Abstract
Recombinant tilapia (Oreochromis mossambicus) fish metallothionein (MT) was used as a surface biosorbent for mercury removal in Escherichia coli. Fish MT conferred better resistance than did mouse or human MT. When tilapia MT (tMT) was fused with an outer-membrane protein, outer membrane protein C (OmpC), the membrane-targeted fusion protein, OmpC–tMT, gave enhanced resistance compared with cytoplasmic tMT expressed in the same host cell. The cytoplasmically expressed tMT showed high mercury adsorption (4.3 ± 0.4 mg/g cell dry weight). The cell surface that expressed E. coli showed about 25% higher adsorption ability (5.6 ± 0.4 mg/g) than the cells expressing cytoplasmic MT, attaining almost twice the level of adsorption of the control plasmid (3.0 ± 0.4 mg/g). As MTs are also known for their ability to scavenge hydroxyl-free radicals, it was also shown that tMT exhibited better radical-scavenging activities than glutathione. These results suggest that fish MT has potential for the development of a bioremediation system for mercury removal that protects the harboring E. coli host by free-radical scavenging.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Metallothionein (MTs) are widely distributed among living organisms and are fairly well conserved in mammals, plants, and fungi. They are heat-stable, low-molecular-weight, cysteine-rich intracellular proteins with high affinity and selectivity for maintaining homeostasis of essential heavy metals, such as Cu2+ and Zn2+, and detoxification of toxic metal cations, such as Cd2+ and Hg2+ (Coyle et al. 2002; Park et al. 2001; Vallee 1995). In addition, MTs possess a defense function against oxidative stress with scavenging activity for reactive oxygen species on the cellular level (Sato and Kondoh 2002). In aqueous environments, tilapia fish, Oreochromis mossambicus, has been considered a biomarker for the contamination level of aqueous pollution based on its resistance to contaminants (Chan 1995; Wong et al. 2000). The ability of tilapia to survive in higher contamination levels prompted us to postulate that tilapia MT (tMT) may serve as a good candidate for both metal-absorbent and reactive-oxygen radicals scavenging.
On the other hand, capacities of bacteria for remediation of heavy metal pollution have been studied through their usage in bioadsorption (Belliveau and Trevors 1989; Volesky and Holan 1995). Advantages of these biomaterials include high specificity and potential for genetic improvement (Valls and de Lorenzo 2002). Various metal-binding peptides and proteins, such as glutathione (GSH), GSH-related phytochelatins, cysteine-rich MTs and synthetic phytochelatins, have been used to enhance the bioaccumulation of heavy metals (Chen and Wilson 1997a; Mejare and Bulow 2001; Volesky and Holan 1995). However, intracellular bioaccumulation of mercury has been problematic because of limited metal uptake by microbe-based biosorbent (Chen and Wilson 1997a). Alternative approaches have been studied for overcoming the limitation of uptake by either co-expressing an Hg2+ transport system (Chen and Wilson 1997b). It has been showed that cell-surface display techniques circumvent the problems associated with intracellular limited accumulation, endowing intact cells with new functionalities that have a vast sphere of new applications (Bae et al. 2001; Saleem et al. 2008; Ueda and Tanaka 2000).
Among many kinds of heavy metals, mercury remains in use in a number of ways, and its toxicity ranking is on top level. It plays no essential biological function (Allen 1994), while it poses a threat to human health via bioaccumulation in the food chain and causes Minamata disease (Harada 1995). In this study, the tMT was fused to the Escherichia coli outer membrane protein C (OmpC) (Jeanteur et al. 1991; Xu and Lee 1999) and the fusion protein was expressed on the surface of E. coli to demonstrate the possibility of its usage for scavenging of radicals in extracellular milieu and increasing the removal ability for mercury contamination.
