Abstract
NAD kinase was overexpressed to enhance the accumulation of poly(3-hydroxybutyrate) (PHB) in recombinant Escherichia coli harboring PHB synthesis pathway via an accelerated supply of NADPH, which is one of the most crucial factors influencing PHB production. A high copy number expression plasmid pE76 led to a stronger NAD kinase activity than that brought about by the low copy number plasmid pELRY. Overexpressing NAD kinase in recombinant E. coli was found not to have a negative effect on cell growth in the absence of PHB synthesis. Shake flask experiments demonstrated that excess NAD kinase in E. coli harboring the PHB synthesis operon could increase the accumulation of PHB to 16–35 wt.% compared with the controls; meanwhile, NADP concentration was enhanced threefold to sixfold. Although the two NAD kinase overexpression recombinants exhibited large disparity on NAD kinase activity, their influence on cell growth and PHB accumulation was not proportional. Under the same growth conditions without process optimization, the NAD kinase-overexpressing recombinant produced 14 g/L PHB compared with 7 g/L produced by the control in a 28-h fermentor study. In addition, substrate to PHB yield Y PHB/glucose showed an increase from 0.08 g PHB/g glucose for the control to 0.15 g PHB/g glucose for the NAD kinase-overexpressing strain, a 76% increase for the Y PHB/glucose. These results clearly showed that the overexpression of NAD kinase could be used to enhance the PHB synthesis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Polyhydroxyalkanoates (PHA) are polyesters of hydroxyalkanoates synthesized as intracellular insoluble granules by a variety of bacteria grown usually under stress conditions, for example, under an excess supply of carbon sources together with limitation on nitrogen or phosphorus nutrients (Anderson and Dawes 1990; Steinbüchel and Valentin 1995). As a biodegradable biopolymer with versatile material properties, PHA have drawn industrial interests and efforts devoted to their production and applications since the first discovery of poly(3-hydroxybutyrate) (PHB) in Bacillus megaterium (Lemoigne 1926; Lee et al. 1999; Steinbüchel and Lutke-Eversloh 2003; Chen and Wu 2005; Lenz and Marchessault 2005). PHB is the most widespread and best studied member of the PHA family. As a model organism, recombinant Escherichia coli harboring the PHB synthesis pathway is regarded as a strong candidate for biopolymer production due to its advantages including the ability to utilize several inexpensive carbon sources, the simplicity of extraction, the ease for genetic manipulation, and the lack of PHA depolymerase (Lee 1997). Since the first introduction of Ralstonia eutropha PHB synthesis pathway into E. coli (Schubert et al. 1988; Slater et al. 1988; Peoples and Sinskey 1989), many strategies have been applied to lower the PHB production cost in E. coli, such as the use of cheap carbon sources (Ahn et al. 2000), introduction of stress-induced system (Kang et al. 2008), and microaerobic process (Wei et al. 2009).
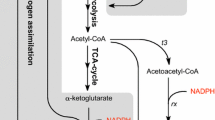
The PHB synthesis pathway in relationship with NADPH in recombinant E. coli harboring the phbCAB operon of R. eutropha was described in Fig. 1. PHB is synthesized from acetyl-CoA through a three-step reaction catalyzed by β-ketothiolase, NADPH-dependent acetoacetyl-CoA reductase, and PHB synthase. The cofactor NADPH plays an important role in the PHB biosynthesis since it is a NADPH-dependent pathway. The supplement of amino acids or oleic acid to defined medium can enhance PHB production because the synthesis of amino acids or oleic acid consumes a lot of NADPH, which competes with PHB accumulation for NADPH (Lee et al. 1995). After studying the effect of nicotinamide nucleotide on the enzyme activities and PHB synthesis in recombinant E. coli, Lee et al. (1996) reported that a high level of NADPH and/or NADPH/NADP ratio plays a critical role in PHB synthesis, which suggested that an efficient NADPH supply could improve the PHB production in E. coli.
