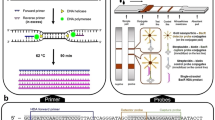
PCR primers and TaqMan probes were designed using computer software (Primer Express v 2.0, Applied Biosystems, Foster City, CA, USA). The TaqMan probes were labeled at the 5′-end with the reporter dye 6-carboxy-fluorescein (6-FAM) for BacHuman and BacBovine assays, and VIC™ (Applied Biosystems) for BacGeneral Assay. All probes were tagged at the 3′-end with a minor groove binder and the non-fluorescent quencher. Specificity of the oligonucleotide primers and probes was verified by performing a BLAST search of GenBank and a Probe Match of the Ribosomal RNA Database Project (Cole et al. 2009). Template genomic DNA, primers, and probes were added to a PCR mastermix to final concentrations of 176 nM for TaqMan probes and 500 nM for each primer. Real-time PCR assays were performed with a temperature profile of 50°C for 2 min and 95°C for 10 min, followed by 40 cycles at 95°C for 15 s and at 60°C for 60 s each.
Nucleic acid extraction
Bacteroidales cells were harvested by passing 100 ml of water samples through a 0.45-μm pore-size mixed cellulose esters membrane filter (Pall Canada Ltd., Ville St. Laurent, QC, Canada) under a partial vacuum. The filter was placed in a sterile disposable tube containing 1.5 ml of 5 M GITC lysis buffer (5 M guanidine isothiocyanate, 100 mM EDTA at pH 8.0, and 0.5% (wt/vol) sarkosyl) (Dick and Field 2004). Cells were separated from the filter by shaking the tube on a sample mixer for 1 h at 70 rpm, and the GITC lysate debris were removed by centrifugation for 10 min at 10,000×g. Genomic DNA was extracted using DNeasy Mini Spin Columns (QIAgen Canada Inc., Mississauga, ON, Canada) according to the manufacturer’s protocol. Fecal samples were homogenized in PBS solution by vortexing and subsequently mixed with GITC lysis buffer at a ratio of 1:2 (vol/vol). The GITC lysate was centrifuged for 10 min at 10,000×g, and DNA was extracted as aforementioned.
Assay calibration and quality control
Assay calibration was conducted using standard quantification plasmids containing the target 16S rRNA gene sequences. The total and host-specific gene markers were amplified with PCR primers Bac32F and Bac708R (Bernhard and Field 2000b) that flanked the target sequences of all three assays. The amplified DNA was cloned into a pTZ57R/T plasmid vector using InsTAclone PCR Cloning Kit (Fermentas, Burlington, ON, Canada), transformed into competent Escherichia coli DH5α host, and selected on LB agar plates containing 50 μg/ml ampicillin. Plasmids were screened for the presence of the target markers by colony PCR.
The standard quantification plasmids that were prepared in a series of 10-fold dilutions served as positive controls for each PCR run. Sample PCR assays were judged to be successful when the positive controls reproduced the known threshold cycles (Ct) along the dilution series. Negative controls, or no template controls, were set up in duplicate for each PCR run by adding TE buffer to the PCR mastermix in place of DNA template.
Fecal indicator bacteria
Viable E. coli cells were enumerated by the membrane filter technique followed by cell cultivation, which was a slight modification of the Standard Method for the Examination of Water and Wastewater, section 9222D (Eaton et al. 2005). A 10-ml aliquot of water samples was passed through a 0.45-μm pore-size membrane filter (Pall Canada). The membrane filters were then placed on mFC agar plates supplemented with 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid (BCIG). Culture plates were incubated at 44.5 ± 0.2°C for 24 h, and E. coli colonies were counted based on the emergence of blue-green color resulting from the hydrolysis of BCIG by β-glucuronidase.
Fecal streptococci were harvested by the membrane filter technique as aforementioned and cultured on mEnterococcus agar according to the Section 9230C of the Standard Methods for the Examination of Water and Wastewater. Culture plates were incubated at 35 ± 0.5°C for 48 h. Fecal streptococcal colonies were counted based on the appearance of red color arising from 2,3,5-triphenyl tetrazolium chloride reduction.
