Abstract
Natural products continue to play a major role in drug discovery and development. However, chemical redundancy is an ongoing problem. Genomic studies indicate that certain groups of bacteria and fungi have dozens of secondary metabolite pathways that are not expressed under standard laboratory growth conditions. One approach to more fully access the metabolic potential of cultivatable microbes is mixed fermentation, where the presence of neighboring microbes may induce secondary metabolite synthesis. Research to date indicates that mixed fermentation can result in increased antibiotic activity in crude extracts, increased yields of previously described metabolites, increased yields of previously undetected metabolites, analogues of known metabolites resulting from combined pathways and, importantly, induction of previously unexpressed pathways for bioactive constituents.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Natural products continue to be an unparalleled resource for pharmaceutical lead discovery. Of all small molecule drugs that were launched between 1981 and 2006, 63% were derived from natural products (Newman and Cragg 2007). Natural products have played a particularly large role in antitumor, antimicrobial, and antihypertensive areas (Banerjee et al. 2008; Newman and Cragg 2007). Although the vast majority of macro and microorganisms have yet to be described (and almost certainly hold a tremendous reservoir of bioactive constituents), there exists today an unfortunate problem with chemical redundancy in natural products drug discovery.
In response to this, there has been a shift to develop strategies to maximize the chemical diversity obtained from each species, particularly microbial species. Environmental conditions are critical to the synthesis of microbial secondary metabolites (Knight et al. 2003). Maximizing the types of samples collected; diversifying the isolation strategies; optimizing nutrients, temperature, pH, aeration, and incubation time; exploiting talented strains; and genetically modifying strains can greatly influence productivity (Knight et al. 2003). Bode et al. (2002) explored the literature regarding alterations in secondary metabolites with varied cultured conditions. Their group, for example, isolated more than 100 compounds from more than 25 structural classes from six different microbes by altering culture conditions (Bode et al. 2002). The fungus Sphaeropsidales sp., which synthesizes the antifungal spirobisnaphthalene cladospirone bisepoxyde, made eight new and six known spironaphthalenes when grown under varied conditions, and new bisnaphthalenes and a rare macrolide when grown in the presence of enzyme inhibitors such as tricyclazole (Bode et al. 2002).
Results of genome analyses reveal why manipulation of microbial culture conditions can be such a fertile avenue for secondary metabolite discovery. Microbial sequencing studies indicate that the biosynthetic potential of many strains is much greater than that observed by fermentation (Bode et al. 2002; Newman and Cragg 2007; Schneider et al. 2008; Udwary et al. 2007). Because the genomes of fungi, actinomycetes, and myxobacteria are much larger than necessary for basic functioning, it was presumed that part of their genomes encoded alternative metabolic pathways (Knight et al. 2003). Indeed, sequencing of Streptomyces avermitilis, a known producer of avermectin, revealed at least 25 gene clusters for siderophores, spore pigments, and secondary metabolites of polyketide or nonribosomal peptide origin (Ōmura et al. 2001). Greater than 6% of the S. avermitilis genome contains genes for secondary metabolite biosynthesis. More than 200 secondary metabolites have been isolated from strains of Streptomyces hygroscopicus (Zhang 2005). Salinispora tropica, a marine actinomycete, devotes a very large portion of its genome, approximately 10%, to natural product assembly (Udwary et al. 2007). The polyketide biosynthetic pathways of S. tropica are of greater variety than any other sequenced bacterial genome, and most of the pathways are novel (Udwary et al. 2007). Aspergilli are another rich source of secondary metabolites (Schneider et al. 2008); they can harbor 30–50 distinct polyketide synthase and nonribosomal peptide synthase gene clusters per species (Payne et al. 2006). Aspergillus niger contains 17 nonribosomal peptide synthase and 34 polyketide synthase-encoding genes; most of which are located in clusters (Pel et al. 2007).
Why such a large genomic investment in secondary metabolites? Microbes interact in their natural environments (Slattery et al. 2001; Wiener 1996), presumably even more so in areas with a high microbial burden, for e.g., soil, terrestrial plant surfaces, and marine plant and animal surfaces. Surviving in such competitive environments likely requires strategies such as the production of bioactive secondary metabolites (Slattery et al. 2001; Wiener 1996). It is now apparent that potentially interesting gene clusters, while possibly expressing metabolites that increase competitiveness in natural environments, can remain silent in the unnatural setting of the microbiology laboratory. Varying culture conditions is one method of turning on these pathways.
Another strategy to mimic more natural microbial environments is to grow them in the presence of other microbes. Such mixed fermentations have been used for centuries in food and beverage production and for decades in industrial enzyme and solvent production and wastewater treatment, to name a few applications (Salmon and Bull 1984). Some results of this approach include decreased costs, increased efficiency, and diversified products.
