Abstract
In a changing scenario of food habits being associated with wellness factors through the concepts of probiotics and prebiotics, an attempt has been made to characterize on molecular basis, the desirable benefits associated with natural isolates of lactic acid bacteria, bifidobacteria, and yeasts. From a diverse range of foods and related samples, based on conventional microbiological protocols, three well-characterized natural isolates of Lactobacillus plantarum MTCC 5422, Bifidobacterium adolescentis MTCC 5423 and Saccharomyces cerevisiae MTCC 5421 were selected. The cultures of L. plantarum and B. adolescentis showed positive polymerase chain reaction (PCR) amplification with oligonucleotide primers targeting genus-specific 16 S rRNA for Lactobacillus and fructose-6-phosphate phosphoketolase for Bifidobacterium. Similarly, species-specific positive amplification in PCR was observed with primers of phytase (acid phosphatase) in S. cerevisiae and α-d-galactosidase and bile salt hydrolase in L. plantarum and B. adolescentis. The cultures of L. plantarum and B. adolescentis exhibited a broad spectrum antibacterial activity against selected foodborne pathogenic bacterial species and tolerance to acid and bile. Gene sequence of respective PCR-amplified products confirmed the genetic identity of the isolated cultures as L. plantarum and B. adolescentis showing 99% homology with the documented sequence of established gene bank.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Use of microorganisms in preparing foods based on milk, cereals, and legumes have been a traditional practice since prehistoric times. In a majority of fermented foods, particularly those of traditional foods of India, which are based on cereals and legumes, the nature of fermentation has been a mixed type with the involvement of lactic acid bacteria (LAB) and yeasts. With the growing interest in health consciousness and idea of health promotion without associated health risks, the concept of probiotic foods has attracted much attention. In this context, probiotics, prebiotics, and synbiotics all have a significant role. Probiotics are live microbial additions to the diet; prebiotics are food components that have a selective metabolism in the hind gut, while synbiotics are combinations of the two approaches. A number of Lactobacillus spp., Bifidobacteria spp., and Saccharomyces spp. have been in use as desirable cultures, so as to act as beneficial food supplements and contribute to a balanced intestinal microflora for human wellness (Ljungh and Wadstrom 2006; Masco et al. 2007; Woodmansey 2007). These beneficial effects have been very much diversified ranging from suppression of harmful bacteria in large intestine to immune system activation effects (Fooks et al. 1999). Considering the potential of beneficial cultures, food industry has been looking toward well-characterized cultures with defined probiotic and other desirable attributes (Masco et al. 2007).
Cereals and legumes are considered as effective substrates for the production of probiotic-incorporated functional food, as they can be used as a source of nondigestible carbohydrates which stimulate the growth of lactobacilli and bifidobacteria, both endogenous and supplemented, and contain water soluble fibres like β-glucan, arabinoxylan, galacto-oligosaccharides, and fructo-oligosaccharides, which are prebiotics (Swennen et al. 2006). They are fermented by several groups of bacteria in the large intestine, yielding a variety of fermentation products, particularly short-chain fatty acids (SCFA). The resulting SCFA are known to provide an acidic environment in the large intestine, which stimulates the proliferation of probiotic cultures (Roberfroid 1997; Macfarlane et al. 2006).
In general, the conventional protocols of morphological, cultural, and biochemical characterization of microbial cultures are in use to identify probiotic microbial cultures. Often, these attributes do not clearly express in the targeted microorganisms due to several causative factors. In such circumstances, focus has been on molecular characterization, particularly by polymerase chain reaction (PCR) methods using primers specific to 16 S rRNA and targeted genes (Matsuki et al. 2003; Mullie et al. 2003; Berthoud et al. 2005; Kwon et al. 2005). At the same time, phylogenetic relatedness among probiotic cultures are being deduced through sequencing of PCR amplifications of target genes of interest (Coeuret et al. 2003; Rekha et al. 2006).
In the background of a predominant mixed-type microbial fermentation, the objective of present study was to assess the occurrence of native isolates of yeasts, LAB, and bifidobacteria in foods and feeds and characterize them on molecular basis for associated beneficial attributes with a focus on phylogenetic relatedness.
Materials and methods
Samples
In all, a total of 65 samples were analyzed in this study, which included juices of grapes, pineapple, apple and tomato, cow milk, curds, cereal/legume-based fermented batters and doughs of traditional foods, concentrated milk sweets, mother’s milk, feces of infants and rabbits. Samples were collected locally under sterile conditions and brought to the laboratory within 60 min of collection and subjected to analysis.
Reference cultures
These included (1) reference cultures of Staphylococcus aureus FRI 722, Bacillus cereus F 4810, Listeria monocytogenes Scott A obtained through courtesy from Dr. S. Notermans, Public Health Laboratory, The Netherlands, Dr. J.H. Kramer, Central Public Health Laboratory, United Kingdom, and Dr. Arun K. Bhunia, Purdue University, USA and (2) strain of Yersinia enterocolitica MTCC 859 procured from Microbial Type Culture Collection, Institute of Microbial Technology, Chandigarh, India. The individual bacterial strains were maintained at 4°C on BHI agar (HiMedia Dehydrated Laboratories, Mumbai, India) slants and subcultured at regular intervals of 15 days. The cultures were propagated in BHI broth at 37°C, prior to use in experiments.
Isolation and characterization of native microbial cultures
Requisite aliquots of appropriate serial dilutions of samples were plated for the respective types of microbial cultures as desired in the present study. For obtaining yeast isolates, dilutions were surface plated on sterile pre-poured plates of potato dextrose agar (HiMedia), and inoculated plates were incubated for 48–72 h at 25°C. For lactic acid bacteria (LAB) isolates, dilutions were pour plated with sterile deMan, Rogosa, and Sharpe (MRS) Lactobacillus agar (HiMedia) and incubated for 24–48 h at 37°C. Similarly, bifidobacteria isolates were obtained by pour plating of sample dilutions with MRS Lactobacillus agar supplemented with 0.05% cysteine hydrochloride and incubated for 48–72 h at 37° under anaerobic condition in anaerobic jar (Don Whitley, UK) using anaerobic gas pack (HiMedia). For isolation of bifidobacteria, the diluent, 0.85% saline was also supplemented with 0.05% cysteine hydrochloride.