NADPH can be generated through the pentose phosphate (PP) pathway in the reactions catalyzed by glucose-6-phosphate dehydrogenase (G6PDH, encoded by zwf in E. coli) and 6-phosphogluconate dehydrogenase (6PGDH, encoded by gnd in E. coli). Overexpression of zwf and gnd promoted PHB production yet cell growth was repressed due to the high level of NADPH (Lim et al. 2002). The overexpression of critical enzyme transketolase (encoded by tktA) and transaldolase (encoded by talA) in the nonoxidative PP pathway also increased PHB accumulation in E. coli (Jung et al. 2004; Song et al. 2006). A soluble pyridine nucleotide transhydrogenase (UdhA) has been employed to improve PHB productivity in E. coli. UdhA catalyzes the reversible transfer of reducing equivalents between NAD and NADP, and the PHB concentration in udhA overexpressing strain was nearly twice that of the control as a result of the increased NADPH availability by high-level expression of UdhA (Sanchez et al. 2006).
NAD kinase catalyzes the phosphorylation of NAD in the presence of Mg2+ and inorganic polyphosphate or adenosine-5′-triphosphate as the phosphoryl donor (Fig. 1). NAD kinase is ubiquitously distributed from bacteria to human cells, and the gene encoding NAD kinase in E. coli (Kawai et al. 2001a), Bacillus subtilis (Garavaglia et al. 2003), Saccharomyces cerevisiae (Kawai et al. 2001b), and humans (Lerner et al. 2001) have been identified and well-studied. It was reported that the NAD kinase was the only enzyme generating the NADP pool in cells, and it played a crucial role in maintaining the intracellular redox balance (Grose et al. 2006; Magni et al. 2006). In this paper, we studied the possible application of NAD kinase in increasing NADP concentration and activating NADPH-dependent anabolic reactions using recombinant E. coli harboring the PHB synthesis pathway as a model system, with an aim to improve PHB production.
Materials and methods
Bacterial strains, primers, and plasmids construction
Bacterial strains, plasmids, and primers were listed in Table 1. E. coli JM109 was used for plasmid construction, shake flask, and fermentation experiments. The native E. coli NAD kinase gene yfjB was cloned by polymerase chain reaction (PCR) from purified E. coli genomic DNA. Plasmids pELRY and pE76 were constructed to express NAD kinase gene. The PCR product by primers yfjB-1F and yfjB-1R was digested with BamHI and SacI and then ligated into the corresponding sites of pLZZH13 to yield pELRY. For pE76 construction, the PCR fragment by primers yfjB-2F and yfjB-2R was cloned into pMD19-T to yield pMD19-T-yfjB, and pyruvate decarboxylase promoter (Ppdc) was amplified using primers pdc-F and pdc-R from Zymomonas mobilis genomic DNA. The PCR product of the pdc promoter was digested with EcoRI and NheI, and then ligated into the EcoRI and XbaI sites of pMD19-T-yfjB to yield pE76. In order to generate plasmid p68E76, the DNA fragment of Ppdc and yfjB was excised from pE76 using EcoRI and HindIII and cloned into pBHR68 cut with the same restriction enzymes. The DNA sequences of the yfjB gene and Ppdc used in this study were confirmed by DNA sequence analysis carried out by AuGCT Biotechnology (Beijing, China).
Cultivation conditions
LB medium was used for seed culture preparation. Shake flask medium contained (in grams per liter): glucose 20, (NH4)2SO4 2.0, MgSO4·7H2O 0.4, Na2HPO4·12H2O 9.65, KH2PO4 1.5, Fe(III)–NH4–citrate 0.05, CaCl2 0.02, and 1 mL/L trace element solution consisted of (in milligrams per liter): ZnSO4·7H2O 100, MnCl2·4H2O 30, H3BO3 300, CoCl2·6H2O 200, CuSO4·5H2O 10, NiCl2·6H2O 20, NaMoO4·2H2O 30, and HCl 0.5 mol/L. The starting fermentation medium was the same as shake flasks except for the addition of 5 g/L yeast extract and higher glucose concentration (30 g/L). When necessary, a final concentration of 50 µg/mL kanamycin and/or 100 µg/mL ampicillin were added to the medium to maintain the plasmid stability.
