Abstract
The cancer-testis (CT) antigen synovial sarcoma X break point 2 (SSX2) was expressed in Pichia pastoris as a means to produce a delayed-type hypersensitivity skin test reagent for monitoring SSX2-specific anti-cancer immune responses. SSX2 was detected intracellularly in P. pastoris despite the addition of the Saccharomyces cerevisiae alpha-mating factor secretion signal. Increasing the SSX2 gene copy number did not improve its secretion but did enhance intracellular SSX2 levels. SSX2 with its C-terminal nuclear localization signal (NLS) deleted (SSX2NORD), however, was secreted. Indirect immunofluorescence indicated that SSX2 containing the NLS did not translocate to the nucleus but accumulated in the endoplasmic reticulum (ER). Experimental results further suggested that SSX2 containing the NLS was misfolded in the ER, while deletion of the NLS facilitated correct folding of SSX2 inside the ER and improved its secretion. Production of SSX2NORD was scaled-up to a 2-L fermentor using a fed-batch protocol to maintain methanol at a concentration of 1 g L−1. Decreasing the cultivation temperature from 25 °C to 16 °C improved protein stability in the culture supernatant. In this process, after 120 h cultivation, the wet cell weight of P. pastoris reached 280 mg mL−1, and the yield of SSX2NORD was 21.6 mg L−1.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The synovial sarcoma X break point 2 or SSX2 gene belongs to a highly homologous family (SSX) that encodes proteins involved in a chromosomal translocation t(X;18)(p11.2;q11.2) characteristically found in 70% of human synovial sarcoma (Clark et al. 1994). The translocation of the synovial sarcoma translocation gene (SYT) on chromosome 18 with an SSX gene (mainly SSX1 and SSX2) creates a SYT-SSX fusion protein which is believed to be a possible transcription activator for aberrant transformation of synovial sarcoma (Nagai et al. 2001). SSX2 is a nuclear protein that contains a bipartite nuclear localization signal (NLS) at its C-terminus that is responsible for the localization of SSX2 and the SYT-SSX2 fusion to the nucleus (dos Santos et al. 2000). SSX2, also referred to as HOM-MEL-40 or CT5.2a, is an immunogenic tumor antigen identified by the serological analysis of recombinant cDNA expression libraries (SEREX) from a melanoma tumor (Tureci et al. 1996). It is expressed in various cancers, while its expression in normal tissue is restricted to the testis. Because of this characteristic expression, SSX2 is also known and classified as a cancer-testis (CT) antigen (Gure et al. 1997).
In cancer cells, endogenous cancer antigens are degraded by cytosolic proteosome, and the resultant peptides can be delivered to endoplasmic reticulum (ER) through the transporter associated with antigen processing (TAP). In ER, these cancer-specific peptides are loaded onto the major histocompatibility complex (MHC) class I molecule and presented on the cancer cell surface by the antigen presenting pathway (Petrovsky and Brusic 2004). Since CT antigens such as SSX2, NY-ESO-1, and MAGE A3 are specifically expressed in cancer and testis cells, and testis germline cells are MHC class I negative, the expression and activation of CT antigen-specific cytotoxic T lymphocytes is highly tumor specific, making CT antigens ideal targets for cancer immunotherapy (Scanlan et al. 2002). The therapeutic strategy includes delivering CT antigens to antigen presenting cells, for example, Dendritic Cells, to induce and activate CT antigen-specific T lymphocytes to kill the cancer cells presenting CT antigen epitopes (van der Bruggen and Van den Eynde 2006). Although, SSX2 is a nuclear protein, it has been shown that nuclear proteins are partly transported to the cytoplasm for degradation (Bader et al. 2007); therefore, the presentation of SSX2-specific epitopes are found in cancer cells (Ayyoub et al. 2003; Bricard et al. 2005). To date, several studies have demonstrated that SSX2 can elicit both humoral and cellular (both CD4+ and CD8+) mediated immune responses to kill cancer cells (Ayyoub et al. 2002; Ayyoub et al. 2004; Scanlan et al. 2002). For these reasons, there is a considerable interest in producing recombinant SSX2 antigen in order to clinically evaluate it as a cancer vaccine. This also includes the production of its delayed-type hypersensitivity (DTH) skin test reagent, which can quickly and accurately measure the immune responses of the immunized cancer patients by dermal injection. However, if the skin test reagent has the same origin with the vaccination reagent, one cannot tell if the positive immune responses are generated from the antigen or the contaminants from the expression host (Davis et al. 2004). To avoid these possible false-positive responses, vaccination and skin test reagent should be derived from different host expression systems.
The methylotrophic yeast Pichia pastoris has been widely used for production of various recombinant proteins by secretion into the supernatant or by intracellular expression (Cereghino et al. 2002). The most commonly used strategy for recombinant protein expression is driven by the alcohol oxidase (AOX1) promoter, which is strongly induced in medium containing methanol as a sole carbon source. This expression system offers several advantages, such as easy manipulation at the genetic level, high stability of transformants, the capability to culture cells at high cell density, and most importantly, the ability to perform higher eukaryotic protein secretion and post-translational modifications (Macauley-Patrick et al. 2005). Because P. pastoris secretes low levels of endogenous proteins, a secreted heterologous protein constitutes the vast majority of the total protein in the culture supernatant (Higgins and Cregg 1998). The predominance of the heterologous protein in the supernatant makes following purification steps easier to implement. However, overexpression of heterologous proteins may saturate the secretory pathway, with proteins accumulating inside the ER where they need to be folded correctly before being transported from the ER to the Golgi apparatus (Damasceno et al. 2007). Additionally, the options for secretion of heterologous proteins are normally limited to foreign proteins which are normally secreted by their native hosts (Higgins and Cregg 1998). Therefore, one of the challenges in using this expression system is to produce and secrete foreign proteins that are not naturally secreted.