The presumptive isolates thus obtained were purified by established protocols using the respective isolation media. Isolated cultures were maintained at 6°C on the slants or broth of respective media with subculturing at regular intervals of 30 days for use in further studies. As a means to select all the presumptive isolates under the scope of present study, initially, conventional method of identification based on morphological, cultural, and biochemical characteristics was followed (Cappuccino and Sherman 2004). This would avoid the probable chances of loosing isolates which may fail to exhibit amplification of specific 16 S rRNA PCR products.
Fructose-6-phosphate phosphoketolase (F-6-ppk) is the key enzyme of the so-called “bifidobacteria shunt,” a unique hexose metabolism that occur via the phosphoketolase pathway in Bifidobacterium is used as a key biochemical character in the identification protocols. Sonicated cells of overnight-grown presumptive native isolates of Bifidobacterium were subjected to the qualitative assay of F-6-ppk following the documented method (Orban and Patterson 2000). Presumptive isolates of LAB and Bifidobacteria were identified to their respective genus and species level according to the Bergey’s Manual of Systematic Bacteriology (Kandler and Weiss 1986). Native isolates of yeasts were identified to their respective genus and species following morphological, cultural, and biochemical characteristics and the identification key described earlier (Deak and Beuchat 1996).
Alpha-d-galactosidase activity
Activity of intracellular α-d-galactosidase in native characterized isolates of LAB and Bifidobacterium spp. was determined in cell-free extract obtained from sonicated cells of 36-h-old MRS Lactobacillus culture broth. α-d-galactosidase activity was assayed in MCIlvaine buffer by measuring the absorbance at 400 nm of the p-nitrophenol (PNP) released by the action of the enzyme upon its specific substrate, P-nitrophenyl-α-d-galactopyranoside (PNPG). One unit of enzyme was defined as the amount of enzyme that released 1 µmol of PNP from the substrate PNPG per milliliter per minute under the assay conditions (Church et al. 1980).
Antibacterial activity
Extracellular culture supernatants of isolates of LAB and bifidobacteria were assayed for antibacterial activity against foodborne pathogenic bacterial cultures of B. cereus, S. aureus, L. monocytogenes, and Y. enterocolitica, individually, by agar well diffusion method as described earlier (Schillinger and Lucke 1989; Varadaraj et al. 1993). Antibacterial activity in the culture supernatants was defined as the reciprocal of the highest dilution showing definite inhibition of indicator bacterial strains and expressed as activity units per millimetre (AU/ml).
Bile salt and acid tolerance
Native isolates of L. plantarum MTCC 5422 and B. adolescentis MTCC 5423 were assessed for their ability to grow in the presence of bile salt concentrations of 0.15%, 0.3%, and 0.45% (w/v) added in MRS Lactobacillus broth (HiMedia) and at pH levels ranging from 2.0 to 5.5 with a difference of 0.5 U using 1 N HCl or 1 N NaOH in MRS broth, according to the method described earlier (Gotcheva et al. 2002). Prepared broth tubes were inoculated with 1% (v/v) cell suspension of 18 h old culture broth, incubated at 37°C for 48 h and absorbance read at 560 nm in a UV-VIS Spectrophotometer (Genesys 5, Milton Roy, USA) at intervals of 12 h. MRS broth without added bile salt and of pH 6.5, respectively, served as controls.
Bile salt hydrolase activity
Bile salt hydrolase activity in the native isolates of L. plantarum MTCC 5422 and B. adolescentis MTCC 5423 was determined in two steps assay (Tanaka et al. 2000), wherein the amount of amino acids liberated from conjugated bile salts used as a substrate. One unit of BSH activity was defined as the amount of enzyme which liberates 1 µmol of amino acids from the substrate per minute.
Phytase activity
Extracellular phytase activity in native identified isolates of yeasts was determined in 48-h-old potato dextrose culture broth supernatants. In the activity assay, inorganic phosphate released from sodium phytate used as a substrate was measured following the earlier described established procedure (Heinonen and Lahti 1981). One unit of the enzyme activity was defined as the amount of phytase required to liberate 1 µmol of inorganic phosphate per minute under defined assay conditions.
PCR detection for species specificity and probiotic and prebiotic attributes
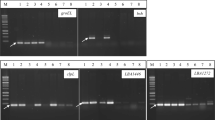
In the present study, the target genes included were those of phytase (pho) for yeasts; 16 S rRNA, α-d-galactosidase (melA), and bile salt hydrolase (bsh) for Lactobacillus spp.; fructose-6-phosphate phosphoketolase (F-6-ppk), bile salt hydrolase (bsh), and α-d-galactosidase (α-D-gal) for Bifidobacterium spp. The nucleotide sequences and amplification conditions of these PCR primers are presented in Table 1. The synthesized primers were obtained from a commercial company (Sigma Aldrich, Bangalore, India). The total genomic DNA of individual isolates was extracted from 1.5-ml aliquots of overnight culture broth by boiling in Triton-X (Wang et al. 1997). The dried DNA was then dissolved in TE buffer (pH 8.0) and used as template DNA for amplifying the target genes using the respective primers. PCR was performed with the individual characterized isolates of yeasts, Lactobacillus and Bifidobacterium according to the procedure described earlier (Klocke and Mundt 2004). PCR amplification was performed in an automated DNA thermal Cycler (Eppendorf, Master Cycler, Cedex, France) following the conditions as detailed in Table 1. The PCR products were run in 1.2% agarose gels in Tris acetate buffer for 1.5 h at 100 V and stained in 0.5 ųg ml−1 ethidium bromide solution (Sambrook and Russel 2001) and documented in Gel Documentation System (Vilber Lourmat, France).