Results
Assay design
The BacGeneral assay was designed based on the consensus 16S rRNA gene sequences of 65 Bacteroidales strains, including 14 human- and 40 bovine-specific sequences (Table 2). The BacBovine assay I and II were derived from two distinct groups of cattle-specific 16S rRNA gene sequences. The primers and probe of the two assays showed substantial differences in their target DNA sequences (Table 1 and 2), and therefore, each assay was unlikely to cross-react with the other targets. Based on this observation, the two assays were combined in a multiplex PCR reaction (as a single “BacBovine assay”) throughout the study. The probe and primers of the BacHuman assay exhibited a perfect sequence homology with 16S rRNA genes of B. fragilis ATCC 25285 and 13 human-specific Bacteroidales strains.
Standard quantification curves and detection sensitivity
Four different standard plasmids, targeted by each of the four Bacteroidales assays, were created as aforementioned. Each standard plasmid was prepared in ten-fold serial dilutions (10−4 to 10−10) and quantified with the corresponding assay in triplicate. Since the target gene (16S rRNA gene) was inserted into the plasmid at a 1:1 ratio, the target copy number was estimated from the number of the standard plasmid DNA. When the measured Ct values were plotted against the logarithm of the target gene copy numbers, all four assays exhibited a strong inverse linear relationship (r 2 > 0.99 for all assays, P < 0.01; regression analysis) (Fig. 1). The result implies that these assays can be reliably applied to quantify the genetic markers within the quantification range of seven orders of magnitude. All four standard quantification curves fell virtually in a single line with very similar slope and Y-intercept values, indicating a comparable PCR efficiency for all assays. The overall detection limit of the assays was determined to be about 6.5 copies of the 16S rRNA gene for BacGeneral and BacHuman assays and 10 copies of the gene for BacBovine assay. Considering that Bacteroides spp. are known to contain four to seven copies of the 16S rRNA gene per cell (Lee et al. 2009), the detection limit was equivalent to about one to two cells per PCR.
PCR inhibition by fecal and environmental samples
The inhibitory effect of fecal and environmental samples on PCR efficiency was tested for each sample in two ways. First, fecal and environmental DNA extracts were spiked with excessive quantity of the standard plasmids (6.5 × 106 copies), and the DNA mixtures were analyzed with the real-time PCR assays. Significant deviations from the known Ct values (i.e., based on 95% confidence limits) were considered as evidence of inhibitory effects. Second, the assays were applied on the 10-fold serial diluents (no dilution, 10−1, 10−2, and 10−3) of fecal or environmental DNA. The Ct values were plotted against the dilution series to detect any deviations from the curve linearity, which was taken as a sign of PCR inhibition. The 10−1 diluents were found to be the least-diluted series with no signs of inhibition and thus selected for the quantitative estimation of the genetic markers.
Specificity of the assays
A total of 70 reference fecal samples were collected from various animal sources, including cattle, pig, horse, deer, dog, cat, gull, goose, raccoon, as well as from human sources (municipal wastewater influents). The reference fecal samples were used to determine specificity of the real-time PCR assays. The three assays positively identified the target DNA sequences (general, human-specific, and cattle-specific markers) in all samples without any false-negative reactions (Table 3). The BacHuman assay produced false-positive signals with fecal samples from pigs (three out of 10 samples), dogs (four out of four), and cats (three out of three), while the BacBovine assay yielded false-positive signal with fecal samples from deer (two out of three) and dogs (three out of four). However, the level of the false-positive reactions (or assays) was much smaller than that of the true-positive ones [only about 1.9 ± 2.1% (mean ± standard deviation) of the true-positive reactions] (Fig. 2), when the reaction level was calculated as the relative number of the human-specific (or bovine-specific) marker as a percentage of the general (total) marker. Based on this finding, we expressed host-specific marker levels of all field samples as a percentage of the general (total) markers to minimize the effect of false-positive reactions on the fecal source identification.