We previously reported the production and purification of SSX2 from Escherichia coli C41(DE3) in a 2-L bench-top fermentor with a yield of 236 mg L−1 of purified SSX2 (Huang et al. 2007). In this study, SSX2 produced from P. pastoris instead of E. coli is for monitoring of SSX2-specific immune responses in a DTH skin tests, thereby avoiding measurement of immunological responses to potential bacterial contaminants in the E. coli-produced SSX2 vaccine protein. We focus on secretion of SSX2 for purification advantages and intend to address the secretion bottlenecks of SSX2 and other nuclear proteins in P. pastoris. In this study, we evaluated SSX2 secretion in P. pastoris and found that SSX2 is minimally secreted and accumulated inside the cell, mainly in the ER. Deletion of its C-terminal NLS improved SSX2 secretion by facilitating its folding inside the ER. We also report a fermentation protocol to scale-up the secretion of SSX2 without the NLS with a yield of 21.6 mg L−1 in the culture media.
Materials and methods
Strains and plasmids construction
The SSX2 gene (GenBank Accession No. NM 175698) was obtained from the New York branch of the Ludwig Institute for Cancer Research, cloned into the plasmid pcDNA 3.1 (Invitrogen, Carlsbad, CA, USA). All strains, plasmids, and constructs created and used in this study are shown in Table 1 and Fig. 1. P. pastoris KM71H and plasmid pPICZαA were purchased from Invitrogen. The SSX2 gene was cloned in frame into pPICZαA which uses the Saccharomyces cerevisiae alpha-mating factor as a secretion signal and the AOX1 promoter, resulting in pPICZαASSX2. The SSX2NORD construct was amplified by oligonucleotides (5′-GAATTCAACGGAGACGACGCCTTTGCAAGGA-3′ and 5′-GCGGCCGCTTATCTCTCGTGAATCTTCTCAGAG-3′) using pPICZαASSX2 as a template and cloned into plasmid pPICZαA to create plasmid pPICZαASSX2NORD. Plasmids were transformed into P. pastoris KM71H and selected on YPDS plates with different zeocin concentrations (100 to 500 μg/ml) to create strains MS03, MS-H, and MN-H. The A33 single chain gene was amplified from plasmid pPIC9K-A33scFv (Damasceno et al. 2004) and cloned into pPICZαA to create pPICZαAA33scFv. The A33scFvNLS construct was amplified by using four oligonucleotides, 5′-CTCGAGAAAAGAGAGGCTGAAGCTGAGCTCCAGA-3′, 5′-TTTGGGTCCAGATGAGGAGACGGTGACCAGGGTG-3′, 5′-ACCGTCTCCTCATCTGGACCCAAAAGGGGGGAACA-3′, and 5′-GGCGGCCGCTTACTCGTCATCTTCCTCAGGGTCG-3′, using a PCR touchdown protocol (Don et al. 1991). A33scFvNLS was later cloned into pPICZαA to create pPICZαAA33scFvNLS. Plasmids of pPICZαAA33scFv and pPICZαAA33scFvNLS were transformed into KM71H to generate strains M33 and M33NLS, respectively.
Illustration of SSX2 and SSX2NORD constructs. The N-terminal white box covers the major histocompatibility complex class one and two epitopes of SSX2 antigen. The C-terminal black box represents the nuclear localization signal of SSX2 at its C-terminus. The two basic amino acid domains of bipartite NLS are underlined
Strain cultivation and recombinant protein induction
P. pastoris colonies were transferred to 10 mL YPD medium (1% yeast extract, 2% peptone, and 2% dextrose) and grown overnight. The cells were then inoculated in 10 mL YPD medium with an initial optical density (OD600) of 0.2 and grown for 24 h. Cells were harvested and resuspended in 10 mL BMMY medium (100 mM potassium phosphate, pH 6, 1.34% (w/v) yeast nitrogen base without amino acids, 4 × 10−5% (w/v) biotin, 1% yeast extract, 2% peptone, and 0.5% methanol). Cultures were then grown in a rotary shaker at 30 °C/250 rpm for 16 h. Samples were centrifuged at 13,000 × g for 10 min, and the culture supernatant and pellet were analyzed.
Southern blot analysis
Transformants, including MS03, MS-H, MN-H, M33, and M33NLS, were grown in YPD medium overnight in a 5-mL culture. Genomic DNA of these transformants was prepared using a Yeaststar genomic DNA kit (Zymo Research, Orange, CA, USA) according to the manufacturer’s instructions. Different concentrations of plasmids (pPICZαASSX2NORD and pPICZα AA33scFv) corresponding to one to ten copies of SSX2NORD and A33scFv genes were calculated according to the ratio of the size of plasmids (4,012 and 4,322 bp, respectively) to the genome size of P. pastoris (9.6 × 106 bp), and used as standards (Hohenblum et al. 2003). Both plasmid and genomic DNA were digested with restriction enzymes, Xho1 and Not1, at 37 °C overnight and run on a 0.8% agarose gel. The separated fragments were transferred to a positively charged nylon membrane (Pall, East Hills, NY, USA) overnight. DIG-labeled probes for both SSX2NORD and A33scFv genes were generated using a PCR DIG Probe Synthesis Kit (Roche Applied Science, Indianapolis, IN, USA). After DNA transfer, the blot was later rinsed, UV-immobilized, and hybridized with DIG-labeled probe at 48 °C for 6 h. After washing, the blot was detected with anti-DIG-Fab alkaline phosphatase conjugate and developed using a DIG nucleic acid detection kit (Roche Applied Science) according to the manufacturer’s instructions. DNA fragments on the developed membrane were then scanned, selected, and plotted by Image J densitometry software (http://rsb.info.nih.gov/ij/), and the copy number was determined by the constructed standard curve.
