Abstract
This paper reports the effects of changing pH (5–7) and temperature (T, 40–60 °C) on the efficiencies of bacterial hydrolysis of suspended organic matter (SOM) in wastewater from food waste recycling (FWR) and the changes in the bacterial community responsible for this hydrolysis. Maximum hydrolysis efficiency (i.e., 50.5% reduction of volatile suspended solids) was predicted to occur at pH 5.7 and T = 44.5 °C. Changes in short-chain volatile organic acid profiles and in acidogenic bacterial communities were investigated under these conditions. Propionic and butyric acids concentrations increased rapidly during the first 2 days of incubation. Several band sequences consistent with Clostridium spp. were detected using denaturing gel gradient electrophoresis. Clostridium thermopalmarium and Clostridium novyi seemed to contribute to butyric acid production during the first 1.5 days of acidification of FWR wastewater, and C. thermopalmarium was a major butyric acid producer afterward. C. novyi was an important propionic acid producer. These two species appear to be important contributors to hydrolysis of SOM in the wastewater. Other acidogenic anaerobes, Aeromonas sharmana, Bacillus coagulans, and Pseudomonas plecoglossicida, were also indentified.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Food losses during storage, distribution, and household generate food waste. Current environmental concerns and regulations in most countries discourage disposal of food wastes in landfills (Liu et al. 2009). In Korea, almost all household food waste is collected and recycled. However, commercial food waste recycling (FWR) generates large quantities of wastewater resulting from the clean-up process and from leachate in storage pits. Disposal of this FWR wastewater causes serious environmental problems mainly because of its volume and high organic material content (Lee et al. 2009). The FWR wastewater has traditionally been managed by ocean disposal, but this is likely to cause a secondary environmental problem of ocean pollution. Ultimately, the FWR wastewater must be collected and treated in an environmentally sound manner.
Anaerobic digestion is a multi-stage process in which complex organic components of the waste are hydrolyzed, broken down, and fermented into intermediate products that are subsequently converted into methane, carbon dioxide, and microbial biomass. Hydrolysis is usually followed by formation of volatile organic acids (VOA, i.e., acidogenesis; Demirel and Scherer 2008; McHugh et al. 2006). These processes involve many different species of microorganisms, which are broadly classified as acidogens, which produce organic acids, and methanogens, which produce methane (Bagge et al. 2008; Yu et al. 2006). Typically, fermentation of FWR wastewater occurs in two steps: the first by acidogens and the second by methanogens.
Acidogens and methanogens differ greatly in their physiology, biokinetics, and growth requirements. Optimization of each stage of anaerobic digestion could enhance the efficiency of waste fermentation if bacterial communities required for acidogenesis and methanogenesis could be maintained optimally in appropriate growth conditions in series (Yu et al. 2006). Because the methanogenic reaction is usually the rate-limiting step in the overall anaerobic digestion, control of the methanogenic stage has been an important factor in the successful operation of most anaerobic digestion processes (Yu et al. 2005). However, overall process enhancement requires understanding of the growth conditions and characteristics of acidogens in a two-stage process because they are important in hydrolyzing suspended organic materials (SOM) and producing short-chain organic acids, which are major substrates for methanogens. Because most bacteria including acidogens cannot digest particulate organic materials, hydrolysis of SOM is necessary to convert these materials to a size and type that can pass through bacterial cell walls for use as energy or nutritional sources. High levels of insoluble suspended particles also cause a serious drawback for the use of high rate anaerobic reactors, such as upflow anaerobic sludge blankets or anaerobic filters, in which case, a pre-hydrolysis/acidification step is often required (Li et al. 2009). Therefore, hydrolysis of particulate organic materials may improve the efficiency of anaerobic digestion. This process has increased the efficiency of volatile suspended solids (VSS) removal in anaerobic hydrolysis of fish-processing wastewater by 58% (Guerrero et al. 1999). Other studies have quantified how different operational conditions affect anaerobic hydrolysis and acidogenesis of various substrates (Gomec et al. 2002; Guerrero et al. 1999; Kim et al. 2003), but the hydrolysis profiles of FWR wastewater have not been studied.