Materials and methods
Construction of plasmids for expression of recombinant proteins in cytosol
Mammalian MT cDNAs were cloned from the pMAL-p2X-MT1A and pMBP-TEV-MT1 plasmids (Kao et al. 2006), which encoded human MT1A (hMT) and mouse MT1 (mMT), respectively. The tMT cDNA was cloned from total cDNA that had been reverse-transcribed from total RNA of tilapia liver. The cDNAs were amplified with the following specific primer pairs (for human mt1A: Hmt-F and Hmt-R; for mouse mt1: Mmt-F and Mmt-R; for tilapia mt: Tmt-F and Tmt-R) (Table 1). The DNA fragments containing the respective mt cDNAs were amplified, flanked by the BglII and SacI restriction sites at the ends, respectively. The resulting 0.2-kbp DNA fragments were cut by BglII and SacI and then ligated to the same sites of digested pET-30a (Novagen, Darmstadt, Germany). The plasmids were designated as pET-Hmt (human mt1A), pET-Mmt (mouse mt1), and pET-Tmt (tilapia mt), respectively.
Construction of plasmids for expression of membrane-targeted fusion proteins
The ompC gene was cloned from total DNA of E. coli K19 with primer pairs OmpC-F and OmpC-R (Table 1) (Jeanteur et al. 1991). The OmpC-F primer contains an NdeI site and OmpC-R primer includes a BamHI site. The DNA fragment encoding OmpC was cloned into the NdeI and BamHI sites of pET-30a to form pET-OmpC. The fragments, containing three species of the mt gene cDNA, were amplified with primers listed in Table 1. By digestion with EcoT22I and dephosphorylation with shrimp alkaline phosphatase (Promega, Madison, WI, USA), the fragments were ligated into the PstI site of pET-OmpC to construct plasmids, pET-OmTmt, pET-OmHmt, and pET-OmMmt, respectively. These plasmids were designed to express outer membrane fusion protein in which the MT was integrated into the seventh outer membrane loop of the OmpC protein.
Expression of cytoplasmic human, mouse, and tMTs in E. coli
Each bacterial strain, harboring pET-Hmt, pET-Mmt, or pET-Tmt, was grown in Luria–Bertani (LB) broth supplemented with 50 μg/mL kanamycin at 37 °C to reach a density (at 600 nm) of about 0.7 (i.e., OD600 = 0.7). Isopropyl-β-d-thiogalactopyranoside (IPTG) (0.6 mM) was then added to the culture media to induce protein expression, which was allowed to progress for 8 h. Cells were harvested by centrifugation at 6,000 rpm (9,000×g) for 5 min. Bacterial cells were resuspended with 10 mM Tris–HCl buffer, pH 7.5, and then disrupted by sonication at 4 °C. The supernatant was collected after centrifugation at 12,000 rpm at 4 °C for 10 min. Equal amounts of the lysates were subjected to SDS–14% PAGE gel to confirm expression of all three species of MT.
Expression of OmpC fusion protein in E. coli
The growth and induction conditions of strains harboring OmpC outer membrane fusion proteins and the control strains (BL21(DE3)/pET-OmpC, BL21(DE3)/pET-30a) were the same as for the other strains. The pellets were washed twice and resuspended in 10 mM Na2HPO4, pH 7.2. After disruption by sonication, the lysates were centrifuged at 12,000 rpm for 2 min to remove cell debris and intact bacterial cells. The membrane fraction was harvested by centrifugation of the supernatant at 12,000 rpm for 30 min. This fraction was dissolved in detergent buffer (0.5% N-lauroylsarcosine sodium salt (Sigma, St. Louis, MO, USA) and 10 mM Na2HPO4, pH 7.2) at 4 °C for 30 min. After treatment, the insoluble fraction was recovered by centrifugation at 12,000 rpm for 30 min in a microfuge at 4 °C (Xu and Lee 1999). The insoluble fraction was washed once with 10 mM Na2HPO4 and dissolved in 10 mM Tris–HCl buffer, pH 7.5. Finally, the membrane fraction was analyzed by electrophoresis through a SDS–14% PAGE gel.