Indirect immunofluorescence and confocal microscopy
Induced P. pastoris cells were fixed in 5 mL 100 mM potassium phosphate buffer (pH 6.5) containing 5% formaldehyde and incubated at room temperature for 1.5 h. Cells were washed twice with solution B (1.2 M sorbitol, 100 mM potassium phosphate, pH 7.4) and converted to spheroplasts by adding 20 μL zymolyase (1 mg mL−1) in 500 μL solution B. Washed cells were placed on L-polylysine coated slides and permeabilized in −20 °C MeOH for 6 min followed by −20 °C acetone for 30 s and blocked in PBS/0.1% bovine serum albumin (BSA) for 20 min. The cells were then incubated with 20 μL of 5 μg mL−1 mouse anti-SSX2 antibody. (A 20-μL of 10 μg mL−1 rabbit anti-Kar2p antibody was also added in confocal microscopy experiment). After 1.5 h incubation at room temperature, cells were washed three times with PBS/0.1% BSA and incubated with 20 μg mL−1 of Alexa546-conjugated goat anti-mouse IgG for 1.5 h for indirect immunofluorescence. For confocal microscopy, slides were incubated with 20 μg mL−1 of Alexa488-conjugated goat anti-mouse IgG and 20 μg mL−1 of Alexa546-conjugated goat anti-rabbit IgG for 1.5 h. Cells were washed three times, and images were examined. For nucleus staining in immunofluorescence, 10 μg mL−1 of Hoechst 33258 dye (Invitrogen) was added into the cells and incubated for 30 min during the secondary antibody incubation. P. pastoris KM71H was used as a negative control in the immunofluorescence experiments. There was no fluorescence detected, indicating no cross-reaction between anti-SSX2 antibodies and the host proteins. Immunofluorescence microscopy was performed using an Olympus BX61 epifluorescent microscope (Olympus America, Center Valley, PA, USA). Images were acquired using a Cooke SensiCam and Slidebook software (Intelligent Imaging, Santa Monica, CA, USA). Confocal microscopy was performed using a Leica TCS SP2 system (Leica Microsystems, Bannockburn, IL, USA) and processed using Leica Lite software. Images of different emission wavelengths were overlaid to examine co-localization.
Protein sample preparations from P. pastoris
A 1-mL aliquot of cells from induced strains (Table 1) was centrifuged at 13,000 × g for 10 min. The culture supernatants were collected, and proteins were precipitated by adding 10% (v/v) trichloroacetic acid (TCA) and incubated on ice for 30 min. Supernatant was centrifuged, carefully removed, and washed with 200 μL cold acetone. Acetone was then removed by centrifugation, and protein pellets were dried at room temperature for 30 min. A 100-μL aliquot of 1X LDS sample buffer (Invitrogen) was added to solubilize the protein pellets, and the resultant protein sample was loaded on the gel. Cytoplasmic (soluble) and membrane-associated (insoluble) fractions were collected as previously described (Damasceno et al. 2007). Briefly, the previous cell pellets were washed in the PBS at pH 7.4 and resuspended in 100 μL of yeast breaking buffer (50 mM sodium phosphate, pH 7.4; 1 mM phenylmethyl sulfonyl fluoride; 1 mM EDTA, and 5% (v/v) glycerol). One hundred microliters of acid-washed glass beads were added and vortexed at maximum speed seven times for 1 min with 1 min intervals on ice to disrupt the cells. The lysate was centrifuged at 14,000 × g for 30 min at 4 °C, and the supernatant was collected as the cytoplasmic protein fraction. The remaining pellet was resuspended in yeast breaking buffer containing 2% sodium dodecyl sulfate (SDS) and centrifuged at 4,000 × g for 5 min. The supernatant was collected as the membrane-associated fraction. For whole cell fraction preparation, 100 μL of lysis buffer (50 mM sodium phosphate, pH 7.4; 1 mM phenylmethyl sulfonyl fluoride; 1 mM EDTA, and 5% (v/v) glycerol; 0.1% Triton X100) and 100 μL of acid-washed glass beads were added and vortexed. The cells were then centrifuged, and the supernatant was collected as the whole cell fraction (Okamura et al. 2000).
SDS-PAGE and immunoblotting
TCA-precipitated culture supernatants, cytoplasmic, membrane-associated, and whole cell fractions were normalized for same cell optical density before analysis. Samples were analyzed by electrophoresis on 12% SDS-polyacrylamide gels (Invitrogen). After electrophoresis, the gel was stained by SimplyBlue SafeStain (Invitrogen). For western blot, the proteins were electroblotted on to a nitrocellulose membrane where SSX2 or SSX2NORD was detected with 2.5 μg of murine anti-human SSX2 monoclonal antibody using a WesternBreeze kit (Invitrogen). Kar2P (BiP) detection was carried out by adding a 1:2,000 dilution of rabbit anti-Kar2p antibody (generous gift from Dr. C. Barlowe). All assays were performed according to manufacturer’s instructions. Protein concentrations and protein level comparisons were estimated from scanned images of the developed western blots with Image J densitometry software (http://rsb.info.nih.gov/ij/). Western blots were scanned, and images of the protein bands were selected, plotted, and compared with standards or a desired band.