In many biological fields, including acidogenic microbial processes, basic knowledge of the phenomenon is insufficient to build a mechanistic model. In such cases, empirical models and statistical analysis are very useful in elucidating basic mechanisms in complex situations and thus providing better process control. Response surface methodology (RSM) is a collection of statistical techniques that is useful for designing experiments, evaluating relative significance of several independent variables, and determining optimum conditions to achieve desired responses (Yang et al. 2003). In most RSM problems, the relationship between the response and the independent variables is unknown. Thus, the first step in RSM is to approximate the response using a low-order polynomial over some range of the independent variables, then to apply higher order polynomials if the simple linear model is not adequate. Once the higher order polynomial is determined, it can be used to locate the optimum set of independent variables by determining where the plane tangent to the response surface is horizontal (Eq. 1).
This study had two objectives: (1) to find the combination of pH and T which maximizes hydrolysis of SOM in FWR wastewater, and (2) to identify the acidogenic bacteria that function at these optimum conditions.
Materials and methods
Experimental materials
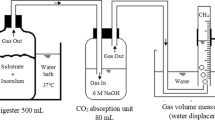
A 120-L batch of FWR wastewater was obtained from a storage pit at a local composting facility. The waste was homogeneously mixed and divided into smaller portions, which were frozen at −25 °C until use. Anaerobic sludge from a local municipal wastewater treatment plant was used as seed inoculum (2% v/v) for all experimental trials.
Conditions of pH and T were varied to determine the combination of these factors at which fermentation of FWR recycling was most efficient. Four identical anaerobic continuously stirred tank reactors with working volumes of 3 L (BioG-M, BioTron, Korea) were used in batch mode in these experiments. All reactors were equipped with pH and temperature controllers. The pH of each batch was adjusted using 6.0 N NaOH. Bromoethane sulfonate (1 mM) was added to inhibit methanogenesis.
Optimization using response surface methodology
RSM was used to determine the relationship of hydrolysis of SOM concentration to combinations of pH and T, which are the factors that most significantly affect microbial growth (Lindstrom et al. 2005; Song et al. 2007). Changes in VSS concentration were used to quantify changes in SOM concentration. RSM was applied to evaluate the relative significance of pH and T and to determine the conditions under which hydrolysis efficiency was maximum, within the experimental range of the independent variables.
To minimize the number of trials needed to obtain statistically valid results, this experiment used a 2 × 2 (T × pH; two levels each) orthogonal central composite cube design (Table 1, Fig. 1; Montgomery 2001). The independent variables were chosen to be 40–60 °C and pH 5.0–7.0, because fermentation of organic solids is most efficient at those temperatures, and because acidogens generally function most effectively under acidic conditions (Gomec et al. 2002; Guerrero et al. 1999; Kim et al. 2003). Treatments at the center point (i.e., 50 °C and pH 6.0) were replicated three times to estimate the experimental error (Montgomery 2001). The least squares method was used to find a higher-degree polynomial equation that approximated the response surface. This equation was then analyzed to locate the optimum condition, which was the set of independent variables such that the plane tangent to the response surface was horizontal. A sequential procedure of collecting data, estimating polynomials, and checking the adequacy of the model was used.
Extraction of DNA
Aliquots (1 mL) of the wastewater sample were centrifuged at 16,000 × g for 10 min, and the supernatant was decanted. The pellets were washed with 1 mL of deionized distilled water (DDW) and centrifuged again under the same conditions. The supernatants were carefully decanted, and the pellets were resuspended in 100 μL of DDW. The genomic DNA was immediately extracted and purified from the suspension using a fully automated nucleic acid extractor (Magtration System 6GC, PSS Co., Japan) with a Magtration Genomic DNA Purification Kit (PSS Co., Japan) using magnetic bead technology. The purified DNA was eluted with 100 μL of Tris–HCl buffer (pH 8.0) and stored at −20 °C until subjected to denaturing gradient gel electrophoresis (DGGE).