Comparison of the level of false-positive reactions with that of true-positive ones. The X- and Y-axes represent fecal samples from various animal hosts and the relative quantity of host-specific Bacteroidales markers as a percentage of the general Bacteroidales marker, respectively. The true-positive reactions (i.e., human feces analyzed with the BacHuman assay, or cattle feces analyzed with the BacBovine assay) exhibited far higher levels than those of the false-positive reactions (i.e., other animal feces reacted with BacHuman and BacBovine assays)
Field study
The MST capability of the BacHuman and BacBovine assays was evaluated in the Grand River and Duffins Creek, which are known for their water quality issues arising from human and bovine fecal pollution, respectively (personal communication with Drs. J. Thomas and T. Howell, Ontario Ministry of the Environment). The number of general Bacteroidales marker, which was estimated from two different sampling occasions at the Grand River, exhibited a drastic increase from about 11,000 ± 2,000 (arithmetic mean ± standard deviation) copy/100 ml at the 15 m upstream location (US) to about 1,200,000 ± 700,000 copy/100 ml at the sewage inflow point (FI) (Fig. 3a). The number decreased to 480,000 ± 290,000 copy/100 ml at the 60 m downstream point (DS). Similarly, the human-specific marker increased from approximately 400 ± 100 copy/100 ml at US to 540,000 ± 300,000 copy/100 ml at FI, and subsequently decreased to about 190,000 ± 120,000 copy/100 ml at DS. When expressed as a percentage of the general marker, the human-specific marker increased from 4% to 43% and remained relatively stable (40%) through the 60-m downstream site (Fig. 3b). The bovine-specific markers remained below 1% of the general marker throughout all three sampling locations.
Host-specific Bacteroidales markers in the Grand River water samples. Marker quantity was expressed in number (a) or as a percentage of the general Bacteroidales marker (b). Samples were collected at where the Waterloo municipal wastewater treatment plant effluent was discharged into the river (fecal inflow, FI), at 15 m upstream of the effluent discharge point (upstream, US), and at 60 m downstream of the effluent discharge point (downstream, DS). Samples were collected on two different days, and each sample was analyzed in triplicate. Bars and error bars represent mean ± SD (n = 6)
The bovine-specific Bacteroidales markers were also enumerated using the samples collected from the Duffins Creek. The general markers increased from about 1,100 ± 100 copy/100 ml at about 120 m upstream location (US) to 140,000 ± 10,000 copy/100 ml at the cattle wastewater inflow point (FI) (Fig. 4a). As observed in the Grand River, the general marker number decreased but remained relatively high (ca. 80,000 ± 4,000 copy/100 ml) through 150 m downstream location (DS). The bovine-specific markers showed a similar increase from an undetectable level at US to about 52,000 ± 3,000 copy/100 ml at FI, and then decreased to 23,000 ± 3,000 copy/100 ml at DS. When expressed as a percentage of the general Bacteroidales marker, bovine-specific Bacteroidales markers increased from 0% at US to 37% at FI, and remained at a relatively high level (30%) through DS. The human-specific Bacteroidales numbers remained below 1% level throughout all three sampling points (Fig. 4b).
Host-specific Bacteroidales markers in the Duffins Creek water samples. Marker quantity was expressed in numbers (a) or as a percentage of the general Bacteroidales marker (b). Samples were collected at where cattle wastewater from adjacent farms was suspected to flow in (fecal inflow, FI), at about 120 m upstream of the inflow point (upstream, US), and at about 150 m downstream of the inflow point (downstream, DS). Bars and error bars represent mean ± SD (n = 3)
Bacteroidales and fecal indicator bacteria
The conventional fecal pollution indicators, E. coli and fecal streptococci, were enumerated from the same stream water samples used in the Bacteroidales assays. The total Bacteroidales cell numbers were estimated by dividing the number of the general Bacteroidales marker (or the 16S rRNA gene copy) by 5.5, which is the median number of 16S rRNA gene copies per cell for many Bacteroides spp. (Lee et al. 2009). When the total Bacteroidales numbers from all sampling locations of the Grand River and Duffins Creek were pooled and plotted against the number of fecal indicator bacteria (FIB), the Bacteroidales numbers exhibited poor to low correlations with the number of E. coli (r 2 = 0.22) and fecal streptococci (r 2 = 0.40), respectively (Fig. 5).