Nucleotide sequence analysis of amplified PCR product and alignment
Considering the positive results with PCR primers and probiotic and prebiotic activities, one isolate each of L. plantarum MTCC 5422 and B. adolescentis MTCC 5423 were selected for this experimental trial. The resultant PCR amplified products with the three respective primers, 16 S rRNA, α-d-gal, and bsh were purified using commercially available PCR purification Spin Kit (HiPur A, HiMedia Laboratories, Mumbai, India) and subjected to sequence analysis by a commercial company (Sigma Aldrich, Bangalore, India). The resulting nucleotide sequences were subjected to BLAST programme of NCBI (online access) to assess the percent homology with closely related strains/variants documented in Gene Bank databases. A similarity network tree was constructed based on force type of neighbour-joining method from the same program.
Results
Native cultures of Lactobacillus sp. and Bifidobacterium sp. with probiotic attributes
Characterization and identification of native isolates
Native isolates of LAB and bifidobacteria were obtained from food and related samples analyzed in this study. In all, from a total of 70 presumptive isolates of LAB and bifidobacteria, 12 isolates were identified to the species of Lactobacillus, Leuconsotoc, and Pediococcus and two isolates to those of Bifidobacterium based on conventional characterization protocols. The remaining native isolates were characterized to their respective genus level. The occurrence of completely identified microbial cultures in different food and related samples is presented in Table 2. Three cultures of LAB were isolated from samples of curd, which is a natural lactic fermented homemade milk product, followed by another three cultures of LAB from naturally fermented batters based on cereal and legumes, which constitute a common base material in the preparation of a variety of traditional foods. Similarly, fruit juices did harbor LAB.
In the background of rare occurrence of bifidobacteria in foods, in the present study, only two well-characterized isolates of Bifidobacterium based on the qualitative assay for fructose-6-phosphate phosphoketolase activity were obtained from feces of breast-fed infant and rabbit. The characterized isolates of LAB and bifidobacteria were designated with specific numbers for convenience of presentation, wherein CFR refers to the host Institute’s name. The native isolates which exhibited potent beneficial attributes were selected and deposited in a publicly accessible culture collection center of India, namely, the Microbial Type Culture Collection at the Institute of Microbial Technology, Chandigarh, India. Among the native isolates obtained in this study, only two isolates, one each of LAB and bifidobacteria were identified as the most potent and used in further experimental trials. These two cultures had the deposition accession details of L. plantarum MTCC 5422 and B. adolescentis MTCC 5423.
As confirmatory evidence and for further selection of isolates, PCR analysis based on 16 S rRNA primers designed for the specificity of genus Lactobacillus resulted in positive amplification with all conventionally characterized Lactobacillus spp. in PCR (data not shown). The same primers failed to show any positive amplification with identified isolates of Leuconostoc mesenteroides subsp. dextranicum CFR 2251, Leuconostoc oenos CFR 2252, and Pediococcus pentosaceus CFR 2253, indicating the high specificity of designed PCR primers for the genus Lactobacillus.
α-d-galactosidase and bile salt hydrolase activities
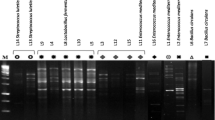
Among the native isolates of LAB assayed for this activity, the only potent culture was that of L. plantarum MTCC 5422, which exhibited an intracellular activity of 6.6 U ml−1, followed by 1.5 U ml−1 by L. fermentum CFR 2196 (Fig. 1). Similarly, another important probiotic property defined for LAB and bifidobacteria has been that of bile salt hydrolase activity. Native isolates of LAB failed to exhibit visible activity in the assay. In order to establish the molecular identity of these two important attributes, all the native well-characterized isolates of LAB were subjected to PCR with primers designed for α-d-galactosidase (melA) and bile salt hydrolase (bsh). The isolate of L. plantarum MTCC 5422 showed positive amplification with melA and bsh in PCR. Similarly, isolates of B. adolescentis MTCC 5423 and B. longum CFR 2202 showed very negligible α-d-galactosidase activity, while bile salt hydrolase activity was to the extent of 0.48 U ml−1. In PCR, the isolate of B. adolescentis MTCC 5423 showed positive amplification with primers designed (Table 1) for fructose-6-phosphate phosphoketolase (F-6-ppk), bile salt hydrolase (bsh), and α-d-galactosidase (aga) in PCR (Fig. 2). In a few of the native isolates of LAB and bifidobacteria, the gel band in PCR was faint for α-d-galactosidase.
Alpha-d-galactosidase activity exhibited by natural isolates of lactic acid bacteria and bifidobacteria 1, L. plantarum MTCC 5422; 2, L. fermentum CFR 2196; 3, L. casei subsp. tolerans CFR 2197; 4, L. casei subsp. pseudoplantarum CFR 2198; 5, L. casei subsp. rhamnosus CFR 2199; 6, L. reuteri CFR 2200; 7, Leu. mesenteroides subsp. dextranicum CFR 2251; 8, Leu. oenos CFR 2252; 9, P. pentosaceus CFR 2253
Antibacterial activity against foodborne pathogenic bacteria
Another important probiotic attribute associated with cultures of LAB and bifidobacteria is that of antibacterial activity toward foodborne pathogenic bacterial species. In the present study, the native isolates exhibited a diversified pattern with respect to antibacterial activity against the four indicator bacterial species (Table 3). The antibacterial activity toward Staphylococcus aureus FRI 722 was very low, and most of the isolates failed to have any positive effect. In terms of activity (AU ml−1), most of the native isolates could inhibit strains of Bacillus cereus F 4810, Listeria monocytogenes Scott A, and Yersinia enterocolitica MTCC 859. Among the native isolates of LAB, L. plantarum MTCC 5422 did exhibit potential antibacterial activity against all the four indicator bacterial strains and could thus be a promising probiotic culture. Similarly, among the two native isolates of Bifidobacterium, B. adolescentis MTCC 5423 appeared to be a potential probiotic culture (Table 3).