Fermentation
A 1-mL of overnight culture of strain MN-H was used to inoculate 100 mL of BMG medium (100 mM potassium phosphate, pH 6, 1.34% (w/v) YNB, 4 × 10−5% (w/v) biotin, and 1% glycerol) and grown at 30 °C for 16 h until the OD600 reached 10. This culture was used to inoculate the fermentor (Bioflo 3000, New Brunswick Scientific, Edison, NJ, USA). The fermentation was carried out in a 2-L bench-top fermentor with an initial volume of 1 L fermentation medium (100 mM potassium phosphate, pH 6, 1.34% (w/v) yeast nitrogen base without amino acids, 4 × 10−5% (w/v) biotin, 1% yeast extract, 2% peptone, and 5% (v/v) glycerol) and supplemented with 4.35 mL PTM1 trace salts (24 mM CuSO4, 0.53 mM NaI, 19.87 mM MnSO4, 0.83 mM Na2MoO4, 0.32 mM boric acid, 2.1 mM CoCl2, 0.15 mM ZnCl2, 0.23 M FeSO4, and 0.82 mM biotin). AFS-Biocommand Bioprocessing software version 2.6 (New Brunswick Scientific) was used for data acquisition and parameter control. During the fermentation, dissolved oxygen was maintained at 40% air saturation and was controlled by a D.O. cascade of agitation followed by pure oxygen sparging when required. At the end of the batch phase, indicated by a D.O. spike, the culture was induced with a methanol feed (100% methanol with 12 mL PTM1 salts/L), and the methanol concentration was controlled at 1 g L−1 throughout the fermentation. A methanol sensor, sensor unit, and PID control was used to monitor and control methanol concentration (Damasceno et al. 2004). Samples were taken for analysis of SSX2NORD and wet cell weight (WCW) during the fermentation. For WCW, a 1-mL of the broth was collected and centrifuged at 13,000 × g for 10 min. The supernatant was collected for SSX2NORD analysis, and the cell pellet was weighed to determine WCW.
Results
SSX2 accumulates intracellularly despite the addition of signal peptide
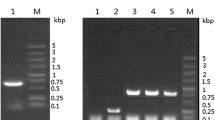
In order to express and secrete SSX2 into the culture media, the SSX2 gene was cloned into the pPICZαA plasmid which carries the S. cerevisiae alpha-mating factor as a secretion signal. The plasmid, pPICZαASSX2, was transformed into competent P. pastoris KM71H cells, and the transformants were selected on YPDS plates containing 100 μg/ml of zeocin. Several transformants including MS03, with one copy of pPICZαA SSX2 integrated, were cultivated, induced, and fractionated into three different cellular fractions. Immunoblot analysis of different fractions revealed that SSX2 was poorly expressed, accumulated in the intracellular membrane-associated fraction, and that the alpha-mating factor signal peptide was not processed. No SSX2 was found in the culture supernatant (Fig. 2). A high copy number strain MS-H, with seven copies of pPICZαA SSX2 integrated, was then selected from a 500 μg/ml zeocin plate to test whether increasing expression of SSX2 would facilitate its secretion. After induction for 16 h, the expression level of SSX2 found in intracellular membrane-associated fraction increased approximately 49-fold when compared with strain MS03. SSX2 was degraded in the soluble cytoplasmic fraction. However, there was only a minute amount of degraded SSX2 detected in the culture supernatant fraction (Fig. 2). Thus, we concluded that SSX2 was barely secreted by P. pastoris.
Secretion of SSX2 at different copy numbers. Western blot of different cell fractions of MS03 and MS-H strains that have been induced for SSX2 secretion in BMMY medium for 16 h. M, molecular weight marker; S, SSX2 standard; 1, membrane-associated fraction; 2, cytoplasmic fraction; 3, culture supernatant fraction
Deletion of nuclear localization signal (NLS) improves SSX2 secretion
Because SSX2 is a nuclear protein, it was hypothesized that SSX2 was translocated to the nucleus, thus inhibiting its secretion. To test this hypothesis, a construct of SSX2 without the NLS was created (SSX2NORD), and the plasmid pPICZαASSX2NORD was transformed into competent P. pastoris KM71H cells. A high copy number strain MN-H, with six copies of pPICZαA SSX2NORD, was selected, and SSX2NORD expression and secretion levels were compared with the native SSX2 expression strain, MS-H. As shown in Fig. 3, the SSX2NORD in the membrane-associated fraction decreased by 80% as compared to the level of SSX2 found in the MS-H strain. There was no sign of SSX2NORD degradation in the soluble cytoplasmic fraction. It is important to note that SSX2NORD was found in the culture supernatant at a concentration of 2.1 mg L−1, compared to results for strain MS-H, which only had a minute amount of degraded SSX2 in its culture supernatant. Therefore, removal of the NLS region led to reduced intracellular accumulation and increased secretion of SSX2 compared to the full-length SSX2 construct.
Effect of deletion of C-terminal NLS on SSX2 secretion. Western blot of different cell fractions of MS-H and MN-H strains that have been induced for SSX2 and SSX2NORD secretion, respectively, in BMMY medium for 16 h. M, molecular weight marker; S, SSX2 standard; 1, membrane-associated fraction; 2, cytoplasmic fraction; 3, culture supernatant fraction
SSX2 is not targeted to the nucleus, but accumulates within the ER
Since there are two competing signal peptides, the N-terminal alpha-mating factor secretion signal and the C-terminal NLS on SSX2, indirect immunofluorescence was performed to localize it and to determine if its translocation to the nucleus was inhibiting secretion. Unexpectedly, as shown in Fig. 4, most of the SSX2 did not co-localize with nucleus, indicating that the C-terminal NLS is not functioning to direct it to the nucleus. In addition, as shown in Fig. 4B, SSX2 accumulated inside the cell, suggesting that SSX2 is possibly retained inside the ER. Confocal microscopy was performed to co-localize intracellular SSX2 with the ER-resident protein Kar2p. SSX2 did co-localize with Kar2p and mainly accumulated inside the ER (Fig. 5). Thus, SSX2 was targeted to the ER by its N-terminal secretion signal. For the MN-H strain, some SSX2NORD also co-localized with Kar2p, but it was not mainly accumulated in the ER and was instead visualized as small foci throughout the cell (Fig. 6). Some of the SSX2NORD on the cell periphery may be within secretory vesicles that are in the process of secretion.