Correlation of the total Bacteroidales population with two fecal indicator bacteria, E. coli and fecal streptococci. The total Bacteroidales numbers from all field sampling locations of the Grand River and Duffins Creek were pooled and plotted against the number of fecal indicators. The Bacteroidales numbers exhibited poor to low correlation with the number of E. coli (r 2 = 0.22) and fecal streptococci (r 2 = 0.40), respectively. n = 16
Discussion
Three TaqMan real-time PCR assays (BacGeneral, BacHuman, and BacBovine) were developed and evaluated for their ability to quantitatively detect the general (total), human-specific, and bovine-specific Bacteroidales 16S rRNA genetic markers. The present human- and bovine-specific assay showed generally a high sensitivity and specificity (see below for details) probably as the result of an extensive sequence search and alignment using the latest gene sequence databases.
Geographic stability
To function as an effective MST tool, Bacteroidales genetic marker sequences should be conserved throughout various geographic locations (i.e., geographic stability) (U.S. Environmental Protection Agency 2005). In the present study, the three assays were designed entirely based on the 16S rRNA gene sequences reported previously (Bernhard and Field 2000b; Jeter et al. 2009; Lamendella et al. 2009; Layton et al. 2006). These 16S rRNA gene sequences provided a sound detection target for Bacteroidales populations found in Ontario, Canada, despite the fact that they were obtained from human and cattle fecal Bacteroidales collected elsewhere in North America (Oregon, Tennessee, Texas, and Pennsylvania, USA). Therefore, these marker sequences appeared to be well conserved across North America. Similarly, pig-specific Bacteroidales markers are conserved in many geographically distant locations, including France, Japan, and the USA (Dick et al. 2005; Mieszkin et al. 2009; Okabe and Shimazu 2007).
Host specificity
Source-tracking tools need to possess a high degree of host specificity to accurately identify the pollution sources (U.S. Environmental Protection Agency 2005). The present BacHuman and BacBovine assays did not cross-react with each other’s target, whereas non-specific reactions with other animal feces were observed in several cases. Human-specific Bacteroidales assays are well known for their cross-reactivity with pigs and domestic animals, especially dogs (Kildare et al. 2007; Layton et al. 2006; Seurinck et al. 2005). The 16S RNA gene sequences of human- and cattle-originated Bacteroidales are closely related to those of pigs and other ruminants, respectively, probably because of similar diet and digestive systems (Bernhard and Field 2000a; Dick et al. 2005; Layton et al. 2006; Mieszkin et al. 2009). Cross-reaction of the human-specific assay with dog feces was explained by a frequent horizontal transmission of fecal bacteria between the two groups due to physical proximity (Dick et al. 2005). Bovine-specific assays also cross-reacted with dog fecal samples in a previous study (Layton et al. 2006).
The application of TaqMan real-time PCR to an MST study allows for enumeration of the host-specific Bacteroidales markers as well as determination of their presence. However, the level of the false-positive reactions (non-specific assays) has been rarely analyzed against that of the true-positive ones (host-specific assays) in previous studies. In the present study, the level of the false-positive reactions was about 50 times smaller than that of the true-positive ones, and therefore, these false-positive reactions were unlikely to cause misidentifications of the human- and bovine-specific gene sequences. It has been known that the total Bacteroidales population in a host is composed of multiple subpopulations (Kildare et al. 2007; Layton et al. 2006). Although the majority of the subpopulations are expected to contain genetic characteristics unique to the host, marginal subpopulations may share their genetic markers with other hosts, thus enabling the low level cross-reactions. The cattle-specific assay exhibited the highest false-positive reaction level (about 7% of the true-positive reaction level) with deer, another ruminant.