Survival/growth in presence of bile salt and acidic conditions
As there is a requirement for probiotic LAB to survive conditions of gastrointestinal tract, simultaneously, the isolates of L. plantarum MTCC 5422 and B. adolescentis MTCC 5423 were being assessed for their ability to survive/grow in presence of bile salts (0.15–0.45%) and under acidic conditions (pH 2.5–4.0). The growth performance of both the native isolates was quite appreciable in the presence of bile salts at concentration levels of 0.15%, 0.3%, and 0.45%, respectively. However, in the presence of 0.45% bile salt, the growth was slightly reduced as against that of control broth medium. Under pH levels of less than 3.5, both the cultures failed to grow, while growth was quite appreciable at pH levels of 4.5 and 5.5 compared with growth at pH 6.5 (Fig. 3 and 4).
Growth response of L. plantarum MTCC 5422 to bile salt concentrations (a) and acid pH levels (b): empty squares MRS broth without bile salt, filled squares with 0.15% bile salt, empty circles with 0.30% bile salt, filled circles with 0.45% bile salt in (a); empty squares MRS broth of pH 6.5; filled squares of pH 5.5; empty circles of pH 4.5; filled circles of pH 3.5 in (b)
Growth response of B. adolescentis MTCC 5423 to bile salt concentrations (a) and acid pH levels (b): empty squares MRS broth without bile salt; filled squares with 0.15% bile salt; empty circles with 0.30% bile salt; filled circles with 0.45% bile salt in (a); empty squares MRS broth of pH 6.5; filled squares of pH 5.5; empty circles of pH 4.5; filled circles of pH 3.5 in (b)
Molecular basis for relatedness/diversity of food isolates of L. plantarum and B. adolescentis
In the present study, resultant partial nucleotide sequences of respective PCR amplicons of 16 S rRNA (385 bases), melA (398 bases), and bsh (422 bases) of L. plantarum MTCC 5422 and F-6-ppk (468 bases) of B. adolescentis MTCC 5423 were screened against BLAST algorithm of NCBI (online access). Based on the resultant outputs, force type neighbour-joining phylogram were generated for partial gene sequences of 16 S rRNA, melA, bsh, and F-6-ppk, respectively (Fig. 5). The phylogenetic tree generated for 16 S rRNA showed 98% homology with other cultures appearing in the generated tree (Fig. 5a). Similarly, the phylogenetic tree generated for the gene of α-d-galactosidase revealed that the native isolate of L. plantarum MTCC 5422 showed 99% homology with a strain of L. plantarum, while it was 78–81% with L. reuteri and L. salivarius (Fig. 5b). However, in case of the tree constructed for bile salt hydrolase revealed 97% homology with three strains of L. plantarum (Fig. 5c). Similarly, the phylogenetic tree of F-6-ppk in Bifidobacterium spp. revealed that the native isolate of B. adolescentis MTCC 5423 had 97% homology with another strain of B. adolescentis and almost 90% with other species of Bifidobacterium (Fig. 5d).
Neighbour-joining phylogenetic tree of representatives of L. plantarum MTCC 5422 based on nucleotide sequence analysis of PCR amplified products with 16 S rRNA primers (a), melA primers (b), and bsh primers (c) and for B. adolescentis MTCC 5423 based on F-6-ppk primers (d). The percent homology expressed is in relation with the two respective native food isolates
Native yeast cultures with phytase activity
In the case of native cultures of yeasts, 22 isolates were obtained on primary isolation and as in case of LAB, conventional characterization by physiological and biochemical attributes resulted in identification of nine cultures to their genus and species level. The remaining isolates were identified to their genus level. Two yeast cultures were isolated from samples of curd and five isolates from naturally fermented batters based on cereal and legumes. Fruit juices did harbor yeasts (Table 2). As the study was aimed at assessing beneficial attributes associated with the microbial cultures, the identified yeast cultures when assayed for phytase activity revealed that an isolate of Saccharomyces cerevisiae MTCC 5421 exhibited 184 U ml−1 of extracellular phytase activity, while isolates of Schizosaccharomyces malidevorans CFR 523 and Sch. octosporus CFR 524 could elaborate 162 and 118 U/ml, respectively. Through the use of acid phosphatase (pho) primers (Table 1), of the nine identified yeast cultures, only six of them gave a positive amplification in PCR. The native cultures of Candida agrestis CFR 527 and Rhododsporidium minuta CFR 529 could not give any positive amplification, although these cultures could elaborate very less quantity of phytase to an extent of 25–28 U ml−1 in assay, as the primers designed was for species-specific phytase gene in S. cerevisiae.
Discussion
Probiotic and prebiotic attributes among LAB and bifidobacteria
Microbial cultures are being increasingly evaluated for their desirable attributes, so as to incorporate in foods to derive benefits and wellness to human health. Mixed fermentation with the involvement of LAB and yeasts are known to occur more commonly in a majority of traditional foods. In the present study, a diverse range of foods and related matter were assessed for the prevalence of lactic acid bacteria and bifidobacteria with probiotic attributes and also for native cultures of yeasts with attributes of reducing nonfunctional food constituents. Further, the native cultures were characterized for the selected attributes by PCR methods and based on these inputs, attempts were made to outline phylogenetic patterns with related genera/species. In the case of identification of bifidobacteria, the assay for F-6-ppk has been a key taxonomic identifying character, so it could be distinguished from other closely related bacterial groups such as lactobacilli, actinomycetes, and corynebacteria (Scardovi 1986; Vlkova et al. 2002). Among the native isolates of LAB and bifidobacteria, based on conventional identification protocols, isolates were characterized to their respective genus and species. Two isolates were selected, which had the deposition details of L. plantarum MTCC 5422 and B. adolescentis MTCC 5423 (Table 2).