Measurement of mercury adsorption
After induction with 0.6 mM IPTG and 8 h of expression, the cultures were harvested by centrifugation and washed twice with 0.05 M Tris–HCl buffer (pH 5.0). The collected cells were suspended in 1 mL of 0.05 M Tris–HCl solution (pH 5.0), including 50 μM Hg(II), to an OD600 of approximately 3.5 (equivalent to 1.0 g/L). Hg(II) adsorption was allowed to occur at 25 °C with 75 rpm agitation for 24 h (Kao et al. 2006). After centrifugation at 13,000 rpm for 5 min, the adsorption value was calculated by determining the concentration of mercury remaining in the supernatant using a Cold Vapor Atomic Absorption Spectrometer (Model 400A Mercury Analyzer, Buck Scientific, East Norwalk, CT, USA). The equilibrium mercury adsorption capacity (q, mg/g cells) was plotted versus the corresponding equilibrium mercury concentration in the aqueous phase (Ce, mg/L) to establish the adsorption isotherm curves. The equilibrium adsorption capacity was calculated as q = (Co − Ce)∕X, where Co denotes the initial mercury concentration (mg/L) and X denotes the initial cell concentration (g/L) (Kao et al. 2006).
Disc assay of heavy metal resistance
Bacteria induced with 0.6 mM IPTG for 8 h were adjusted to a concentration of 0.68 ± 0.08 g/L. Each strain was plated on LB agar containing kanamycin (50 mg/mL) and 0.1 mM IPTG. Each disc (Creative Microbiologicals, Taipei, Taiwan) received 20 μL of 500 ppm Hg2+, 1,000 ppm Zn2+, Pb2+, Cu2+, Cd2+, or Hg2+ solution. These heavy metal standard solutions were obtained from Merck (Darmstadt, Germany). After incubation at 37 °C overnight, the diameter of the growth inhibition zone was measured with five repeats in three independent experiments.
Purification of tMT protein by liquid chromatography
Strain BL21(DE3)/pET-Tmt was grown in LB broth supplemented with 50 μg/mL kanamycin at 37 °C to an OD600 of approximately 0.7. IPTG (0.6 mM) was then added to induce recombinant protein expression. After 8 h of recombinant protein expression, the cells were harvested by centrifugation and disrupted by sonication at 4 °C. tMT protein was purified by liquid chromatography using a diethylamino ethanol column and a gel filtration column (binding buffer: 10 mM Tris–HCl buffer, pH 7.5, 1% β-mercaptoethanol; elution buffer: 600 mM Tris–HCl buffer, pH 7.5, 1% β-mercaptoethanol).
Measurement of ability to scavenge free-radicals
The protocol of preparing 2,2-diphenyil-1-picrylhydrazyl (DPPH●) and 2,2-azinobis (3-ethylbenzothiazoline-6-sulfonic acid) diammonium salt (ABTS●+) solution followed that previously described (Brand-Williams et al. 1995; Re et al. 1999). The strains, after induction for 8 h with IPTG, were harvested by centrifugation and washed twice with 0.01 M Tri–HCl buffer (pH 7.0). The cells were suspended in 1 mL of DPPH● or ABTS●+ solution to reach concentrations of 0.16 ± 0.01, 0.33 ± 0.07, and 0.68 ± 0.09 g/L. Besides, the purified tMT protein or GSH (Sigma) samples were adjusted to 0, 10, 20, 30, 40, and 50 μM in 100 μL 10 mM Tris–HCl buffer (pH 7.5). Each sample was mixed with 1 mL of either DPPH● or ABTS●+ solution and placed in the dark at room temperature. After 15 min (DPPH●) or 2 min (ABTS●+) of reaction, the absorbance at specific wavelength of 515 or 734 nm was measured by photometer (Brand-Williams et al. 1995; Re et al. 1999). The absorbance values were expected to decrease if DPPH● or ABTS●+ was diminished by the radical-scavenging ability of the sample.