Field MST capacity
The present Bacteroidales assays were tested for source-identifying capability using water samples from two local freshwater streams that were polluted by human and cattle feces, respectively. For each stream, we have purposely selected a section of the stream receiving fecal pollutants from a single dominant source. During the field study, the MST capability of the assays was verified from three different aspects. Firstly, the assays were capable of accurately ascertaining the inflow of fecal pollutants into relatively clean water. For instance, the relative fraction of the human-specific marker increased considerably from 4% at US to 43% at FI in the Grand River. Similarly, the fraction of bovine-specific Bacteroidales markers increased from 0% to 37% within the corresponding section of the Duffins Creek. Secondly, the relative quantity of Bacteroidales markers remained generally stable towards the downstream area. For example, the absolute number of the human-specific and bovine-specific markers decreased substantially between FI and DS in the Grand River and the Duffins Creek, respectively (see “Results” for detail). At the same sampling locations, however, the relative quantity of the human-specific and bovine-specific markers decreased in smaller scales from 43% to 40% and from 37% to 30%, respectively, of the general Bacteroidales marker. The characteristics may play an important role in a field MST study because we may still be able to determine the fecal pollution source in a water body where Bacteroidales markers exist in relatively small numbers due to dilution and loss during transport along the water stream (e.g., the present downstream areas). Lastly, these assays were capable of specifically differentiating the target Bacteroidales marker from the background signals, which might help to accurately determine the fecal pollutant contribution by sources. The background markers (i.e., the bovine-specific markers in the Grand River and the human-specific marker in the Duffins Creek) remained at very low levels (0% to 1% of the general Bacteroidales marker) throughout the three sampling points due to the absence, or marginal presence, of corresponding pollution sources. In contrast, the target markers (i.e., the human-specific marker in the Grand River and the bovine-specific markers in the Duffins Creek) underwent drastic changes in number because of fecal pollutant inputs.
In the present study, the mean proportions of human- and bovine-specific Bacteroidales markers in feces (44% and 45%, respectively) may be considered as the “baseline ratio” that represents the maximum relative quantity of the host-specific Bacteroidales marker in environmental samples (Kildare et al. 2007). In the Grand River samples, the proportions of human-specific marker (i.e., 40% to 43% of the general marker) were similar to the baseline ratio, and this suggests that water pollution at FI and DS sites were mostly accounted for by human feces. Similarly, the Duffins Creek water samples were found to be polluted mainly by cattle feces because the cattle-specific markers (30% to 37% of the general marker) also reached close to the maximum level.
Strategies for MST capability testing
Many previous studies fell short of successfully demonstrating the field MST capability of Bacteroidales-based assays probably because of the following two reasons. Firstly, they attempted to determine fecal pollution sources based only on the presence of the host-specific markers rather than fully taking advantage of the quantitative outcome (Lamendella et al. 2009; Walters et al. 2007). Host specificity of a Bacteroidales assay can be determined quantitatively by reporting the relative quantity of the host-specific markers. These data can provide reference information for estimating the relative fecal pollutant contribution by individual sources, or for the quantitative identification of fecal sources. The present assays were validated for their quantitative host specificity and utilized to identify fecal sources in the field. Secondly, despite the application of quantitative PCR tools, many previous assays were not capable of unequivocally attributing fecal pollution to specific hosts in the field tests (Kildare et al. 2007; Mieszkin et al. 2009; Okabe and Shimazu 2007; Savichtcheva et al. 2007). This might have resulted from the fact that the assays were not evaluated under clearly defined conditions in field tests, which included applying the assays on water bodies with multiple fecal pollutant sources. Although this condition may be environmentally relevant, it can also make it difficult to interpret the source-tracking results, especially if the method is still at the development stage. To minimize the confounding effect of mixed-source fecal pollution, the present assays were tested on a section of a freshwater stream where the water quality was affected by a single, dominant fecal source (i.e., wastewater effluent in the Grand River, and cattle farm wastewater inflow in the Duffins Creek). In addition, the MST capability of the assays was demonstrated under environmentally relevant conditions through a series of tests at the downstream areas where the target genetic markers were expected to be present in diluted forms over a relatively wide area.
We should note that there is a potential limitation in the current quantitative approach. For many environmental samples, only a fraction of Bacteroidales strains can be attributed to a specific host(s) and a larger proportion of Bacteroidales strains remain as originating from “unknown hosts.” This knowledge gap may limit our ability to clearly identify a major fecal pollution source. Additional assays are being developed to increase the number of strains that can be positively identified.
Bacteroidales as fecal indicator bacteria
The present study found only poor to low correlations between the number of FIB and the host-specific Bacteroidales markers. Similarly, previous studies also reported no correlation (r 2 = 0.11 to 0.15) between the number of FIB and Bacteroidales markers when tested with river waters (Mieszkin et al. 2009; Okabe and Shimazu 2007). Fecal indicator bacteria have been known to behave differently from Bacteroidales 16S rRNA markers in environmental waters. For example, fecal enterococcal levels vary with host (human or cattle) whereas host-specific Bacteroidales levels were similar in both hosts (Walters and Field 2009). In addition, the environmental persistence of fecal indicator bacteria and Bacteroidales markers are affected differently by factors such as light, salinity, and temperature (Okabe and Shimazu 2007; Walters and Field 2009). The discrepancy could have also resulted in part from the application of two different detection techniques (i.e., plate counting for E. coli and TaqMan PCR for Bacteroidales). Bacteroidales 16S rRNA genetic markers are known to be more accurate than conventional FIB in terms of detecting recent fecal pollution and indicating the presence of waterborne pathogens, including E. coli O157:H7, Salmonella spp., and Campylobacter spp. (Fremaux et al. 2009; Savichtcheva et al. 2007; Walters et al. 2007).