Confocal microscopy analysis of the co-localization of SSX2 and ER-luminal protein Kar2p in strain MS-H. SSX2 was stained with Alexa flour 488 conjugated with goat anti-mouse antibody (a) and Kar2p was stained with Alexa flour 546 conjugated with goat anti-rabbit antibody (b). Images were overlapped in c
Confocal microscopy analysis of the co-localization of SSX2NORD and ER-luminal protein Kar2p in strain MN-H. SSX2NORD was stained with Alexa flour 488 conjugated with goat anti-mouse antibody (A) and Kar2p was stained with Alexa flour 546 conjugated with goat anti-rabbit antibody (B). Images were overlapped in C
Unfolded protein response is induced in both SSX2 and SSX2NORD expression strains
It is known that accumulation of unfolded and misfolded proteins in the ER induces an unfolded protein response (UPR), triggering up-regulation of various ER-resident proteins, including Kar2p, calnexin, and protein disulfide isomerase (PDI) (Shamu et al. 1994; Chawla and Niwa 2005). Cultures of MS-H, MN-H, KM71 H strain, and KM71H with addition of 10 mM Dithiothreitol (DTT) were induced in BMMY medium for 16 h, and intracellular Kar2p levels were compared. DTT is an ER stressor that inhibits correct disulfide bond formation, thereby inducing the UPR in cells (Resina et al. 2007) and was used as a positive control. After induction, Kar2p levels in MS-H and MN-H strains were 3.5- and 2.3-fold higher than observed in the negative control (KM71H), respectively, indicating that the UPR was initiated in both strains (Fig. 7). Nevertheless, UPR was more intensively induced in the MS-H strain. The Kar2p level in the MS-H strain was similar to the positive control where its Kar2p level was 3.9 fold higher than the negative control.
Intracellular membrane-associated Kar2p levels in MS-H, MH-H, and KM71H strains after methanol induction for 16 h. Cells were normalized before loading on immunoblots. Image analysis was performed using immunoblots from three independent experiments. Kar2p level of KM71H strain was normalized as one fold and compared its level with other strains. Addition of 10 mM DTT to KM71 strain was used as a positive control for measuring UPR response
Fusion of the SSX2 NLS inhibits the secretion of protein in P. pastoris
To further characterize the role of the NLS in recombinant secretion in P. pastoris, a single chain antibody, A33scFv, which is normally secreted, was tagged with SSX2 NLS at its C-terminus (A33scFvNLS). Its secretion was compared with the native A33scFv after induction in BMMY medium for 16 h. Both strains have one expression cassette integrated (Table 1). As indicated in Fig. 8, the secretion of A33scFv decreased by 70% when tagged with NLS. These data suggested that the NLS of SSX2 may decrease recombinant protein secretion in P. pastoris.
Tagging NLS to A33scFv reduced its secretion. After induction for 16 h in BMMY medium, culture supernatant of M33 and M33NLS were collected, analyzed, and compared on the Coomassie-stained sodium dodecyl sulfate-polyacrylamide gel electrophoresis. A33scFv level of M33 strain was normalized as one fold and compared its level with A33scFvNLS of M33NLS strain
Scale-up production of SSX2 without NLS
SSX2NORD production in P. pastoris was scaled to a 2-L fermentor. However, when the MN-H strain was cultivated at 25 °C, pH 6, and induced for 48 h, less than 1.3 mg mL−1 of SSX2NORD was found in the culture supernatant, and SSX2NORD was detected intracellularly (Fig. 9A). It was possible that secreted SSX2NORD was not stable in the culture supernatant and was susceptible to degradation under the aforementioned conditions. To improve SSX2NORD stability in the culture supernatant, the cultivation temperature was changed from 25 °C to 16 °C to decrease the protease activity in the culture media (Jahic et al. 2006). The corresponding SDS-polyacrylamide gel electrophoresis and immunoblot of the samples from these fermentation conditions are shown in Fig. 9B. The stability of SSX2NORD in the culture supernatant was improved significantly by lowering the cultivation temperature. The production yields of SSX2NORD in the culture supernatant at 72, 102, and 120 h were 3.5, 28.6, and 21.6 mg L−1, respectively. The fermentation profile of SSX2NORD at 16 °C, pH 6.0 is shown in Fig. 10. The batch phase was continued for 48 h until the glycerol was exhausted, which was indicated by a sharp rise in the dissolved oxygen and a decrease in agitation. The methanol feed then started at a steady-state concentration of 1 g L−1 until the end of the fermentation. In this process, after 120 h cultivation, the WCW of strain MN-H reached 280 mg mL−1, and the SSX2NORD was produced at a yield of 21.6 mg L−1 in the culture media.
Effect of different cultivation temperatures on SSX2NORD secretion in fermentor cultures. a Western blot of different cell fractions of MN-H cultivated at 25 °C after induction for 48 h. b Coomassie-stained sodium dodecyl sulfate-polyacrylamide gel electrophoresis and (c) western blot of culture supernatant of strain MN-H cultivated at 16 °C. M, molecular weight marker; S, SSX2 standard; 1, membrane-associated fraction; 2, cytoplasmic fraction; 3, supernatant. In (b) and (c), lane 3 to 6 indicate samples taken at different time points
Fermentation profile of strain MN-H cultivated at 16 °C using a 2-L fermentor. During the fermentation, dissolved oxygen (D.O.) was maintained over 40% of air saturation, and pH was maintained around 6. After induction, methanol concentration was maintained at 1 g L−1. Agit, agitation speed; wcw, wet cell weight
Discussion
P. pastoris is a robust system to express and secrete recombinant proteins. However, many studies including our unpublished data have suggested that some proteins are poorly secreted by P. pastoris (Pechan et al. 2004; Vassileva et al. 2001). In this study, when SSX2, a nuclear protein and cancer antigen, was expressed and secreted in P. pastoris, the protein accumulated intracellularly even with the addition of the alpha-mating factor secretion signal. Several approaches have been explored in our lab to improve SSX2 secretion including changes in cultivation conditions (e.g., different cultivation temperature, media, and pH) and different secretion signals. However, none of these approaches improved SSX2 secretion (data not shown), indicating that the apparent poor secretion is inherent to the protein itself. In addition, increased copy number only led to more SSX2 accumulation inside the cells, thus showing that SSX2 is also one of the recombinant proteins that is poorly secreted by P. pastoris despite the addition of a secretion signal.