The seed culture was grown at 37°C in LB medium for 12 h at 200 rpm on a rotary shaker. For shake flask cultures, seed culture was inoculated into 500 mL shake flasks containing 50 mL cultivation medium at an inoculation volume of 4%. For fermentor cultures, seed culture was inoculated into a 6-L fermentor (NBS 3000, New Brunswick, USA) at 10% inoculation volume with an operating volume of 3 L. The pH in the fermentor culture was maintained at 7.0 by automatic addition of 5 M NaOH and 5 M H2SO4. Dissolved oxygen was provided by injecting filtered air at a flow rate of 3 L min−1 and was maintained at 20% of air saturation by automatically adjusting the agitation rate from 200 to 800 rpm. When glucose concentration was lower than 10 g/L, a fed nutrition solution (20 g/L glucose, 0.4 g/L MgSO4·7H2O, 2.0 g/L (NH4)2SO4) was added into the culture.
Measurement of NAD kinase activity
Bacterial cells were harvested and disrupted by sonication (JY92-2D, SEIENTZ, Ningbo, China) as reported previously (Liu et al. 2007). The concentration of total cell proteins was determined by the method of Bradford with bovine serum albumin as standards. NAD kinase activity was assayed at 30°C by a two-step method described previously with minor modifications (Ochiai et al. 2004). The formation of NADPH was continuously determined by a spectrophotometer (UV2450, Shimadzu, Japan) through the absorbance at 340 nm. One unit of enzyme activity was defined as 1.0 nmol NADP produced in 1 min, and specific activity was expressed in units per milligram of protein.
Study of intracellular NADP and NADPH concentrations
NADP and NADPH were extracted from bacterial cells using HCl and NaOH based on the method reported by Lee et al. (1996) with minor modifications. Concentrations of the intracellular NADP and NADPH were measured by a spectrophotometric enzymatic cycling method described previously (Zerez et al. 1987). The reaction mixture contained 5.0 mM Na4EDTA, 2.0 mM phenazine ethosulfate, 0.5 mM thiazolyl blue, 1.3 U G6PDH, 1.0 mM glucose-6-phosphate (G6P), and appropriate volume of cell extract in 100 mM Tris–HCl (pH 8.0). Reactions were conducted at 37°C and started by G6P addition. The absorbance at 570 nm was followed for 5 min to determine the NADP and NADPH concentrations.
Analytical techniques
Bacteria were harvested by centrifugation at 8,000×g for 10 min and then washed with distilled water. The supernatant was filtered through a 0.2-µm syringe filter and stored chilled for high-performance liquid chromatography (HPLC) analysis. Cell dry weight (CDW) was measured after vacuum lyophilization. PHB content was analyzed by gas chromatography (Hewlett-Packard model 6890) after methanolysis of lyophilized cells in chloroform. The concentration of glucose and acetate was determined using HPLC (P2000, AS3000, Thermo Spectra System, USA), equipped with an ion exchange column (Aminex® HPX-87H, 7.8 × 300 mm, BioRad) and a refractive index detector (RI-150, Thermo Spectra System, USA). A mobile phase of 5 mM H2SO4 at a 0.5 mL/min flow rate was used.