Bacterial DGGE analysis and DNA sequencing
Bacterial 16S rRNA gene was amplified by polymerase chain reaction (PCR) using the primers BAC 338F (5′-ACTCCTACGGGAGGCAG-3′) and BAC 805R (5′-GACTACCAGGGTATCTAATCC-3′; Yu et al. 2005). A 40-bp GC-clamp (5′-CGCCCGCCGCGCGCGGCGGGCGGGGCGGGGGCACG GGGGG -3′) was added at the 5′ end of BAC 338F to stabilize the melting behavior of the PCR fragments (Lee et al. 2008).
A touchdown PCR was initially conducted in a thermal cycler (PTC-100, MJ research Inc., Waltham, MA, USA), according to the following protocol: initial denaturation at 94 °C for 10 min; 20 cycles of denaturation at 94 °C for 30 s, annealing at 65 to 55 °C (reducing the temperature by 0.5 °C per cycle), and extension at 72 °C for 1 min; 15 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 1 min; and final extension at 72 °C for 7 min.
DGGE was performed using a D-code system (BioRad Inc., Hercules, CA, USA). Archaeal PCR product (16 μL) was loaded onto 8% (w/v) acrylamide gel containing a 30–60% denaturant gradient, where 100% was defined as 7 M urea with 40% (v/v) formamide. Electrophoresis was run at 85 V for 16 h in 1 × TAE buffer. After DGGE, the gel was stained with ethidium bromide and scanned under UV illumination. DNA bands were excised directly from the gel and eluted into 40 μL DDW. A 2-μL sample of the eluted solution was further amplified using BAC 338F and BAC 805R primers without attaching a GC-clamp. The PCR products were purified from a 1% (w/v) low melting-point agarose gel and cloned into the pGEM-T Easy vector (Promega, UAS). The cloned 16S rRNA gene fragments were sequenced using a 3730XL DNA analyzer (Applied Biosystems, Foster City, CA, USA) using the T7 primer. The sequencing results were compared with reference sequences in the GenBank database (http://blast.ncbi.nlm.nih.gov/). Sequence alignment and phylogenetic analysis were performed using DNAMAN software (version 5.2.2, Lynnon Biosoft, Canada). The phylogenetic tree was constructed in DNAMAN using a neighbor-joining method.
Analytical methods
Chemical oxygen demand (COD) was measured using closed reflux colorimetry. Solids were analyzed according to the procedures in Standard Methods (APHA-AWWA-WEF 2005). Total carbohydrate was analyzed by the phenol-sulfuric acid method (Dubois et al. 1956). Optical density at 600 nm was measured using a spectrophotometer (UV-2401PC, Shimadzu, Japan). The amounts of ammonia, organic nitrogen, and total Kjeldahl nitrogen were measured using the Kjeldahl method (APHA-AWWA-WEF 2005). A gas chromatograph (6890+, Agilent, Palo Alto, CA, USA) equipped with an Innowax capillary column (Agilent) and a flame ionization detector was used to quantify short-chain VOAs (C2-C4) and ethanol. The same gas chromatograph with an HP-5 capillary column and a thermal conductivity detector was used to analyze the gas composition. Helium was used as the carrier gas at flow rates of 2.5 mL/min for the analysis of organic acids and 8 mL/min for the analysis of gas composition.
Results
Estimating optimum condition of maximum hydrolysis of SOM
Most of the organic materials in the FWR wastewater samples used in this study occurred in the suspended form (Table 2). The concentration of total volatile solid (TVS), which is potentially biodegradable, was 110.6 g/L, i.e., 86.4% of the total solid (TS). Approximately 61% of the TVS (53% of TS) occurred in the suspended form. These suspended forms must be broken down to their monomers to achieve efficient anaerobic digestion.
A total of 12 trials, including a center point, were conducted to estimate the response surface for solubilization of the SOM in FWR (Table 1). To find the maximum reduction in VSS concentration, increasingly complex equations from linear to partial cubic were sequentially tested to model the data. Regression was significant (p < 0.05), and lack of fit was not significant (p > 0.05), only for a partial cubic model (Eq. 2):
where η is VSS reduction efficiency (%), x 1 is pH, and x 2 is T. The regression coefficient and residual standard deviation of the partial cubic model were 0.91 and 4.0, respectively.