It has been demonstrated that SSX2 has a bipartite NLS at its C-terminus which is responsible for its translocation from the cytoplasm to the nucleus similar to that observed for the translocation for the SYT-SSX2 fusion protein (dos Santos et al. 2000). A study that investigated secretion of human tumor suppressor p53 in P. pastoris has hypothesized that the NLS at the C-terminus of p53 might cause its intracellular accumulation (Abdelmoula-Souissi et al. 2007). It is reasonable to speculate that the C-terminal NLS of SSX2 may inhibit its secretion by translocation of SSX2 to the nucleus. The SSX2 gene lacking NLS region (SSX2NORD) was constructed, expressed, and secreted in P. pastoris. Deletion of the NLS improved SSX2 secretion from no observed secretion to 2.1 mg L−1 (Fig. 4). Immunofluorescence, however, showed that intracellular SSX2 did not localize in the nucleus, suggesting that the NLS does not function to direct it there. In fact, most of the SSX2 was translocated to and accumulated intracellularly mainly in the ER (Fig. 5). The mechanism of improved secretion of SSX2NORD was further investigated.
Overexpression of heterologous proteins in eukaryotic cells can cause saturation of the secretory pathway, mostly accumulating proteins inside the ER (Smith and Robinson 2002). The accumulation of misfolded proteins inside the ER or an increase in protein traffic through the ER will induce the UPR to help the cells to cope with the ER stress by up-regulating levels of ER chaperones (e.g., Kar2P or BiP, calreticulin, and calnexin), and protein-folding enzymes (e.g., PDI and FKB2; Chawla and Niwa 2005). Therefore, an increase in Kar2p level is a marker for UPR activation in yeast and mammalian cells (Kaufman 1999). We found that intracellular Kar2p levels in MS-H were 3.5-fold higher than in the KM71H strain, indicating that UPR was initiated to help cells cope with secretion stress. In addition, degraded SSX2 was found in the cytoplasmic fraction (Fig. 2) of MS-H strain, presumably by ER-associated degradation (ERAD) which dislocates misfolded protein across the ER membrane for degradation by cytosolic proteasomes (Sitia and Braakman 2003). Together, these results suggested while expressing SSX2, it was misfolded and accumulated in the ER, alarming cell to initiate UPR and ERAD responses.
On the other hand, while compared to SSX2, the expression and secretion of SSX2NORD in MN-H strain displayed different cellular responses. Deletion of the NLS improved SSX2 secretion and decreased intracellular SSX2NORD inside the cells. UPR was also induced to a lesser extent in SSX2NORD expressing cells, 2.3-fold higher than the control strain, which is possibly caused by intracellularly accumulated SSX2NORD (Fig. 3). These results indicated that deletion of the NLS improved SSX2 folding in the ER, thus facilitating its secretion. Nevertheless, significant amount of SSX2NORD was still found in the intracellular fraction. This suggests that even though the secretion was improved in SSX2NORD, ER folding or post-ER translocation could still be a limiting step for SSX2NORD secretion. This idea is also supported by the confocal microscopy, where some of the SSX2NORD accumulated in cells may be in vesicles. These vesicles might be secretory vesicles waiting for secretion. However, it is also possible that some vesicles were targeted to the vacuole for degradation, as seen in other recombinant protein secretion (Hong et al. 1996) which explains the low level secretion of SSX2NORD.
To further characterize the role of the NLS in recombinant protein secretion in P. pastoris, the NLS of SSX2 was tagged to a well-secreted protein, single chain antibody A33scFV, which has been demonstrated to reach a level of 4 g L−1 in fermentor cultures (Damasceno et al. 2007). The fusion of the SSX2 NLS reduced the secretion of A33scFv by 70%, compared to the native A33scFv. The mRNA expression levels of A33scFv and A33scFvNLS in M33 and M33NLS strains did not have appreciable difference (data not shown). We conclude that the NLS plays a negative role in recombinant protein secretion in P. pastoris. It is known that the NLS of nuclear proteins are exposed on the protein surface in order to interact and be recognized by importin alpha and importin beta for successful nuclear transportation (Boulikas 1993; Gorlich 1997). In our case, it is possible that exposed NLS on the surface inhibits SSX2 and A33scFvNLS folding in the ER, thus preventing or decreasing their secretion, respectively.
The SSX2NORD produced in this study can be used as a DTH skin test reagent as well as another format of SSX2 cancer vaccine. Several studies have identified that the important epitopes of SSX2 are located at the N-terminus of the protein. The region of 41–49 amino acid residues of the SSX2 protein is recognized by the tumor reactive CD8+ T lymphocytes in association with MHC class I allele HLA-A2. The region of 19–59 amino acid residues of SSX2 was later identified to be recognized by specific CD4+ T lymphocytes in association with MHC class II alleles HLA-DR3, 4 and 11, and DPI (Ayyoub et al. 2002, 2004, 2005; Neumann et al. 2004). Because the NLS region is downstream of the two important epitopes described above, its removal should not impact its utility as another format of a SSX2 cancer vaccine.