References
Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL (2008) GenBank. Nucl Acids Res 36:D25–D30
Bernhard AE, Field KG (2000a) A PCR assay to discriminate human and ruminant feces on the basis of host differences in Bacteroides–Prevotella genes encoding 16S rRNA. Appl Environ Microbiol 66:4571–4574
Bernhard AE, Field KG (2000b) Identification of nonpoint sources of fecal pollution in coastal waters by using host-specific 16S ribosomal DNA genetic markers from fecal anaerobes. Appl Environ Microbiol 66:1587–1594
Centers for Disease Control and Prevention (2008) Surveillance for waterborne disease and outbreaks—United States 2005–2006. Centers for Disease Control and Prevention, MMWR 57(SS-9)
Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM (2009) The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucl Acids Res 37:D141–D145
Craun GF, Calderon RL, Craun MF (2004) Waterborne outbreaks caused by zoonotic pathogens in the USA. In: Cotruvo JA, Dufour A, Rees G, Bartman J, Carr R, Cliver DO, Craun GF, Fayer R, Ganon VPJ (eds) Waterborne zoonoses—identification, causes, and control. World Health Organization, IWA, London, pp 120–135
Dick LK, Field KG (2004) Rapid estimation of numbers of fecal Bacteroidetes by use of a quantitative PCR assay for 16S rRNA genes. Appl Environ Microbiol 70:5695–5697
Dick LK, Bernhard AE, Brodeur TJ, Santo Domingo JW, Simpson JM, Walters SP, Field KG (2005) Host distributions of uncultivated fecal Bacteroidales bacteria reveal genetic markers for fecal source identification. Appl Environ Microbiol 71:3184–3191
Dorfman M, Stoner N, Merkel M (2004) Swimming in sewage. Natural Resources Defense Council and Environmental Integrity Project, New York
Eaton AD, Clesceri LS, Rice EW, Greenberg AE, Franson MAH (2005) Standard methods for examination of water and wastewater, 21st edn. American Public Health Association, Washington
Elshorbagy A, Teegavarapu RSV, Ormsbee L (2005) Total maximum daily load (TMDL) approach to surface water quality management: concepts, issues, and applications. Can J Civ Eng 32:442–448
Field KG, Bernhard AE, Brodeur TJ (2003) Molecular approaches to microbiological monitoring: fecal source detection. Environ Monit Assess 81:313–326
Fremaux B, Gritzfeld J, Boa T, Yost CK (2009) Evaluation of host-specific Bacteroidales 16S rRNA gene markers as a complementary tool for detecting fecal pollution in a prarie watershed. Water Res 43:4838–4849
Hold G, Pryde SE, Russell VJ, Furrie E, Flint HJ (2002) Assessment of microbial diversity in human colonic samples by 16S rDNA sequence analysis. FEMS Microbiol Ecol 39:33–39
Jeter SN, McDermot CM, Bower PA, Kinzelman JL, Bootsma MJ, Goetz GW, McLellan SL (2009) Bacteroidales diversity in ring-billed gulls (Laurus delawarensis) residing at Lake Michigan beaches. Appl Environ Microbiol 75:1525–1533
Kildare BJ, Leutenegger CM, McSwain BS, Bambic DG, Rajal VB, Wuertz S (2007) 16S rRNA-based assays for quantitative detection of universal, human-, cow-, and dog-specific fecal Bacteroidales: A Bayesian approach. Water Res 41:3701–3715
Kreader CA (1998) Persistence of PCR-detectable Bacteroides distasonis from human feces in river water. Appl Environ Microbiol 64:4103–4105
Lamendella R, Santo Domingo JW, Yannarell AC, Ghosh S, Di Giovanni G, Mackie RI, Oerther DB (2009) Evaluation of swine-specific PCR assays used for fecal source tracking and analysis of molecular diversity of swine-specific “Bacteroidales” populations. Appl Environ Microbiol 75:5787–5796