Analysis of variance using Eq. 2 was initially performed to investigate possible interaction between the variables (Table 1). The possible two-way interaction among the variables (i.e., pH × T) was not significant at the 5% α level. This meant that the two independent variables (pH and T at two levels each) were not interdependent. Further statistical inspection showed that pH and T affected VSS reduction efficiency significantly at 5% α level each. The residual plots for the model and the experimental data set showed no patterns or trends (data not shown). Therefore, the model accurately predicted the effects of pH and T on VSS reduction efficiency by acidogens in FWR wastewater.
This equation was then used to determine conditions that would maximize VSS reduction by setting to zero the partial derivatives of the equation with respect to pH and T. The RSM model estimated a maximal VSS reduction efficiency (50.5%) at pH = 5.7 and T = 44.5 °C.
Two- and three-dimensional response surfaces of the partial cubic model for VSS reduction efficiency (Fig. 2) showed a clear peak at the lower left region; therefore, the optimum condition was within the design boundary. In the contour surfaces, the effects of independent variables on the response were evaluated using the grade of the contour lines along transects from the optimum condition toward the design boundary from Eq. 2. A saddle region, where the degree of change in the response with respect to the pH and temperature is minimal, as well as the region of the steepest decline in the hydrolysis efficiency can be seen in the response surface (Fig. 2). Therefore, VSS reduction efficiency decreased sharply if pH increased above 6.5 or if T decreased below 45 °C.
An additional batch experiment (Table 1, trial 10) was conducted to quantify VOA production at the predicted optimum conditions. Propionic and butyric acids were the major products; acetic acid and ethanol did not increase significantly (Fig. 3). Propionic and butyric acid accounted for 31.1% and 58.9% of the sum of C2–C4 VOAs and ethanol concentration, respectively. Propionic acid concentration increased sharply after the initial 1.5 days of incubation and stabilized at 9.1 g/L after 2 days of incubation. Butyric acid concentration also increased rapidly (i.e., to 6.3 g/L) during the first 2 days of incubation but continued to increase gradually to 16.6 g/L at the end of incubation (i.e., 8 days).
DGGE and phylogenetic analysis
DNAs extracted, with respect to the increase in the product concentrations, were used to investigate changes in bacterial community structure during incubation. In total, 19 DGGE bands, designated B1 to B19, were detected (Fig. 4) and excised from the gel for sequencing. The affiliations of the 16S rRNA gene sequences were determined by comparing them to the GenBank database (Table 3). All band sequences belonged to two phyla, Proteobacteria and Firmicutes (Fig. 5).
B9 was dominant during the acidogenic period (Fig. 4). B9, B6, B7, and B10 were 99.8% similar to Clostridium thermopalmarium, which is an obligately anaerobic, moderately thermophilic species that grows best when 6.0 ≤ pH ≤ 8.2 (optimum approximately 6.6) and when 50 ≤ T ≤ 55 °C (upper growth limit, 60 °C; Soh et al. 1991). B6, B7, and B10 were also >99% similar to C. thermopalmarium.
B8 was also common at 1.5 days incubation. B8, B2, B3, B4, and B5 were all >99% similar to Clostridium novyi, which is an obligate anaerobe (Dixit et al. 2005) that grows best when pH = 7.0 and 20 ≤ T ≤ 45 °C (optimum 30–35 °C).
B12 was 100% similar to Clostridium sporogenes, which is an obligate anaerobe which grows best when 5.0 ≤ pH ≤ 7.0 and has an optimum growth temperature of 37 °C (Boone 2001). C. sporogenes consumes glucose and amino acids and produces organic acids, H2, and CO2.
B17 was 99.6% similar to Bacillus coagulans, which is a facultative anaerobe which grows best when 5.5 ≤ pH ≤ 6.5 and 35 ≤ T ≤ 50 °C (Boone 2001). B. coagulans mainly uses carbohydrates and amino acids for growth and produces mainly lactic acid, with small amounts of other organic acids, H2, and CO2.