Immunofluorescence labeling
Cells were fixed with 4% formaldehyde and treated with lysozyme for detecting cytoplasmic tMT. Cells for detecting surface tMT were incubated directly in blocking buffer (2% horse blood serum in phosphate-buffered saline (PBS) buffer, pH 7.2, with 0.05% Tween20) without fixation and lyozyme prior to immunostaining. The rabbit anti- tMT IgG (Wu et al. 2000) was used as primary antibody at the concentration of 1 mg/ml. The mixture of cells and antibody were incubated at 4 °C overnight and the cells were washed with PBST buffer (PBS buffer, pH 7.2, with 0.05% Tween20) three times. The Alexa Flour 488 donkey anti-rabbit IgG antibody and Alexa Fluor 594 donkey anti-rabbit IgG antibody (Invitrogen, Carlsbad, CA, USA) were used for detecting surface and cytoplasmic tMT and were reacted with cells at room temperature for 1 h. After the cells were washed with PBST buffer and stained with either 4′,6-diamidino-2-phenylindole (DAPI) or Nile red (Sigma) solution to identify nuclei and membrane of bacterial cell. The immunofluorescence was viewed with Nikon ECLIPSE 80I (Nikon, Tokyo, Japan).
Results
Expression of MT proteins and the OmpC-MT fusion proteins
The recombinant cell lysates separated on SDS–PAGE showed a major band that cytosolic MTs were ranged within 12–14 kDa (Fig. 1a). As the prediction of human MT monomer (6.1 kDa) is slightly bigger than the other two MTs (6.0 kDa), the result from SDS–PAGE showed that hMT dimmers migrated less than tMT and mMT dimmers. On the other hand, since the cDNA of MTs was integrated into loop seven of the E. coli ompC gene, the molecular weights of the outer membrane fusion proteins were higher than OmpC (Fig. 1b). Furthermore, as the samples containing the fusion proteins were extracted from the cell membrane fractions (Fig. 1b), the result indicated that OmpC fusion proteins have being properly localized to the cell outer membrane.
Protein expression in E. coli by SDS–PAGE: a Protein expression was examined by using E. coli clones harboring pET-30a encoding no MT (lane 1), pET-Hmt (lane 2), pET-Mmt (lane 3), or pET-Tmt (lane 4). The cell lysates showed that the hMT (upper arrow) migrated less than mMT and tMT (lower arrows). b Protein expression of recombinant outer membrane fusion protein: BL21(DE3)/pET-30a (lane 1) was used as the control. Escherichia coli clones harboring pET-OmpC (lane 2), pET-OmTmt (lane 3), pET-OmHmt (lane 4), or pET-OmMmt (lane 5) after induction with IPTG (0.6 mM). Arrows indicate the position of OmpC protein at about 38 kDa and show that the OmpC fusion protein was shifted to a higher position
Cellular location of cytosolic and outer-membrane-targeted tMT
To identify the location of cytosolic and outer-membrane-targeted tMT, the anti-tMT IgG was used to detect tMT through immunofluoresence labeling in BL21(DE3)/pET-Tmt and BL21(DE3)/pET-OmTmt. When outer-membrane-targeted tMT of the BL21(DE3)/pET-OmTmt was expressed and exposed to extracellular, it could be directly detected without lysozyme treatment (Fig. 2a). On the contrary, no signal could be detected from strains either without tMT or with cytosolically expressed tMT (Fig. 2a). Meanwhile, the cytosolic tMT (BL21(DE3/pET-Tmt)) could be labeled in red fluorescence while it was treated with lysozyme for primary antibody to pass through the membrane (Fig. 2b).