Although isolates of LAB were characterized by conventional protocols of cultural and biochemical attributes, the use of molecular-based approach in targeting the specific gene of importance gives an impetus to the study which focuses on obtaining cultures with defined characteristics. Earlier studies have focused on isolating specific cultures of Lactobacillus, wherein genomic DNA of test cultures was amplified using 16 S rRNA genus specific primers. In recent years, 16 S rRNA-targeted PCR primers enable rapid and specific detection of bacterial genera (Kaufmann et al. 1997). In a study with fermentative process, isolates of L. plantarum and L. fermentum were found to occur as predominant cultures based on PCR amplification of the intergenic segment between 16 S and 23 S rDNA subunits, followed by restriction enzyme digestion specific for Lactobacillus (Tannock et al. 1999). In a broader perspective, research studies have shown the usefulness of 16 S rRNA targeted primers to identify the genera/species of Lactobacillus and Bifidobacterium through PCR (Klocke and Mundt 2004; Mangin et al. 2006; Mignard and Flandrois 2006; Tamang et al. 2008). Molecular characterization using RAPD-PCR with M13 primers revealed the predominance of populations of L. curvatus, L. plantarum, and L. sakei during natural fermentation of sausages (Rantsiou et al. 2006). In a similar approach, PCR using 16 S rDNA specific primers confirmed six bacteriocinogenic strains as cultures of L. plantarum (Omar et al. 2006). In our study, the samples assessed for LAB showed the predominance of L. plantarum, followed by L. casei. The use of 16 S rRNA primers is more useful in characterization of genus and not to distinguish very closely related species and/or subspecies of a single species. This was evident in case of Lactococcus lactis subsp. cremoris and L. lactis subsp. lactis, wherein 16 S rRNA gene sequences of these two subspecies had significant similarities (Maruo et al. 2006). A similar observation did exist in our study, wherein the 16 S rRNA primers specifically designed to identify Lactobacillus genus, failed to show any amplification in PCR with isolates conventionally identified as Leuconostoc and Pediococcus.
The cultures of LAB and bifidobacteria are known more for their probiotic attributes. However, these cultures can also function in a manner to make available prebiotic substrate through α-d-galactosidase activity. Although cultures of Lactobacillus spp. are known to exhibit α-d-galactosidase activity, most of the research investigations have focused on L. fermentum and L. plantarum, wherein quantification assays have resulted in diversified units of activity. The first genetic characterization of α-d-galactosidase was established by cloning and expressing melA gene from L. plantarum in Escherichia coli yielding an active α-Gal (Silvestroni et al. 2002). The behavior of L. fermentum CRL 722 and CRL 251 evaluated under different pH conditions of 6.0 to 4.5 revealed an activity of 5 U ml−1 at pH 5.5 (Le Blanc et al. 2004). In our study, the native isolate of L. plantarum MTCC 5422 could elaborate a slightly higher activity of 6.6 U ml−1. In general, bile salt hydrolase activity is being included as one of the probiotic attributes of desirable microbial cultures, which are nonpathogenic/toxigenic/virulent.
Initially, Bifidobacterium sp. was identified by the presence of F-6-ppk in cellular extracts, more so as a qualitative approach (Scardovi 1986). This was later confirmed in PCR by amplification of Bifidobacterium targeted gene sequence for xylulose-6-phosphate phosphoketolase (xfp) primers (Matsuki et al. 2003; Berthoud et al. 2005). Besides, researchers have identified bifidobacteria of human origin as well as those exhibiting probiotic properties using 16 S rRNA-targeted primers (Mullie et al. 2003; Masco et al. 2007) and species-specific primers based on region extending from 16 S rRNA through 23 S rRNA (Shuhaimi et al. 2004; Kwon et al. 2005). In the present study, initially, we could classify the native isolate as B. adolescentis based on carbohydrate fermentation profile and later by confirmation in PCR (Fig. 2). The identified culture exhibited almost all the major phenotypic characteristics reported for this species. Interestingly, strains of Bifidobacterium have shown higher bsh activity as against those reported for other bacterial groups. Research investigations have focused on biochemical and genetic characterization of bsh of B. longum (Tanaka et al. 2000; Shuhaimi et al. 2001). Similar characterization studies were established with B. bifidum strains and B. adolescentis (Kim et al. 2004, 2005). In a similar manner, in the present study, only the presence of bsh gene in a native isolate of B. adolescentis MTCC 5423 was established in PCR (Fig. 2). Although no definite relationship can be attributed to activity in assay system and PCR amplification for target genes in the microbial cultures, it is appropriate to mention that PCR primers designed in the present study were species specific, as it was the case in α-d-galactosidase activity and PCR amplification for the same.
Another important probiotic attribute associated with cultures of LAB and bifidobacteria is that of antibacterial activity toward foodborne pathogenic bacterial species. Besides, this probiotic property becomes more evident with the cultures exhibiting appreciable growth performance in the presence bile salts and at low pH levels. In the present study, both the isolates of L. plantarum MTCC 5422 and B. adolescentis MTCC 5423 revealed appreciable antibacterial activity (Table 3) and survival/growth in the presence of bile salts and low pH conditions (Fig. 3 and 4). These potent beneficial attributes of LAB and bifidobacteria have found applications in food fermentation processes, a well established fact over the centuries, more particularly with that of intestinal species like L. acidophilus and/or B. bifidum in preparation of fermented products (Driessen and Boer 1989). In this context, an increasing interest exists for milk-based foods with a focus on those bacterial species providing health-improving properties (Portier et al. 1993). The interest in application of these microorganisms and their metabolites toward wellness factor for human health has taken a major leap over the last decade (Ljungh and Wadstrom 2006; Masco et al. 2007; Woodmansey 2007). The species of Lactobacillus and Bifidobacterium are well known to have health promoting effects in human and animal intestinal tract, for which they are termed as probiotics. Though a matter of debate, it is considered that these beneficial microbial cultures are to be present as live cell populations of 106 CFU ml−1 or g−1 of product to have the exact meaning of a probiotic (Kailasapathy and Chin 2000). As the culture-dependent methods have limitations of reliability and sensitivity in terms of specific characterization for probiotic attributes, PCR-based methods are being established as determinants of probiotic attributes. In the present study, probiotic attributes associated with selected cultures of L. plantarum and B. adolescentis were evaluated by culture-dependent methods. Further, the same attributes were supported on a molecular basis with the positive amplification for specific and targeted genes being evidenced in PCR.