B13 was 99.6% similar to Aeromonas sharmana, and B14 was 99.8% similar to Pseudomonas plecoglossicida. Those species, probably did not contribute to acidogenesis in FWR wastewater because these bands were not clearly visible during incubation (Fig. 4).
Several bands could not be identified with certainty. B1 was not closely related (<96.0% similarity) to any known species. B11, B15, B16, B18, and B19 were closely related to uncultured (i.e., unidentified) clones and were found in seed sludge.
Discussion
Among the species of the genus Clostridium, several strains produce butyric acid as the main product of sugar metabolism (Soh et al. 1991). C. thermopalmarium uses various carbohydrates including glucose, xylose, and sucrose. Butyric acid is likely to be its main end product of fermentation (approximately 1 mol butyric acid produced per mole of glucose consumed); it also produces H2, CO2, and small amounts of acetate, ethanol, and lactate. C. thermopalmarium does not reduce sulfate, thiosulfate, and sulfite to H2S (Soh et al. 1991).
C. novyi is also capable of utilizing carbohydrates including raffinose, glucose, melibiose, cellobiose, and ribose and produces acetate, propionate, isobutyrate, butyrate, isovalerate, hydrogen, and CO2; but this species cannot ferment some common carbohydrates such as sucrose, mannose, lactose, maltose, glycogen, and galactose. Furthermore, it can use only five amino acids (alanine, arginine, glutamine, histidine, and proline) as substrates for growth. It can also ferment some complex substrates like yeast extract, cassaminoacids, peptone, and gelatin (Dixit et al. 2005).
C. thermopalmarium (B6, B7, B9, and B10) and C. novyi (B2, B4-5, and B8) were first observed at day 1.5 (Fig. 4), and this point coincided with the increase of both propionic and butyric acids concentrations (Fig. 3). C. thermopalmarium-related bands (B9 and B10) remained at constant intensity to the end of incubation, whereas those of C. novyi-related bands (B2, B4, B5, and B8) gradually faded away after 1.5 days of incubation. During the initial 1.5 days of incubation, 8.1 g/L of propionic acid was produced (i.e., 88.8% of total propionic acid accumulated during fermentation). Butyric acid production during the first 1.5 days was 6.3 g/L (i.e., 38.2% of overall accumulation). Approximately 1.0 g/L of propionic acid was additionally formed during 1.5 to 2 days of incubation, but none was produced after 2 days (Fig. 3). Based on the similarity between propionic acid accumulation and the changes in intensity of bands corresponding to C. novyi, this species was probably the main producer of propionic acid.
Butyric acid concentration increased to 6.3 g/L by 1.5 days of fermentation then sharply increased to 12.8 g/L by the second day (Fig. 3). Although both C. thermopalmarium and C. novyi seemed to contribute to butyric acid production in an early stage (i.e., up to 1.5 days) of acidification of FWR wastewater, C. thermopalmarium was clearly the major producer of butyric acid after 1.5 days of incubation (Fig. 4).
The putative functions and the pH and T preferences of the microorganisms deduced from the DGGE band sequences corresponded well with the reactor conditions and chemical profiles. Therefore, C. thermopalmarium and C. novyi were likely the most important acidogenic producers of propionic and butyric acids in this experiment. They were also likely to the main species that solubilized SOM in FWR wastewater.