Immunofluorescence labeling of cellular location of tMTs. Escherichia coli clones harboring pET-30a (control), pET-OmpC (E. coli K12 outer membrane protein C), pET-OmTmt (OmpC-tMT fusion protein), pET-Tmt (tMT). a The strains treated without lyozyme were stained membrane by Nile red and reacted with Alexa Flour 488 donkey anti-rabbit IgG antibody. b The strains treated with lyozyme were stained nuclei by DAPI and reacted with Alexa Flour 594 donkey anti-rabbit IgG antibody. Bars = 5 μm
Increased mercury resistance of host cells by membrane-targeted MT
A disk assay for the evaluation of heavy metal resistance was performed using the recombinant bacteria (Table 2). The inhibition zones show no distinct difference among those recombinants that performed disk assay with heavy metals such as lead, copper, and zinc. On the other hand, disk assay with either cadmium or mercury showed more effects on their differences of inhibition zones. For cytosolically expressed MT strains, BL21(DE3)/pET-Mmt showed higher mercury resistance than BL21(DE3)/pET-Hmt and BL21(DE3)/pET-Tmt, while BL21(DE3)/pET-Tmt showed the lowest resistance to mercury. The trend for their mercury resistance was the same when 1,000 ppm mercury was used for the disk assay (data not shown). Furthermore, when tMT was targeted to the outer membrane, the strain BL21(DE3)/pET-OmTmt showed the highest resistance among those recombinants. It is interesting that control strain BL21(DE3)/pET30a possessed higher resistance to Hg2+ than strains harboring recombinant MTs in cytosol (Table 2).
Mercury adsorption by MT-expressed recombinants
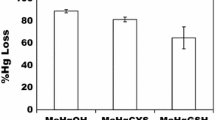
Though MTs from different species are highly conserved in their amino acid sequences, they performed different level of mercury resistance in bacterial cells. The concentrations of residual mercury of recombinant cells after the adsorption reactions were 30 μM (pET-30a), 32.7 μM (pET-Mmt), 28.05 μM (pET-Tmt), 31.5 μM (pET-Hmt), 34.4 μM (pET-OmpC), 29.75 μM (pET-OmMmt), 22.6 μM (pET-OmTmt), and 27.25 μM (pET-OmHmt), respectively. Their adsorption ability was showed (Fig. 3). For cytosolically expressed MTs, tMT (4.3 ± 0.4 mg/g cell dry weight) harboring strain accumulated a higher amount of mercury than hMT (3.8 ± 0.6 mg/g) and mMT (3.3 ± 0.5 mg/g). The mMT-harboring strain accumulated less than the tMT- and hMT-harboring strains, and it had the lowest mercury accumulation. It is interesting to know that the higher the amount of mercury accumulated, the lower resistance performance was for those cytosolically expressed MTs.
Mercury adsorption by recombinant strains with expression of MT. Escherichia coli BL21(DE3) harboring pET-30a ( ) series plasmids were exposed for 24 h in 0.05 M Tris–HCl buffer containing 50 μM Hg2+. pET-Tmt Tilapia MT (
) series plasmids were exposed for 24 h in 0.05 M Tris–HCl buffer containing 50 μM Hg2+. pET-Tmt Tilapia MT ( ), pET-Hmt human MT1A (
), pET-Hmt human MT1A ( ), pET-Mmt mouse MT1 (
), pET-Mmt mouse MT1 ( ), pET-OmpC E. coli K12 outer membrane protein C (
), pET-OmpC E. coli K12 outer membrane protein C ( ), pET-OmTmt OmpC-tMT (
), pET-OmTmt OmpC-tMT ( ) fusion protein, pET-OmHmt OmpC-hMT (
) fusion protein, pET-OmHmt OmpC-hMT ( ) fusion protein, and pET-OmMmt OmpC-mMT (
) fusion protein, and pET-OmMmt OmpC-mMT ( ) fusion protein. Data were means from three independent experiments
) fusion protein. Data were means from three independent experiments
On the other hand, all of the outer-membrane-targeted MTs, including OmpC-tMT (5.6 ± 0.4 mg/g), OmpC-hMT (4.6 ± 0.5 mg/g), and OmpC-mMT (4.0 ± 0.3 mg/g), showed higher Hg2+ accumulation than that of cytosolically expressed MTs (Fig. 3). Furthermore, the strain harboring OmpC-tMT accumulated almost twice as much as the cells harboring the control vector (3.0 ± 0.4 mg/g) (Fig. 3).