Since there are two potential applications of SSX2NORD, the production of SSX2NORD was scaled-up in a 2-L bench-top fermentor. Surprisingly, there was only a minute amount of SSX2NORD found in the culture supernatant when cells were cultivated at 25 °C, pH 6, while intracellular SSX2NORD was observed at all times. It is well known that high cell density fermentation of P. pastoris is accompanied by significant cell lysis, releasing intracellular proteins into the culture medium, including vacuolar proteases that cause secreted recombinant protein degradation (Jahic et al. 2006). It is possible that secreted SSX2NORD was degraded during the fermentation process. To minimize proteolytic instability of secreted SSX2NORD, the MN-H strain was cultivated at 16 °C to decrease protease activity (Jahic et al. 2003). By changing the cultivation temperature from 25 °C to 16 °C, the stability of SSX2NORD in the supernatant was improved significantly; however, signs of degradation were still observed as SSX2 degradation products were observed in the western blot (Fig. 9C). These data indicate that decreasing the culture temperature can improve SSX2NORD stability, which is likely due to decreasing protease activity in the culture supernatant. In this process, it was estimated that a yield of 21.6 mg L−1 of SSX2NORD was secreted in the media.
In this study, we have demonstrated that SSX2 containing NLS is minimally secreted in P. pastoris but mainly accumulates in the ER, and it is not translocated to the nucleus. Deletion of the NLS from SSX2 improves its folding, thus improving its secretion. Even though the secretion of SSX2NORD is improved, folding inside the ER or post-ER translocation might still be a limiting step for its secretion. Finally, a protocol to produce SSX2NORD in a 2-L fermentor was developed, and the secreted SSX2NORD can be used as a DTH skin test reagent of a SSX2 cancer vaccine.
References
Abdelmoula-Souissi S, Rekik L, Gargouri A, Mokdad-Gargouri R (2007) High-level expression of human tumour suppressor P53 in the methylotrophic yeast: Pichia pastoris. Protein Expr Purif 54:283–288
Ayyoub M, Stevanovic S, Sahin U, Guillaume P, Servis C, Rimoldi D, Valmori D, Romero P, Cerottini JC, Rammensee HG, Pfreundschuh M, Speiser D, Levy F (2002) Proteasome-assisted identification of a SSX-2-derived epitope recognized by tumor-reactive CTL infiltrating metastatic melanoma. J Immunol 168:1717–1722
Ayyoub M, Rimoldi D, Guillaume P, Romero P, Cerottini JC, Valmori D, Speiser D (2003) Tumor-reactive, SSX-2-specific CD8+ T cells are selectively expanded during immune responses to antigen-expressing tumors in melanoma patients. Cancer Res 63:5601–5606
Ayyoub M, Hesdorffer CS, Montes M, Merlo A, Speiser D, Rimoldi D, Cerottini JC, Ritter G, Scanlan M, Old LJ, Valmori D (2004) An immunodominant SSX-2-derived epitope recognized by CD4+ T cells in association with HLA-DR. J Clin Invest 113:1225–1233
Ayyoub M, Merlo A, Hesdorffer CS, Speiser D, Rimoldi D, Cerottini JC, Ritter G, Chen YT, Old LJ, Stevanovic S, Valmori D (2005) Distinct but overlapping T helper epitopes in the 37–58 region of SSX-2. Clin Immunol 114:70–78
Bader N, Jung T, Grune T (2007) The proteasome and its role in nuclear protein maintenance. Exp Gerontol 42:864–870
Boulikas T (1993) Nuclear localization signals (NLS). Crit Rev Eukaryot Gene Expr 3:193–227
Bricard G, Bouzourene H, Martinet O, Rimoldi D, Halkic N, Gillet M, Chaubert P, Macdonald HR, Romero P, Cerottini JC, Speiser DE (2005) Naturally acquired MAGE-A10- and SSX-2-specific CD8+ T cell responses in patients with hepatocellular carcinoma. J Immunol 174:1709–1716
Cereghino GP, Cereghino JL, Ilgen C, Cregg JM (2002) Production of recombinant proteins in fermenter cultures of the yeast Pichia pastoris. Curr Opin Biotechnol 13:329–332
Chawla A, Niwa M (2005) The unfolded protein response. Curr Biol 15:R907
Clark J, Rocques PJ, Crew AJ, Gill S, Shipley J, Chan AM, Gusterson BA, Cooper CS (1994) Identification of novel genes, SYT and SSX, involved in the t(X;18)(p11.2;q11.2) translocation found in human synovial sarcoma. Nat Genet 7:502–508
Damasceno LM, Pla I, Chang HJ, Cohen L, Ritter G, Old LJ, Batt CA (2004) An optimized fermentation process for high-level production of a single-chain Fv antibody fragment in Pichia pastoris. Protein Expr Purif 37:18–26
Damasceno LM, Anderson KA, Ritter G, Cregg JM, Old LJ, Batt CA (2007) Cooverexpression of chaperones for enhanced secretion of a single-chain antibody fragment in Pichia pastoris. Appl Microbiol Biotechnol 74:381–389
Davis ID, Chen W, Jackson H, Parente P, Shackleton M, Hopkins W, Chen Q, Dimopoulos N, Luke T, Murphy R, Scott AM, Maraskovsky E, McArthur G, MacGregor D, Sturrock S, Tai TY, Green S, Cuthbertson A, Maher D, Miloradovic L, Mitchell SV, Ritter G, Jungbluth AA, Chen YT, Gnjatic S, Hoffman EW, Old LJ, Cebon JS (2004) Recombinant NY-ESO-1 protein with ISCOMATRIX adjuvant induces broad integrated antibody and CD4+ and CD8+ T cell responses in humans. Proc Natl Acad Sci U S A 101:10697–10702