Application of mixed fermentation to natural product drug discovery seems an obvious extension, but the field is in its infancy, probably because of early fears of lack of reproducibility. Results of mixed fermentation include increased antibiotic activity in crude extracts, increased yields of previously described metabolites, increased yields of previously undetected metabolites, analogues of known metabolites resulting from combined pathways, and induction of previously unexpressed pathways for bioactive constituents.
Mixed fermentation as a strategy for natural product drug discovery
In an early report of mixed fungal fermentation, Gloeophyllum abietinum increased its synthesis of six toxins, including oosponol and oospoglycol, when grown in liquid culture with Heterobasidion annosum (Sonnenbichler et al. 1994; Table 1). Growth retardation of G. abietinum was noted concomitant with enhanced toxin synthesis. Stimulation of toxin synthesis also occurred with cell-free extracts of H. annosum, but was not induced by H. annosum cell walls, nor by polypeptides or polysaccharides.
In another early report of mixed fungal fermentation, tremendous increases in Monascus pigment production and in Monascus growth were noted in solid medium when co-cultivated with either Saccharomyces cerevisiae or Aspergillus oryzae (Shin et al. 1998; Table 1). For these experiments, inducer organisms were added 1 day after Monascus was inoculated. Increased pigment production was also noted in liquid culture, but was much less than that on solid medium. No increased pigment production or cell growth occurred upon cultivation of Monascus with the bacterium Bacillus cereus. Culture filtrates of S. cerevisiae were also effective in inducing pigment production in Monascus, suggesting that enhanced pigment production was not due to cell-to-cell contact or limitation of components due to consumption by S. cerevisiae. Further experiments revealed that hydrolytic enzymes (e.g., chitinase) from a variety of fungi were effective in enhancing Monascus pigment production, while those from bacteria were not. The authors suggest that hydrolysis of Monascus cell walls stimulates a defense mechanism resulting in overproduction of hydrophobic substances such as pigments, in order to block enzyme attack. Co-cultured Monascus exhibited increased numbers and sizes of vacuoles filled with electron dense substances, likely pigments (Suh and Shin 2000a). Increased cell mass and pigment production did not occur in the presence of protein kinase C inhibitors, and protein kinase C activity was only detected in mixed cultures (Suh and Shin 2000b).
Unidentified, surface-associated marine bacteria increased their synthesis of antibiotics when cultured with Staphylococcus aureus, Pseudomonas aeruginosa, or Escherichia coli (Mearns-Spragg et al. 1998). Five-day-old cultures of the terrestrial inducer strains were placed inside semi-permeable dialysis tubing and added to liquid cultures of the marine bacteria. Relative amounts of antibiotics in culture supernatants were determined after 24 h. Heat-killed cells of S. aureus were also capable of inducing antibiotic production in certain marine strains. Using an even larger collection of surface-associated marine bacteria, these researchers later demonstrated that an unexpectedly large percentage of their collection (35%) synthesized antibiotics in the presence of P. aeruginosa, E. coli, Bacillus subtilis, other surface-associated marine bacteria, or cell-free culture supernatants (Burgess et al. 1999). Further confirmation that bacterial competition is potentially very common in marine environments was provided by Long and Azam (2001). Eighty-six marine bacteria were examined for their inhibition of 85 other isolates by agar diffusion assays. More than half of the isolates produced inhibitory compounds. Significantly more of the attached bacteria than the free-living bacteria produced inhibitory compounds. The attached bacteria also inhibited more isolates than the free-living bacteria. Surface-associated bacteria live in a highly competitive environment. Synthesis of bioactive secondary metabolites may be common in this environment, and these in vitro studies support this idea. Surface-associated marine microbes may be a valuable source of biologically active natural products.
When 76 species of Streptomyces were plated in pairs 1 cm apart, 34% of the strains stimulated antibiotic production in 13% of the strains (Ueda et al. 2000). Synthetic A factor, γ-lactone, and butanolides, which are known to stimulate antibiotic production in Streptomyces, did not stimulate antibiotic production in these strains.
Fifty-three species of surface-associated marine bacteria were cultured in liquid media with the marine bacterium Streptomyces tenjimariensis, a known producer of the antibiotics istamycin A and B (Slattery et al. 2001). The inducing strains were selected based on growth rates similar to S. tenjimariensis. Co-culture with 22.6% of the marine isolates resulted in an approximate doubling of the istamycin levels found in S. tenjimariensis monocultures (Table 1). The inducing strains included Gram-positive, Gram-negative, and Gram-variable isolates. Pre-establishment of S. tenjimariensis (24 h) was critical to enhanced antibiotic synthesis. Co-inoculation or pre-establishment of the inducing bacteria resulted in a significant reduction in istamycin levels compared to monocultures.