tMT protein shows scavenging ability of extracellular free radicals
As MTs are antioxidants that are able to scavenge reactive oxygen radicals (Sato and Kondoh 2002), tMT was examined for its scavenging ability. tMT was purified and compared with GSH, a well-studied antioxidant. As shown in Fig. 4, for all of the tested concentrations, the purified tMT showed better scavenging ability than GSH. For example, in 50 μM of tMT concentration, the scavenging ratio of purified tMT and GSH was 39.6% versus 21.0% for DPPH●, while it was 98.7% versus 71.1% for ABTS●+, respectively. The tMT showed a higher ability than GSH to scavenge both DPPH● and ABTS●+. Furthermore, for the strains harboring recombinant ompC gene, expressing the outer membrane fusion proteins showed scavenging ability for extracellular radicals, especially for ABTS●+ (Table 3).
Scavenging free radical ability with purified tMT. The assay of decolorization technique was applied to the measurement of total antioxidant activity of target substances, a DPPH● and b ABTS●+. Scavenging was determined by treatment with 0 to 50 μM tMT protein (black bars) or glutathione (GSH) (gray bars). Scavenging ratio \( \left( \% \right) = {{\left( {{\hbox{O}}{{\hbox{D}}_{\rm{Control}}} - {\hbox{O}}{{\hbox{D}}_{\rm{Sample}}}} \right)} \mathord{\left/{\vphantom {{\left( {{\hbox{O}}{{\hbox{D}}_{\rm{Control}}} - {\hbox{O}}{{\hbox{D}}_{\rm{Sample}}}} \right)} {{\hbox{O}}{{\hbox{D}}_{\rm{Control}}} \times {1}00.{\hbox{ O}}{{\hbox{D}}_{\rm{Control}}}}}} \right.} {{\hbox{O}}{{\hbox{D}}_{\rm{Control}}} \times {1}00.{\hbox{ O}}{{\hbox{D}}_{\rm{Control}}}}} \) indicated the absorbance from DPPH● or ABTS●+ solution without adding MT protein or glutathione. Data were M ± SD collected from three independent experiments
Discussion
In the mercury adsorption experiments, the E. coli host-expressed cytosolic tMT yielded about 10–20% better sorption than cells expressing either hMT or mMT. In the meantime, the same host possessed the lowest resistance among three cytosolically expressed MTs in the disc assay (Table 2). The results suggested that the greater the amount of intracellular mercury accumulated, the greater was toxicity shown and the lower was resistance. It has been showed that intracellular accumulation of heavy metals, such mercuric ion, often encounter some problems in the host cell, such as minimal recycling on account of the slow release of accumulated metals, and the disturbance of cysteine-rich protein with the redox pathways in microbial cells (Bardwell 1994; Gadd and White 1993). Our results showed that the limitation of intracellular accumulation could be overcome by anchoring the metal-binding proteins directly to the cell membrane for improving adsorption (Fig. 3) and reducing the toxicity (Table 2) (Bae et al. 2001; Kotrba et al. 1999; Saleem et al. 2008; Sousa et al. 1998; Valls et al. 1998). On the other hand, the significant differences of resistance levels among those recombinants with specific heavy metals, such as Hg2+ and Zn2+, were shown in our study (Table 2). This may be due to the involvement of some natural resistance mechanisms, such as CzcCBA (Silver and le Phung 2005), of a heavy metal efflux system in host strain.
The protein sequences of the tilapia, human, and mouse MTs show that 19 of 20 cysteines were conserved at the same positions of these three MTs (Scudiero et al. 2005). However, the ninth cysteine residue located in the alpha-domain of tMT is shifted two residues earlier than that of mammalian MTs (Scudiero et al. 2005). The CXC motif of the mammalian MTs is converted into CXXXC in the fish counterpart (Scudiero et al. 2005). This peculiarity has a marked effect on the structure of the fish MT, as determined by nuclear magnetic resonance (NMR) studies (Capasso et al. 2003). Moreover, the structure of the fish MT presents short alpha-helical stretches in both alpha and beta domains, in contrast with mammalian MT, and shows a different number of acidic residues in the polypeptide chain (Capasso et al. 2002). It has been suggested that a close relationship between structural diversity and protein reactivity is related to the significant differences in cysteine reactivity and metal exchangeability among those MTs (Capasso et al. 2003). However, the changes of residues in tMT and the variance of CXCC to CXXXCC may play important roles for its binding and scavenging ability.