Don RH, Cox PT, Wainwright BJ, Baker K, Mattick JS (1991) ‘Touchdown’ PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res 19:4008
dos Santos NR, de Bruijn DR, Kater-Baats E, Otte AP, van Kessel AG (2000) Delineation of the protein domains responsible for SYT, SSX, and SYT-SSX nuclear localization. Exp Cell Res 256:192–202
Gorlich D (1997) Nuclear protein import. Curr Opin Cell Biol 9:412–419
Gure AO, Tureci O, Sahin U, Tsang S, Scanlan MJ, Jager E, Knuth A, Pfreundschuh M, Old LJ, Chen YT (1997) SSX: a multigene family with several members transcribed in normal testis and human cancer. Int J Cancer 72:965–971
Higgins DR, Cregg JM (1998) Introduction to Pichia pastoris. Methods Mol Biol 103:1–15
Hohenblum H, Borth N, Mattanovich D (2003) Assessing viability and cell-associated product of recombinant protein producing Pichia pastoris with flow cytometry. J Biotechnol 102:281–290
Hong E, Davidson AR, Kaiser CA (1996) A pathway for targeting soluble misfolded proteins to the yeast vacuole. J Cell Biol 135:623–633
Huang C, Chen RH, Vannelli T, Lee F, Ritter E, Ritter G, Old LJ, Batt CA (2007) Expression and purification of the cancer antigen SSX2: a potential cancer vaccine. Protein Expr Purif 56:212–219
Jahic M, Wallberg F, Bollok M, Garcia P, Enfors SO (2003) Temperature limited fed-batch technique for control of proteolysis in Pichia pastoris bioreactor cultures. Microb Cell Fact 2:6
Jahic M, Veide A, Charoenrat T, Teeri T, Enfors SO (2006) Process technology for production and recovery of heterologous proteins with Pichia pastoris. Biotechnol Prog 22:1465–1473
Kaufman RJ (1999) Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev 13:1211–1233
Macauley-Patrick S, Fazenda ML, McNeil B, Harvey LM (2005) Heterologous protein production using the Pichia pastoris expression system. Yeast 22:249–270
Nagai M, Tanaka S, Tsuda M, Endo S, Kato H, Sonobe H, Minami A, Hiraga H, Nishihara H, Sawa H, Nagashima K (2001) Analysis of transforming activity of human synovial sarcoma-associated chimeric protein SYT-SSX1 bound to chromatin remodeling factor hBRM/hSNF2 alpha. Proc Natl Acad Sci U S A 98:3843–3848
Neumann F, Wagner C, Stevanovic S, Kubuschok B, Schormann C, Mischo A, Ertan K, Schmidt W, Pfreundschuh M (2004) Identification of an HLA-DR-restricted peptide epitope with a promiscuous binding pattern derived from the cancer testis antigen HOM-MEL-40/SSX2. Int J Cancer 112:661–668
Okamura K, Kimata Y, Higashio H, Tsuru A, Kohno K (2000) Dissociation of Kar2p/BiP from an ER sensory molecule, Ire1p, triggers the unfolded protein response in yeast. Biochem Biophys Res Commun 279:445–450
Pechan T, Ma PW, Luthe DS (2004) Heterologous expression of maize (Zea mays L.) Mir1 cysteine proteinase in eukaryotic and prokaryotic expression systems. Protein Expr Purif 34:134–141
Petrovsky N, Brusic V (2004) Virtual models of the HLA class I antigen processing pathway. Methods 34:429–435
Resina D, Bollok M, Khatri NK, Valero F, Neubauer P, Ferrer P (2007) Transcriptional response of P. pastoris in fed-batch cultivations to Rhizopus oryzae lipase production reveals UPR induction. Microb Cell Fact 6:21
Scanlan MJ, Gure AO, Jungbluth AA, Old LJ, Chen YT (2002) Cancer/testis antigens: an expanding family of targets for cancer immunotherapy. Immunol Rev 188:22–32
Shamu CE, Cox JS, Walter P (1994) The unfolded-protein-response pathway in yeast. Trends Cell Biol 4:56–60
Sitia R, Braakman I (2003) Quality control in the endoplasmic reticulum protein factory. Nature 426:891–894
Smith JD, Robinson AS (2002) Overexpression of an archaeal protein in yeast: secretion bottleneck at the ER. Biotechnol Bioeng 79:713–723
Tureci O, Sahin U, Schobert I, Koslowski M, Scmitt H, Schild HJ, Stenner F, Seitz G, Rammensee HG, Pfreundschuh M (1996) The SSX-2 gene, which is involved in the t(X;18) translocation of synovial sarcomas, codes for the human tumor antigen HOM-MEL-40. Cancer Res 56:4766–4772
van der Bruggen P, Van den Eynde BJ (2006) Processing and presentation of tumor antigens and vaccination strategies. Curr Opin Immunol 18:98–104
Vassileva A, Chugh DA, Swaminathan S, Khanna N (2001) Expression of hepatitis B surface antigen in the methylotrophic yeast Pichia pastoris using the GAP promoter. J Biotechnol 88:21–35
Acknowledgment
We would like to thank Dr. Charles K. Barlowe from Dartmouth College for providing the anti-Kar2p for our Kar2p analyses. This project was funded by the Ludwig Institute for Cancer Research.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Huang, CJ., Anderson, K.A., Damasceno, L.M. et al. Improved secretion of the cancer-testis antigen SSX2 in Pichia pastoris by deletion of its nuclear localization signal. Appl Microbiol Biotechnol 86, 243–253 (2010). https://doi.org/10.1007/s00253-009-2275-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-009-2275-2