Results
Overexpression of NAD kinase and its effects on cell growth
E. coli NAD kinase gene yfjB was inserted into vectors pUC19 and pLZZH13 under the control of Z. mobilis pyruvate decarboxylase promoter (Ppdc) and phbCAB operon promoter, respectively, resulting in plasmids pE76 and pELRY. Four recombinant E. coli harboring the four plasmids were cultivated in mineral medium supplemented with 20 g/L glucose at 37°C for 24 h. Strong NAD kinase activity reaching 2,008 U was detected in recombinant E. coli containing the high copy number expression plasmid pE76, while 18 U NAD kinase activity was also recorded in recombinant E. coli containing the low copy number plasmid pELRY. In contrast, two recombinant E. coli without NAD kinase overexpression produced minimum activity of only 3 U (Table 2). Previous studies showed that the two promoters can be expressed constitutively with similar activities in E. coli (Lai and Chen 2006). The differences of NAD kinase activity produced by plasmids pE76- and pELRY-carrying strains can be regarded as the result of the difference in plasmid copy numbers. Plasmid pE76 derived from pMD19-T could reach 500 copies per cell in E. coli (Yanisch-Perron et al. 1985), while pELRY derived from pBBR1MCS-2 replicates up to tens of copies per cell (Kovach et al. 1995). Besides the difference in enzyme activity, there was no obvious difference among recombinant E. coli with and without yfjB overexpression in terms of their cell growth (CDW), glucose consumption, and acetate production (Table 2), suggesting that overexpression of NAD kinase did not have a negative impact on the growth behavior of the four recombinant E. coli strains.
Influence of NAD kinase overexpression on PHB production in shake flask cultures
To study the influence of NAD kinase overexpression on PHB production, yfjB gene along with Ppdc was excised from pE76 and inserted into pBHR68 to yield a coexpression plasmid for PHB and NAD kinase production. Two groups of shake flask experiments employing single or double plasmid systems were designed. PHB synthesis operon phbCAB and NAD kinase gene yfjB were coexpressed by a single plasmid (p68E76) or by two compatible plasmids (pBHR68 and pELRY). As a control, recombinant E. coli carrying only phbCAB without the yfjB gene was used. When cultivated in mineral medium supplemented with 20 g/L glucose at 37°C for 48 h, NAD kinase activity, CDW, and acetate and PHB production were studied (Fig. 2). All recombinants harboring the yfjB gene produced more CDW, PHB concentration, and NAD kinase activity than those without the yfjB gene did, except acetate which was reduced in yfjB-expressing recombinants (Fig. 2). In detail, recombinant strains harboring NAD kinase overexpression plasmid p68E76 produced 3.4 g/L PHB, higher than that of the control of 2.9 g/L. In the double plasmid system, PHB concentration increased from 2.6 g/L for the control to 3.5 g/L for the yfjB-expressing recombinant. In both single and double plasmid systems, acetate production showed a 50% reduction in NAD kinase-overexpressing strains compared with that of their controls. However, although PHB concentrations were found similar despite of the difference in NAD kinase activities produced by the two NAD kinase overexpression strains, all yfjB-expressing recombinants showed improvements on PHB yield (Y PHB/glucose) compared with their controls, indicating that more carbon sources were rerouted to PHB accumulation in these NAD kinase overexpression strains (Fig. 2).
CDW, PHB concentration, PHB content, glucose substrate to PHB yield, acetate production, and NAD kinase activity profile of E. coli harboring the phbCAB operon with or without NAD kinase overexpression in shake flask cultures. The recombinants harboring different plasmids were cultivated at 37°C for 48 h as described in the “Materials and methods” section. The data shown are the average and standard deviations of three parallel experiments
Effect of NAD kinase overexpression on intracellular NADP and NADPH concentrations
Recombinant E. coli strains harboring NAD kinase-overexpressing plasmids and their controls were cultivated in mineral medium supplemented with 20 g/L glucose at 37°C for 24 h. The intracellular NADP and NADPH concentrations were studied (Table 3). The yfjB-expressing recombinants showed a remarkable increase on NADP concentration, indicating the functional expression of NAD kinase. In detail, NADP concentration in the strain harboring p68E76 (containing yfjB) was approximately six times that of the strain harboring pBHR68. In the dual plasmid system, NADP concentration produced by the yfjB-overexpressing strain was three times more compared with the control. The NADPH/NADP ratio was decreased due to the enhanced concentration of NADP in NAD kinase overexpression strains.