Often, conventional methods of identification of microbial species based on phenotypic characteristics lead to uncertain conclusions (Bernardeau et al. 2006). The use of molecular-based approaches has added newer dimensions to characterize the desirable species of Lactobacillus and Bifidobacterium with focus on targeted attributes (Dellaglio et al. 1991). These methods mostly target rRNA genes as a primary means to identify species of Lactobacillus. Although intergenic 16 S-23S rRNA spacer region has been commonly targeted in a majority of the investigations (Hayashi et al. 2002; Vizoso Pinto et al. 2006), there appears to be certain inconclusive and nondistinguishable similarities among closely related species. As a means to achieve a clear understanding of the interrelationship, phylogenetic determinations have been generated based on sequence analysis data for PCR amplified product with specific target genes of interest (Chavagnat et al. 2002; Ventura et al. 2003; Blaiotta et al. 2008).
Lactobacillus with nearly 80 recognized species is a highly diverse group as could be evidenced from a complex phylogeny. This diversity of the genus Lactobacillus was evidenced in a study relating to comparative genomics of five species of Lactobacillus, namely, L. salivarius, L. plantarum, L. acidophilus, L. johnsonii, and L. sakei. Phylogeny based on whole genome alignment suggested that L. salivarius was closer to L. plantarum than to L. sakei (Canchaya et al. 2006). Although, it is believed that during evolution, the major genes are conserved between related bacterial species, the phylogenetic trees reconstructed for individual genes may differ from this general viewpoint, and they may give an indication of horizontal transfer. Individual trees are often not resolved with clarity to bring in meaningful presentation of analysis (Nicolas et al. 2007).
Studies focusing on evolutionary trends in microbial species are gaining importance in the broad area of “microbial diversity,” which becomes evident by molecular biology approaches. In the present study, attempts were made to generate phylogeny tree for L. plantarum based on genus specific 16 S rRNA primers as well as individual genetic characters such as α-d-galactosidase and bile salt hydrolase, wherein bsh-based tree exhibited almost complete homology with the same species and in melA-based tree, a lower percent homology existed with L. salivarius and L. reuteri (Fig. 5b and 5c). The appearance of few strains of L. plantarum in the tree may be attributed to the low prevalence of this gene among species of Lactobacillus. In our study, it was very much evident that gene for bile salt hydrolase though detected in native isolates of Lactobacillus spp. by PCR, quantification of activity was not very much appreciable, which may be due to certain parameters in PCR analysis. Similarly, based on 16 S rRNA generated tree, the native isolate of L. plantarum MTCC 5422 did exhibit 98% homology with other cultures of Lactobacillus sp. (Fig. 5a). On the other hand, phylogenetic tree generated for B. adolescentis based on F-6-ppk showed the appearance of several species of Bifidobacterium, as this attribute being the major character of all Bifidobacterium spp. would definitely enable most of the species to be interrelated (Fig. 5d).
Native yeast cultures with phytase activity
Research studies have focused on biochemical and molecular characterization of acid phosphatases of yeasts (Kaur et al. 2007). These enzymes reside on the cell surface and catalyze the release of inorganic phosphate from different phosphorylated organic compounds. In S. cerevisiae, the acid phosphatase encoding genes pho3, pho5, pho11, and pho12 have been isolated and characterized (Vogel and Hinnen 1990). Genetic studies with some of the yeast phosphatase genes have indicated their role as phytases. These genes were pho1 and pho4 from Schizosaccharomyces pombe and pho3 from S. cerevisiae (Wodzinski and Ullah 1996). As S. cerevisiae and its by-products have long been known to be safe for human and animal consumption, the phytases elaborated by this culture have found commercial applications to overcome the problems of antinutritional factor in cereal/legume-based foods. Studies with different yeast cultures have revealed a diverse range of phytase activity ranging from 77 to 176 U g−1 of dry biomass (Bindu et al. 1998; Vohra and Satyanarayana 2004). A similar situation prevailed in the present study, wherein isolates of Saccharomyces spp., Candida spp., Schizosaccharomyces spp., and Rhodosporidium sp. elaborated phytase activity ranging from a lowest of 25 to a highest of 228 U ml−1. As the PCR primers designed were species specific to S. cerevisiae, a few of the yeast cultures failed to show positive amplification in PCR with these primers. Further, it is likely that the targeted phytase gene in our study may not be either present in other yeast cultures or not related genetically.
The findings of this study have conclusively established that microbial cultures with genetically stable desirable attributes are present in diversified habitats, and they need to be well characterized and exploited commercially toward enriching wellness factor in human health mediated through traditional fermented and/or nonfermented foods.