Layton A, McKay L, Williams D, Garrett V, Gentry R, Sayler G (2006) Development of Bacteroides 16S rRNA gene TaqMan-based real-time PCR assays for estimation of total, human, and bovine fecal pollution in water. Appl Environ Microbiol 72:4214–4224
Lee ZM-P, Bussema C III, Schmidt TM (2009) rrnDB: documenting the number of rRNA and tRNA genes in bacteria and archaea. Nucl Acids Res 37:D489–D493
Mieszkin S, Furet JP, Corthier G, Gourmelon M (2009) Estimation of pig fecal contamination in a river catchment by real-time PCR using two pig-specific Bacteroidales 16S rRNA genetic markers. Appl Environ Microbiol 75:3045–3054
Okabe S, Shimazu Y (2007) Persistence of host-specific Bacteroides–Prevotella 16S rRNA genetic markers in environmental waters: effects of temperature and salinity. Appl Microbiol Biotechnol 76:935–944
Rabinovici SJM, Bernknopf RL, Wein AM, Coursey DL, Whitman RL (2004) Economic and health risk trade-offs of swim closures at a Lake Michigan beach. Environ Sci Technol 38:2737–2745
Reischer GH, Kasper DC, Steinborn R, Mach RL, Farnleitner AH (2006) Quantitative PCR method for sensitive detection of ruminant fecal pollution in freshwater and evaluation of this method in alpine karstic regions. Appl Environ Microbiol 72:5610–5614
Savichtcheva O, Okayama N, Okabe S (2007) Relationships between Bacteroides 16S rRNA genetic markers and presence of bacterial enteric pathogens and conventional fecal indicators. Water Res 41:3615–3628
Seurinck S, Defoirdt T, Verstraete W, Siciliano SD (2005) Detection and quantification of the human-specific HF183 Bacteroides 16S rRNA genetic marker with real-time PCR for assessment of human faecal pollution in freshwater. Environ Microbiol 7:249–259
Sghir A, Gramet G, Suau A, Rochet V, Pochart P, Dore J (2000) Quantification of bacterial groups within human fecal flora by oligonucleotide probe hybridization. Appl Environ Microbiol 66:2263–2266
Simpson JM, Santo Domingo JW, Reasoner DJ (2002) Microbial source tracking: state of the science. Environ Sci Technol 36:5279–5288
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Till D, Field K, Dufour A (2004) Managing risk of waterborne zoonotic disease through water quality surveillance. In: Cotruvo JA, Dufour A, Rees G, Bartman J, Carr R, Cliver DO, Craun GF, Fayer R, Ganon VPJ (eds) Waterborne zoonoses—identification, causes, and control. World Health Organization, IWA, London, pp 338–348
U.S. Environmental Protection Agency (2005) Microbial source tracking document. U.S. Environmental Protection Agency, Cincinnati
Walters SP, Field KG (2009) Survival and persistence of human and ruminant-specific faecal Bacteroidales in freshwater microcosms. Environ Microbiol 11:1410–1421
Walters SP, Gannon VPJ, Field KG (2007) Detection of Bacteroidales fecal indicators and the zoonotic pathogens E. coli O157:H7, Salmonella, and Campylobacter in river water. Environ Sci Technol 41:1856–1862
Acknowledgements
This research was supported by the Ontario Ministry of the Environment Best in Science program. We sincerely thank Drs. J. Thomas and T. Howell (Ontario Ministry of the Environment) as well as Mr. T. Rance (Toronto and Region Conservation Authority) and Mr. P. Wu (Ontario Ministry of Agriculture, Food and Rural Affairs) for their field sampling work in support of our research.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Lee, DY., Weir, S.C., Lee, H. et al. Quantitative identification of fecal water pollution sources by TaqMan real-time PCR assays using Bacteroidales 16S rRNA genetic markers. Appl Microbiol Biotechnol 88, 1373–1383 (2010). https://doi.org/10.1007/s00253-010-2880-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-010-2880-0