Fermentor study of the influence of NAD kinase overexpression on PHB production
Recombinant E. coli strains harboring p68E76 and pBHR68, respectively, were grown in a 6-L NBS fermentor for 28 h under the same growth conditions without process optimization. The NAD kinase-overexpressing recombinant produced 28 g/L CDW containing 50% PHB after consuming 92 g/L glucose. In comparison, the control strain harboring only phbCAB produced 21 g/L CDW containing 34% PHB after 85 g/L glucose was used. It appears that the yfjB-carrying strain was more productive than the strain without yfjB. In terms of substrate to PHB yield Y PHB/glucose, the yfjB expressing strain was 0.15 g PHB/g glucose, significantly higher than 0.08 g PHB/g glucose of the control without yfjB overexpression. In the future, an optimized process employing yfjB overexpression recombinant should be able to reach a new high level for CDW and PHB production.
Discussion
Enhancing the carbon flux through the PP pathway has been proved to be effective in increasing NADPH concentration for PHB production (Lim et al. 2002; Jung et al. 2004; Song et al. 2006). Recently, a soluble pyridine nucleotide transhydrogenase has been applied to improve PHB productivity in E. coli (Sanchez et al. 2006). It was, therefore, proposed that NAD kinase which can catalyze the formation of NADP from NAD may increase NADP(H) concentrations and help improve PHB production in the presence of excess NADPH. To study the effect of overexpressing NAD kinase on PHB accumulation, E. coli NAD kinase gene yfjB was cloned and placed into two expression plasmids with different promoters and copy numbers.
In the absence of PHB production, no significant difference was found between the controls and the NAD kinase-overexpressing strains in terms of CDW, glucose consumption, and acetate production (Table 2), clearly suggesting that a high expression level of NAD kinase could not affect cell growth. This result was consistent with the previous study in Salmonella enterica (Grose et al. 2006). NAD kinase was found to be an allosteric enzyme, and E. coli NAD kinase is inhibited by the allosteric modifiers NADH and NADPH so that [NADPH]/[NADP] and [NAD]/[NADH] could be maintained at high levels (Kawai et al. 2001a). Under normal aerobic growth conditions, NAD kinase may be strongly inhibited (Grose et al. 2006). Excess amount of NAD kinase can be repressed by the regulation mechanism. All of these may contribute to the limited influence of NAD kinase overexpression on cell growth. Phosphoglucose isomerase gene pgi could be inactivated to direct carbon flux to PP pathway, resulting in the overproduction of NADPH, which in turn strongly suppressed the bacterial growth. This suppression could be partially removed by introducing a NADPH consumption pathway, such as PHB synthesis (Shi et al. 1999; Kabir and Shimizu 2003). Overexpression of NAD kinase which can regulate cofactor NADPH concentration described in this study seemed to be a much better approach to release the negative impact on cell growth in that the NAD kinase activity could be allosterically regulated to ensure intracellular redox balance (Kawai et al. 2001a).
NAD kinase overexpression in PHB production strains resulted in improved cell growth and PHB production, accompanied by a reduced acetate production compared with that of the corresponding controls (Fig. 2). Under the same growth conditions without process optimization, recombinant E. coli strains (p68E76) overexpressing NAD kinase gene grew 31% better than that of the control without NAD kinase in a 28-h study conducted in a 6-L NBS fermentor (Fig. 3). PHB content was also increased from 34% for the control to 50% for the NAD kinase-overexpressing strain. In addition, substrate to PHB yield Y PHB/glucose showed an increase from 0.08 for the control to 0.15 for the NAD kinase-overexpressing strain, a 76% increase for the Y PHB/glucose.
PHB synthesis is a NADPH-dependent process and may cause a decrease in NADPH pool (Fig. 1), leading to the reduced inhibition of NAD kinase to some extent. Excess amount of NAD kinase can quicken the conversion from NAD to NADP, thus enhance the NADP concentration. In a 24-h shake flasks study, NADP concentrations in NAD kinase-overexpressing strains were increased threefold to sixfold compared with the controls. NADP can be transformed into NADPH via three ways including the PP pathway, isocitrate dehydrogenase, and pyridine nucleotide transhydrogenase (Sauer et al. 2004). High NADP concentration may result in efficient supply of NADPH, which can promote PHB synthesis. Owing to the more effective NADP(H) supply, more carbon flux will run into the PHB biosynthesis pathway, resulting in the reduced acetate production and increased Y PHB/glucose observed (Figs. 2 and 3).