References
Bernardeau M, Guguen M, Vernoux JP (2006) Beneficial lactobacilli in food and feed: long term use, biodiversity and proposals for specific and realistic safety assessments. FEMS Microbiol Rev 30:487–513
Berthoud H, Chavagnat F, Haueter M, Casey MG (2005) Comparison of partial gene sequences encoding a phosphoketolase for the identification of bifidobacteria. Lebensm Wiss U Technol 38:101–105
Bindu S, Somashekar D, Joseph R (1998) A comparative study on permeabilization treatments for in situ determination of phytase of Rhodotorula gracilis. Lett Appl Microbiol 27:336–340
Blaiotta G, Fusco V, Ercolini D, Aponte M, Pepe O, Villani F (2008) Lactobacillus strain diversity based on partial hsp60 gene sequences and design of PCR restriction fragment length polymorphism assays for species identification and differentiation. Appl Environ Microbiol 74:208–215
Canchaya C, Claesson MJ, Fitzgerald GF, van Sinderen D, O’Toole PW (2006) Diversity of the genus Lactobacillus revealed by comparative genomics of five species. Microbiol 152:3185–3196
Cappuccino JG, Sherman N (2004) Microbiology: A Laboratory Manual, 6th edn. Pearson Education, Singapore
Chavagnat F, Haueter M, Jimeno J, Casey MG (2002) Comparison of partial tuf gene sequences for the identification of lactobacilli. FEMS Microbiol Lett 217:177–183
Church FC, Meyers SP, Srinivasan VR (1980) Isolation and characterization of alpha-galactosidase from Pichia guilliermondii. In: Underkofler LA, Wulf ML (eds) Developments in Industrial Microbiology. SIM, Arlington, VA, pp 339–348
Coeuret V, Dubernet S, Bernardeau M, Gueguen M, Vernoux JP (2003) Isolation, characterization and identification of Lactobacilli focusing mainly on cheeses and other dairy products. Lait 83:269–306
Deak T, Beuchat LR (1996) Handbook of food spoilage yeasts, 2nd edn. CRC, Boca Raton, p 350
Dellaglio F, Dicks LMT, du Toit M, Torriani S (1991) Designation of ATCC 334 in place of ATCC 393 (NCDO 161) as the neotype strain of Lactobacillus casei subsp. casei and rejection of the name of Lactobacillus paracasei. Intl J Syst Bacteriol 41:340–342
Driessen F, Boer R (1989) Fermented milks with selected intestinal bacteria, a healthy trend in new products. Netherlands Milk Dairy J 43:369–382
Fooks LJ, Fuller R, Gibson GR (1999) Prebiotics, probiotics and human gut microbiology. Intl Dairy J 9:53–61
Gotcheva V, Hristozova E, Hristozova T, Guo M, Roshkova Z, Angelov A (2002) Assessment of potential probiotic properties of lactic acid bacteria and yeast strains. Food Biotechnol 16:211–225
Hayashi H, Sakamoto M, Benno Y (2002) Phylognetic analysis of the human gut microbiota using 16 S rDNA clone libraries and strictly anaerobic culture based methods. Microbiol Immunol 46:535–548
Heinonen JK, Lahti RJ (1981) A new and convenient colorimetric determination of inorganic orthophosphate and its application to the assay of inorganic pyrophosphatase. Anal Biochem 113:313–317
Kailasapathy K, Chin J (2000) Survival and therapeutic potential of probiotic organisms with reference to Lactobacillus acidophilus and Bifidobacterium spp. Immunol Cell Biol 78:80–88
Kandler O, Weiss N (1986) Lactic acid bacteria In: De Vos P, Garrity G, Jones D, Krieg NR, Ludwig W, Rainey FA, Schleifer KH, Whitman, WB (eds) Bergey’s Manual of Systematic Bacteriology, 2nd edn, WW, Baltimore, pp 1330
Kaufmann P, Pfefferkorn A, Teuber M, Meile L (1997) Identification and quantification of Bifidobacterium species isolated from foods with genus-specific 16 S rRNA-targeted probes by colony hybridization and PCR. Appl Environ Microbiol 63:1268–1273
Kaur P, Lingner A, Singh B, Boer E, Polajeva J, Steinborn G, Bode R, Gellissen G, Satyanarayana T, Kunze G (2007) APHO1 from the yeast Arxula adeninivorans encodes an acid phosphatase of broad substrate specificity. Antonie Van Leeuwenhoek 91:45–55
Kim GB, Miyamoto CM, Meighen EA, Lee BH (2004) Cloning and characterization of the bile salt hydrolase genes (bsh) from Bifidobacterium bifidum strains. Appl Environ Microbiol 70:5603–5612
Kim GB, Brochet M, Lee BH (2005) Cloning and characterization of a bile salt hydrolase (bsh) from Bifidobacterium adolescentis. Biotechnol Lett 27:817–822
Klocke M, Mundt K (2004) Development of a 16 S rDNA-targeted PCR assay for monitoring of Lactobacillus plantarum and Lact. rhamnosus during co-cultivation for production of inoculants for silages. Lett Appl Microbiol 39:267–273
Kwon HS, Yang EH, Lee SH, Yeon SW, Kang BH, Kim TY (2005) Rapid identification of potentially probiotic Bifidobacterium species by multiplex PCR using species-specific primers based on the region extending from 16 S rRNA through 23 S rRNA. FEMS Microbiol Lett 250:55–62
LeBlanc JG, Garro MS, Savoy de Giori G (2004) Effect of pH on Lactobacillus fermentum growth, raffinose removal, α-galactosidase activity and fermentation products. Appl Microbiol Biotechnol 65:119–123
Ljungh A, Wadstrom T (2006) Lactic acid bacteria as probiotics. Curr Issues Intest Microbiol 7:73–90
Macfarlane S, Macfarlane GT, Cummings JH (2006) Prebiotics in the gastrointestinal tract. Aliment Pharmacol Ther 24:701–714
Mangin I, Suau A, Magne F, Garrido D, Gotteland M, Neut C, Pochart P (2006) Characterization of human intestinal bifidobacteria using competitive PCR and PCR-TTGE. FEMS Microbiol 55:28–37
Maruo T, Sakamoto M, Toda T, Benno Y (2006) Monitoring the cell number of Lactococcus lactis subsp. cremoris FC in human faeces by real-time PCR with strain-specific primers designed using the RAPD technique. Intl J Food Microbiol 110:69–76
Masco L, Vanhoutte T, Temmerman R, Swings J, Huys G (2007) Evaluation of real-time PCR targeting the 16 S rRNA and recA genes for the enumeration of bifidobacteria in probiotic products. Intl J Food Microbiol 113:351–357