At the extracellular environment, xenobiotics may result in macromolecular damage of oxidative stress and, in addition, free radicals could directly attack membrane by lipid peroxidation (Jones 2008). In Fig. 4, tMT protein exhibit better scavenging ability of ABTS●+ than DPPH●, while it is known that MT antioxidation activity was target to reactive oxygen radicals (Sato and Kondoh 2002). Therefore, the cells possessing MT outer membrane fusion protein showed scavenging ability for extracellular radicals, especially ABTS●+ (Table 2), and could prevent damage from oxidative stress. These results suggest that outer-membrane-targeted tMT protein could act as an antioxidant and the harboring strain may play dual functions in bioremediation of mercury and free radical scavenging.
Reference
Allen P (1994) Mercury accumulation profiles and their modification by interaction with cadmium and lead in the soft tissues of the cichlid Oreochromis aureus during chronic exposure. Bull Environ Contam Toxicol 53:684–692
Bae W, Mehra RK, Mulchandani A, Chen W (2001) Genetic engineering of Escherichia coli for enhanced uptake and bioaccumulation of mercury. Appl Environ Microbiol 67:5335–5338
Bardwell JC (1994) Building bridges: disulphide bond formation in the cell. Mol Microbiol 14:199–205
Belliveau BH, Trevors JT (1989) Mercury resistance and detoxification in bacteria. Appl Organomet Chem 3:283–294
Brand-Williams W, Cuvelier ME, Berset C (1995) Use of a free radical method to evaluate antioxidant activity. Food Sci Technol Res 28:25–30
Capasso C, Abugo O, Tanfani F, Scire A, Carginale V, Scudiero R, Parisi E, D'Auria S (2002) Stability and conformational dynamics of metallothioneins from the antarctic fish Notothenia coriiceps and mouse. Proteins 46:259–267
Capasso C, Carginale V, Crescenzi O, Di Maro D, Parisi E, Spadaccini R, Temussi PA (2003) Solution structure of MT_nc, a novel metallothionein from the Antarctic fish Notothenia coriiceps. Structure 11:435–443
Chan KM (1995) Metallothionein: Potential biomarker for monitoring heavy metal pollution in fish around Hong Kong. Mar Pollut Bull 31:411–415
Chen S, Wilson DB (1997a) Construction and characterization of Escherichia coli genetically engineered for bioremediation of Hg(2+)-contaminated environments. Appl Environ Microbiol 63:2442–2445
Chen S, Wilson DB (1997b) Genetic engineering of bacteria and their potential for Hg2+ bioremediation. Biodegradation 8:97–103
Coyle P, Philcox JC, Carey LC, Rofe AM (2002) Metallothionein: the multipurpose protein. Cell Mol Life Sci 59:627–647
Gadd GM, White C (1993) Microbial treatment of metal pollution—a working biotechnology? Trends Biotechnol 11:353–359
Harada M (1995) Minamata disease: methylmercury poisoning in Japan caused by environmental pollution. Crit Rev Toxicol 25:1–24
Jeanteur D, Lakey JH, Pattus F (1991) The bacterial porin superfamily: sequence alignment and structure prediction. Mol Microbiol 5:2153–2164
Jones DP (2008) Radical-free biology of oxidative stress. Am J Physiol Cell Physiol 295:C849–C868
Kao WC, Chiu YP, Chang CC, Chang JS (2006) Localization effect on the metal biosorption capability of recombinant mammalian and fish metallothioneins in Escherichia coli. Biotechnol Prog 22:1256–1264
Kotrba P, Pospisil P, de Lorenzo V, Ruml T (1999) Enhanced metallosorption of Escherichia coli cells due to surface display of beta- and alpha-domains of mammalian metallothionein as a fusion to LamB protein. J Recept Signal Transduct Res 19:703–715