The nucleotides including NADP or NADPH can be easily oxidized, reduced, or broken down during the process of cell lysis and extraction. Published cofactor pool sizes varied significantly depending on different extraction and analysis methods. Moreover, cofactor pools can be changed quickly to meet the changes of cell environments; it is, therefore, hard to present the exact values of a cofactor pool in shake flasks (Sanchez et al. 2006). Based on the above considerations, the calculation of real-time intracellular cofactor concentrations was avoided. Instead, we measured the NADP and NADPH concentrations in 24-h shake flask studies using HCl (for NADP), NaOH (for NADPH) extraction methods, and an enzymatic cycling detection procedure (Zerez et al. 1987). Moreover, NAD kinase activity assays were performed to elucidate the enzyme performances, too. The NAD kinase activity in the strain harboring a high copy number plasmid was about 50 times higher than that of strains harboring a low copy number plasmid; also, the NADP concentration was increased approximately 70%. However, the PHB production was similar in the two NAD kinase-overexpressing recombinants (Fig. 2). Previous study showed that the addition of an appropriate amount of oleic acid to defined medium enhanced PHB production, yet excessive oleic acid did not lead to further PHB content increase (Lee et al. 1995). The reason for this oleic acid influence was given as follows: although NADPH plays an important role in PHB synthesis, it is not the only key factor. The supply of acetyl-CoA was also critical to PHB production. Moreover, considering the regulation mechanism of NAD kinase, an excessive amount of enzyme may be inhibited by other intracellular cofactors. Thus, the enhanced PHB production induced by NAD kinase overexpression was not without its limitation.
In cofactor assay experiments, the NADPH concentration of strains harboring pBHR68 and p68E76 was similar. However, a high NADPH concentration was observed in the NAD kinase overexpression dual plasmid system (Table 3). Reasons could be the different growth status of the strains since cofactor concentrations depend on growth conditions and growth stages. In the NAD kinase-overexpressing strains, the NADPH/NADP ratio was decreased compared to the controls due to the elevated NADP concentrations (Table 3). A high level of NADPH and/or NADPH/NADP ratio was known to be important for PHB synthesis in E. coli (Lee et al. 1996). It was recently reported that the NADPH/NADP ratio was lower in the pyridine nucleotide transhydrogenase gene (udhA) overexpressing strain than that in the control strain during the maximal PHB synthesis rate, and PHB accumulation was promoted with udhA overexpression (Sanchez et al. 2006). Sanchez et al. (2006), therefore, proposed that a drop in the NADPH/NADP ratio suggests a depletion of the NADPH pool to meet the demand of PHB synthesis. In this study of NAD kinase overexpression, the NADPH/NADP ratio was decreased due to the increase of NADP. However, the PHB yield from glucose was enhanced. The promotion of PHB synthesis could be regarded as an effective supply NADP(H). Thus, the availability of NADPH seems more important than high NADPH/NADP ratio in improving PHB productivity.
In this study, we demonstrated that NAD kinase overexpression enhanced PHB production in recombinant E. coli harboring the PHB synthesis operon, probably due to the effective supply of cofactors NADP(H). Our results could be extended to include the overexpression of NAD kinase as a method to enhance NADP(H)-dependent anabolic reactions.