References
APHA-AWWA-WEF (2005) Standard methods for the examination of water and wastewater. American Public Health Association, Washington DC
Bagge E, Sahlstrom L, Albihn A (2008) The effect of hygienic treatment on the microbial flora of biowaste at biogas plants. Water Res 39:4879–4886
Boone DR (2001) Bergey's manual of systematic bacteriology. Springer, New York
Demirel B, Scherer P (2008) The roles of acetotrophic and hydrogenotrophic methanogens during anaerobic conversion of biomass to methane: a review. Rev Environ Sci Biotechnol 7:173–190
Dixit A, Dhaked RK, Alam SI, Singh L (2005) Characterization of Clostridium sp. RKD producing botulinum-like neurotoxin. Syst Appl Microbiol 28:405–414
Dubois M, Gilles KA, Hamilton JK, Rebers PA, Smith F (1956) Colorimetric methods for determination of sugars and related substances. Anal Chem 28:350–356
Gomec CY, Kim M, Ahn Y, Speece RE (2002) The role of pH in mesophilic anaerobic sludge solubilization. J Environ Sci Health A Tox Hazard Subst Environ Eng 37:1871–1878
Guerrero L, Omil F, Méndez R, Lema JM (1999) Anaerobic hydrolysis and acidogenesis of wastewaters from food industries with high content of organic solids and protein. Water Res 33:3281–3290
Kim M, Gomec CY, Ahn Y, Speece RE (2003) Hydrolysis and acidogenesis of particulate organic material in mesophilic and thermophilic anaerobic digestion. Environ Technol 24:1183–1190
Lee C, Kim J, Shin SG, Hwang S (2008) Monitoring bacterial and archaeal community shifts in a mesophilic anaerobic batch reactor treating a high-strength organic wastewater. FEMS Microbiol Ecol 65:544–554
Lee DH, Behera SK, Kim JW, Park HS (2009) Methane production potential of leachate generated from Korean food waste recycling facilities: a lab-scale study. Waste Manag 29:876–882
Li D, Zhou T, Chen L, Jiang W, Cheng F, Li B, Kitamura Y (2009) Using porphyritic as a new additive improving hydrolysis and acidogenesis of solid organic wastes. Bioresour Technol 100:5594–5599
Lindstrom ES, Agterveld MPKV, Zwart G (2005) Distribution of typical freschwater bacterial groups is associated with pH, temperature, and lake water retention time. Appl Environ Microbiol 71:8201–8206
Liu G, Zhang R, El-Mashad HM, Dong R (2009) Effect of feed to inoculum ratios on biogas yields of food and green wastes. Bioresour Technol 100:5103–5108
McHugh S, Carton M, Mahony T, O'Flaherty V (2006) Methanogenic population structure in a variety of anaerobic bioreactors. FEMS Microbiol Lett 219:297–304
Montgomery DC (2001) Design and analysis of experiments. Wiley, New York
Soh ALA, Ralambotiana H, Ollivier B, Prensier G, Tine E, Garcia JL (1991) Clostridium thermopalmarium sp. nov., a moderately thermophilic butyrate-producing bacterium isolated from palm wine a Senegal. Syst Appl Microbiol 14:135–139
Song M, Kim N, Lee S, HWang S (2007) Use of whey permeate for cultivating Ganoderma lucidum mycelia. J Dairy Sci 90:2141–2146
Yang K, Yu Y, Hwang S (2003) Selective optimization in thermophilic acidogenesis of cheese-whey wastewater to acetic and butyric acids: partial acidification and methanation. Water Res 37:2467–2477
Yu Y, Lee C, Kim J, Hwang S (2005) Group-specific primer and probe sets to detect methanogenic communities using quantitative real-time polymerase chain reaction. Biotechnol Bioeng 89:670–679
Yu Y, Kim J, Hwang S (2006) Use of real-time PCR for group-specific quantification of aceticlastic methanogens in anaerobic processes: population dynamics and community structures. Biotechnol Bioeng 93:424–433
Acknowledgements
This research was supported by the Korea Ministry of Knowledge and Economy (MKE) as the Manpower Development Program for Energy and Resources and the Ministry of Environment (MOE) as the Human Resource Development Project for Waste to Energy. This work was also supported by the Korea Ministry of Education, Science, and Technology (MEST) through the BK-21 program.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kim, M.D., Song, M., Jo, M. et al. Growth condition and bacterial community for maximum hydrolysis of suspended organic materials in anaerobic digestion of food waste-recycling wastewater. Appl Microbiol Biotechnol 85, 1611–1618 (2010). https://doi.org/10.1007/s00253-009-2316-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-009-2316-x