Mejare M, Bulow L (2001) Metal-binding proteins and peptides in bioremediation and phytoremediation of heavy metals. Trends Biotechnol 19:67–73
Park JD, Liu Y, Klaassen CD (2001) Protective effect of metallothionein against the toxicity of cadmium and other metals. Toxicology 163:93–100
Re R, Pellegrini N, Proteggente A, Pannala A, Yang M, Rice-Evans C (1999) Antioxidant activity applying an improved ABTS radical cation decolorization assay. Free Radic Biol Med 26:1231–1237
Saleem M, Brim H, Hussain S, Arshad M, Leigh MB, Zia ul H (2008) Perspectives on microbial cell surface display in bioremediation. Biotechnol Adv 26:151–161
Sato M, Kondoh M (2002) Recent studies on metallothionein: protection against toxicity of heavy metals and oxygen free radicals. Tohoku J Exp Med 196:9–22
Scudiero R, Temussi PA, Parisi E (2005) Fish and mammalian metallothioneins: a comparative study. Gene 345:21–26
Silver S, le Phung T (2005) A bacterial view of the periodic table: genes and proteins for toxic inorganic ions. J Ind Microbiol Biotech 32:587–605
Sousa C, Kotrba P, Ruml T, Cebolla A, De Lorenzo V (1998) Metalloadsorption by Escherichia coli cells displaying yeast and mammalian metallothioneins anchored to the outer membrane protein LamB. J Bacteriol 180:2280–2284
Ueda M, Tanaka A (2000) Genetic immobilization of proteins on the yeast cell surface. Biotechnol Adv 18:121–140
Vallee BL (1995) The function of metallothionein. Neurochem Int 27:23–33
Valls M, de Lorenzo V (2002) Exploiting the genetic and biochemical capacities of bacteria for the remediation of heavy metal pollution. FEMS Microbiol Rev 26:327–338
Valls M, Gonzalez-Duarte R, Atrian S, De Lorenzo V (1998) Bioaccumulation of heavy metals with protein fusions of metallothionein to bacterial OMPs. Biochimie 80:855–856
Volesky B, Holan ZR (1995) Biosorption of heavy metals. Biotechnol Prog 11:235–250
Wong CK, Yeung HY, Cheung RY, Yung KK, Wong MH (2000) Ecotoxicological assessment of persistent organic and heavy metal contamination in Hong Kong coastal sediment. Arch Environ Contam Toxicol 38:486–493
Wu SM, Weng CF, Hwang JC, Huang CJ, Hwang PP (2000) Metallothionein induction in early larval stages of tilapia (Oreochromis mossambicus). Physiol Biochem Zool 73:531–537
Xu Z, Lee SY (1999) Display of polyhistidine peptides on the Escherichia coli cell surface by using outer membrane protein C as an anchoring motif. Appl Environ Microbiol 65:5142–5147
Acknowledgements
We thank Professor Jo-Shu Chang (National Cheng Kung University, Taiwan) for his kind gift of the pMAL-p2X-MT1A and pMBP-TEV-MT1 plasmids containing human MT1A and mouse MT1 cDNA. We also thank Prof. Su-Mei Wu (National Chia Yi University, Taiwan) for antibody of the rabbit anti-tMT IgG. This work was supported by the grants (NSC 96-3114-P-001-004-Y and NSC 97-3114-P-001-001) from the National Science Council, Taiwan, People’s Republic of China.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lin, KH., Chien, MF., Hsieh, JL. et al. Mercury resistance and accumulation in Escherichia coli with cell surface expression of fish metallothionein. Appl Microbiol Biotechnol 87, 561–569 (2010). https://doi.org/10.1007/s00253-010-2466-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-010-2466-x