References
Ahn WS, Park SJ, Lee SY (2000) Production of poly(3-hydroxybutyrate) by fed-batch culture of recombinant Escherichia coli with a highly concentrated whey solution. Appl Environ Microbiol 66:3624–3627
Anderson AJ, Dawes EA (1990) Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol Rev 54:450–472
Chen GQ, Wu Q (2005) The application of polyhydroxyalkanoates as tissue engineering materials. Biomaterials 26:6565–6578
Garavaglia S, Galizzi A, Rizzi M (2003) Allosteric regulation of Bacillus subtilis NAD kinase by quinolinic acid. J Bacteriol 185:4844–4850
Grose JH, Joss L, Velick SF, Roth JR (2006) Evidence that feedback inhibition of NAD kinase controls responses to oxidative stress. Proc Natl Acad Sci U S A 103:7601–7606
Jung YM, Lee JN, Shin HD, Lee YH (2004) Role of tktA gene in pentose phosphate pathway on odd-ball biosynthesis of poly-beta-hydroxybutyrate in transformant Escherichia coli harboring phbCAB operon. J Biosci Bioeng 98:224–227
Kabir MM, Shimizu K (2003) Fermentation characteristics and protein expression patterns in a recombinant Escherichia coli mutant lacking phosphoglucose isomerase for poly(3-hydroxybutyrate) production. Appl Microbiol Biotechnol 62:244–255
Kang Z, Wang Q, Zhang HJ, Qi QS (2008) Construction of a stress-induced system in Escherichia coli for efficient polyhydroxyalkanoates production. Appl Microbiol Biotechnol 79:203–208
Kawai S, Mori S, Mukai T, Hashimoto W, Murata K (2001a) Molecular characterization of Escherichia coli NAD kinase. Eur J Biochem 268:4359–4365
Kawai S, Suzuki S, Mori S, Murata K (2001b) Molecular cloning and identification of UTR1 of a yeast Saccharomyces cerevisiae as a gene encoding an NAD kinase. FEMS Microbiol Lett 200:181–184
Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RMII, Peterson KM (1995) Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176
Lai WJ, Chen GQ (2006) Polyhydroxybutyrate synthesis in recombinant Zymomonas mobilis affected ethanol production. China Biotechnology 26:52–56
Lee SY (1997) E. coli moves into the plastic age. Nat Biotechnol 15:17–18
Lee SY, Lee YK, Chang HN (1995) Stimulatory effects of amino-acids and oleic-acid on poly(3-hydroxybutyric acid) synthesis by recombinant Escherichia coli. J Ferment Bioeng 79:177–180
Lee IY, Kim MK, Park YH, Lee SY (1996) Regulatory effects of cellular nicotinamide nucleotides and enzyme activities on poly(3-hydroxybutyrate) synthesis in recombinant Escherichia coli. Biotechnol Bioeng 52:707–712
Lee SY, Choi JI, Wong HH (1999) Recent advances in polyhydroxyalkanoate production by bacterial fermentation: mini-review. Int J Biol Macromol 25:31–36
Lemoigne M (1926) Products of dehydration and of polymerization of β-hydroxybutyric acid. Bull Soc Chim Biol 8:770–782
Lenz RW, Marchessault RH (2005) Bacterial polyesters: biosynthesis, biodegradable plastics and biotechnology. Biomacromolecules 6:1–8
Lerner F, Niere M, Ludwig A, Ziegler M (2001) Structural and functional characterization of human NAD kinase. Biochem Biophys Res Commun 288:69–74
Lim SJ, Jung YM, Shin HD, Lee YH (2002) Amplification of the NADPH-related genes zwf and gnd for the oddball biosynthesis of PHB in an E. coli transformant harboring a cloned phbCAB operon. J Biosci Bioeng 93:543–549
Liu Q, Ouyang SP, Chung A, Wu Q, Chen GQ (2007) Microbial production of R-3-hydroxybutyric acid by recombinant E. coli harboring genes of phbA, phbB, and tesB. Appl Microbiol Biotechnol 76:811–818
Magni G, Orsomando G, Raffaelli N (2006) Structural and functional properties of NAD kinase, a key enzyme in NADP biosynthesis. Mini Rev Med Chem 6:739–746