Matsuki T, Watanabe K, Tanaka R (2003) Genus- and species-specific PCR primers for the detection and identification of bifidobacteria. Curr Issues Intest Microbiol 4:61–69
Mignard S, Flandrois JP (2006) 16 S rRNA sequencing in routine bacterial identification: A 30-month experiment. J Microbiological Methods 67:574–581
Mitsuoka T (1992) The human gastrointestinal tract. In: Wood BJB (ed) The Lactic Acid Bacteria in Health and Disease. Elsevier Appl Sci, London, pp 69–114
Mullie C, Odou MF, Singer E, Romond MB, Izard D (2003) Multiplex PCR using 16 S rRNA gene-targeted primers for the identification of bifidobacteria from human origin. FEMS Microbiol Lett 222:129–136
Nicolas P, Bessieres P, Ehrlich DS, Maguin E, Guchte MV (2007) Extensive horizontal transfer of core genome genes between two Lactobacillus species found in the gastrointestinal tract. BMC Evol Biol 7:141–154
Omar NB, Abriouel H, Lucas R, Canamero MM, Guyot JP, Galvez A (2006) Isolation of bacteriocinogenic Lactobacillus plantarum strains from ben saalga, a traditional fermented gruel from Burkina Faso. Intl J Food Microbiol 112:44–50
Orban JI, Patterson JA (2000) Modification of the phosphoketolase assay for rapid identification of bifidobacteria. J Microbiol Methods 40:221–224
Portier A, Boyaka P, Bougoudoga F, Dubarry M, Huneau F, Tome D, Dodin A, Coste M (1993) Fermented milks and increased antibody responses against cholera in mice. Intl J Immunotherapy 94:217–224
Rantsiou K, Drosinos EH, Gialitaki M, Metaxopoulos I, Comi G, Cocolin L (2006) Use of molecular tools to characterize Lactobacillus spp. isolated from Greek traditional fermented sausages. Intl J Food Microbiol 112:215–222
Roberfroid M (1997) Health benefits of non-digestible oligosaccharides. In: Kritchevsky D, Bonfield C (eds) Dietary Fiber in Health and Disease. Plenum, New York, pp 211–219
Rekha R, Rizvi MA, Jaishree P (2006) Designing and validation of genus-specific primers for human gut flora Study. Electronic J Biotechnol 9:505–511
Sambrook J, Russel DW (2001) Molecular Cloning: A Laboratory Manual, 3 rdth edn. Cold Spring Harbor Laboratory Press, New York
Scardovi V (1986) Genus Bifidobacterium Orla-Jensen 1974, 472AL. In: Sneath PHA, Mair NS, Sharpe ME, Holt JG (eds) Bergey’s Manual of Systematic Bacteriology. WW, Baltimore, pp 1418–1434
Schillinger U, Lucke FK (1989) Antibacterial activity of Lactobacillus sake isolated from meat. Appl Environ Microbiol 55:1901–1906
Shuhaimi M, Ali AM, Saleh NM, Yazid AM (2001) Cloning and sequence analysis of bile salt hydrolase (bsh) gene from Bifidobacterium longum. Biotechnol Lett 23:1775–1780
Shuhaimi M, Ali AM, Norjihan A, Saleh NM, Yazid AM (2004) Characterization of Bifidobacterium species—a review. Biosci Microflora 23:81–92
Silvestroni A, Connes C, Sesma F, Savoy de Giori G, Piard JC (2002) Characterization of the melA locus for α-galactosidase in Lactobacillus plantarum. Appl Environ Microbiol 68:5464–5471
Swennen K, Courtin CM, Delcour JA (2006) Non-digestible oligosaccharides with prebiotic properties. Critical Rev Food Sci Nutr 46:459–471
Tamang B, Tamang JP, Schillinger U, Franz CMAP, Gores M, Holzapfel WH (2008) Phenotypic and genotypic identification of lactic acid bacteria isolated from ethnic fermented bamboo tender shoots of North East India. Intl J Food Microbiol 121:35–40
Tanaka H, Hashiba H, Kok J, Mierau I (2000) Bile salt hydrolase of Bifidobacterium longum: biochemical and genetic characterization. Appl Environ Microbiol 66:2502–2512
Tannock GW, Tilsala-Timisjarvi A, Rodtong S, NG J, Munro K, Alatossava T (1999) Identification of Lactobacillus isolates from the gastrointestinal tract, silage, and yoghurt by 16 S–23S rRNA gene intergenic spacer region sequence comparisons. Appl Environ Microbiol 65:4264–4267
Varadaraj MC, Devi N, Keshava N, Manjrekar SP (1993) Antimicrobial activity of neutralized extracellular culture filtrates of lactic acid bacteria isolated from a cultured Indian milk product (dahi). Intl J Food Microbiol 20:259–267
Ventura M, Canchaya C, Meylan V, Klaenhammer TR, Zink R (2003) Analysis, characterization and loci of the tuf genes in Lactobacillus and Bifidobacterium species and their direct application for species identification. Appl Environ Microbiol 69:6908–6922
Vizoso Pinto MG, Franz CMAP, Schillinger U, Holzapfel WH (2006) Lactobacillus spp. with in vitro probiotic properties from human faeces and traditional fermented products. Intl J Food Microbiol 109:205–214
Vlkova E, Medkova J, Rada V (2002) Comparison of four methods for identification of Bifidobacteria to the genus level. Czech J Food Sci 20:171–174
Vogel K, Hinnen A (1990) The yeast phosphatase system. Mol Microbiol 4:2013–2017
Vohra A, Satyanarayana T (2004) A cost-effective cane molasses mediu for enhanced cell bound phytase production by Pichia anomola. J Appl Microbiol 97:471–476
Wang RF, Cao WW, Cerniglia CE (1997) A universal protocol for PCR detection of 13 species of foodborne pathogens in foods. J Appl Microbiol 83:727–736
Wodzinski RJ, Ullah AHJ (1996) Phytase. Adv Appl Microbiol 42:263–302
Woodmansey EJ (2007) Intestinal bacteria and ageing. J Appl Microbiol 102:1178–1186
Acknowledgements
The authors are thankful to Dr. V. Prakash, Director, CFTRI, Mysore, India for providing the facilities and interest in present work. The first author is grateful to Council of Scientific and Industrial Research, New Delhi, India for awarding the Senior Research Fellowship.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Roopashri, A.N., Varadaraj, M.C. Molecular characterization of native isolates of lactic acid bacteria, bifidobacteria and yeasts for beneficial attributes. Appl Microbiol Biotechnol 83, 1115–1126 (2009). https://doi.org/10.1007/s00253-009-1991-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-009-1991-y