Ochiai A, Mori S, Kawai S, Murata K (2004) Overexpression, purification, and characterization of ATP-NAD kinase of Sphingomonas sp A1. Protein Expr Purif 36:124–130
Peoples OP, Sinskey AJ (1989) Poly-beta-hydroxybutyrate (PHB) biosynthesis in Alcaligenes eutrophus H16. Identification and characterization of the PHB polymerase gene (phbC). J Biol Chem 264:15298–15303
Sanchez AM, Andrews J, Hussein I, Bennett GN, San KY (2006) Effect of overexpression of a soluble pyridine nucleotide transhydrogenase (UdhA) on the production of poly(3-hydroxybutyrate) in Escherichia coli. Biotechnol Prog 22:420–425
Sauer U, Canonaco F, Heri S, Perrenoud A, Fischer E (2004) The soluble and membrane-bound transhydrogenases UdhA and PntAB have divergent functions in NADPH metabolism of Escherichia coli. J Biol Chem 279:6613–6619
Schubert P, Steinbüchel A, Schlegel HG (1988) Cloning of the Alcaligenes eutrophus genes for synthesis of poly-beta-hydroxybutyric acid (PHB) and synthesis of PHB in Escherichia coli. J Bacteriol 170:5837–5847
Shi HD, Nikawa J, Shimizu K (1999) Effect of modifying metabolic network on poly-3-hydroxybutyrate biosynthesis in recombinant Escherichia coli. J Biosci Bioeng 87:666–677
Slater SC, Voige WH, Dennis DE (1988) Cloning and expression in Escherichia coli of the Alcaligenes eutrophus H16 poly-beta-hydroxybutyrate biosynthetic pathway. J Bacteriol 170:4431–4436
Song BG, Kim TK, Jung YM, Lee YH (2006) Modulation of talA gene in pentose phosphate pathway for overproduction of poly-beta-hydroxybutyrate in transformant Escherichia coli harboring phbCAB operon. J Biosci Bioeng 102:237–240
Spiekermann P, Rehm BH, Kalscheuer R, Baumeister D, Steinbüchel A (1999) A sensitive, viable-colony staining method using Nile red for direct screening of bacteria that accumulate polyhydroxyalkanoic acids and other lipid storage compounds. Arch Microbiol 171:73–80
Steinbüchel A, Valentin HE (1995) Diversity of bacterial polyhydroxyalkanoic acids. FEMS Microbiol Lett 128:219–228
Steinbüchel A, Lutke-Eversloh T (2003) Metabolic engineering and pathway construction for biotechnological production of relevant polyhydroxyalkanoates in microorganisms. Biochem Eng J 16:81–96
Wei XX, Shi ZY, Yuan MQ, Chen GQ (2009) Effect of anaerobic promoters on the microaerobic production of polyhydroxybutyrate (PHB) in recombinant Escherichia coli. Appl Microbiol Biotechnol. doi:https://doi.org/10.1007/s00253-008-1816-4
Yanisch-Perron C, Vieira J, Messing J (1985) Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103–119
Zerez CR, Lee SJ, Tanaka KR (1987) Spectrophotometric determination of oxidized and reduced pyridine nucleotides in erythrocytes using a single extraction procedure. Anal Biochem 164:367–73
Zheng Z, Li M, Xue XJ, Tian HL, Li Z, Chen GQ (2006) Mutation on N-terminus of polyhydroxybutyrate synthase of Ralstonia eutropha enhanced PHB accumulation. Appl Microbiol Biotechnol 72:896–905
Acknowledgements
We are grateful to Prof. Alexander Steinbüchel of the University of Münster in Germany for the generous donation of plasmid pBHR68. The research was supported by the China National High Tech 863 Grants (Project Nos. 2006AA02Z242 and 2006AA020104), as well as the State Basic Science Foundation 973 (2007CB707804).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Li, ZJ., Cai, L., Wu, Q. et al. Overexpression of NAD kinase in recombinant Escherichia coli harboring the phbCAB operon improves poly(3-hydroxybutyrate) production. Appl Microbiol Biotechnol 83, 939–947 (2009). https://doi.org/10.1007/s00253-009-1943-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-009-1943-6