New lipoaminopeptides thought to be the result of biosynthetic pathways from two different fungi were produced in solid co-culture. The fungal parasite Mycogone rosea was cultured with the endophytic fungus Acremonium sp., and acremostatins A, B, and C were detected (Degenkolb et al. 2002; Table 1). Acremonium sp. was established for 7 days prior to inoculation with a 14-day culture of M. rosea, and then co-cultured for 14 days. The authors postulate that M. rosea contributes 2-methyl-decanoic acid to the terminus of the Acremonium products leucinostatins A, B, and K, resulting in acremostatins A, B, and C. The new derivatives were not detected in monocultures of either fungus. Thus, chimeric pathways resulting from co-culture can also lead to new natural products.
New aminoglycoside antibiotics that may be the result of combined biosynthetic pathways were produced by Rhodococcus fascians when cultured with Streptomyces padanus (Kurosawa et al. 2008; Table 1). Rhodostreptomycins A and B were not detected in pure cultures of either bacterium, and both compounds had broad-spectrum antibacterial activity. There was a correlation between production of rhodostreptomycins and the presence of S. padanus DNA in R. fascians. The transferred DNA may directly induce aminoglycoside production, or it may contain genes encoding enzymes for aminoglycoside synthesis. Alternatively, combined pathways from both microbes’ genomes could result in rhodostreptomycin synthesis. Hopefully, details of the molecular biology of the transferred DNA are forthcoming.
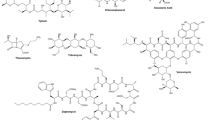
In 2001, Cueto et al. reported that a surface-associated marine Pestalotia synthesized a new antibiotic only when cultured with an unidentified Gram-negative marine bacterium (Table 1). Pestalone (Fig. 1), a new benzophenone antibiotic, was undetectable in pure cultures of the fungus Pestalotia. Pestalone had potent activity against methicillin-resistant S. aureus and vancomycin-resistant Enterococcus faecium and marginal activity against the National Cancer Institute’s 60 human tumor cell line screen. In these experiments, the bacterium was added 24 h after Pestalotia inoculation, and cultures extracted 6 days later. Neither organic extracts nor cell-free culture supernatants of the marine bacterium induced pestalone synthesis. Interestingly, low yields of pestalone could be induced by ethanol in pure cultures of Pestalotia.
A second example of a previously unexpressed pathway induced by microbial competition was provided by Fenical’s group (Oh et al. 2005). Four new diterpenoids, libertellenones A–D (Table 1, Fig. 1), were detected when the marine fungus Libertella sp. was cultured with an unidentified marine bacterium (the same bacterium used to induce pestalone synthesis). Libertella was established in liquid culture for 3 days prior to adding the marine bacterium, and co-culture extractions performed 2 days later. No new metabolites were detected in 49 other marine fungal strains co-cultured with the bacterium. The libertellenones were not detected in pure cultures of the fungus or the bacterium. Heat-killed bacterial cells, cell-free culture supernatants, or ethyl acetate extracts of a viable bacterial culture did not induce diterpenoid synthesis. The authors suggest that diterpenoid induction is controlled by cell–cell interactions and not signaling molecules. Indirect evidence is provided that the libertellenones are Libertella products. The diterpenoids had no obvious antibacterial activity (including no activity against the inducing marine bacterium), but had potent activity against a human colon carcinoma cell line.
Further demonstration that mixed fermentation represents a potentially valuable strategy for discovery of novel metabolites was the report of marinamide A and B production in co-culture of two marine fungi (Zhu and Lin 2006; Table 1, Fig. 1). The two unidentified, plant-associated fungi were inoculated simultaneously into liquid media and cultured for 12 days. Marinamide A, a novel 1-isoquinolone alkaloid, and its methyl ester, marinamide B, were not obtained in pure culture of either fungus. Both compounds had activity against E. coli, Pseudomonas pyocyanea, and S. aureus. Additional, previously undetected metabolites have since been found in co-cultures of these two marine fungi (Zhu et al. 2007; Table 1). The diketopiperazine cyclo-(phe-phe) and 6-methyl-salicylic acid inhibited growth of the plant pest Heliothis armigera and the parasitic copepod Sinergasilus sp. The metabolites were not obtained in pure culture of either fungus.
Examples of bioactive microbial constituents that were unknown until their production was enhanced 100-fold in mixed fermentation are the cyclic depsipeptides emericellamides A and B (Oh et al. 2007; Table 1, Fig. 1). This report provides another example of a cross-kingdom mixed fermentation. A surface-associated marine fungus, Emericella sp., was grown in liquid culture for 3 days prior to inoculation of the marine actinomycete Salinispora arenicola and incubated for two more days. Emericellamides A and B had moderate activity against methicillin-resistant S. aureus. Once the depsipeptides were isolated from the mixed cultures, the researchers found that the new compounds were indeed produced by Emericella sp. in pure culture, but in amounts undetectable by normal LC–MS analysis.
Conclusions and prospects for the future
To date, at least nine new compounds with biological activity have been identified by mixed fermentation, pestalone, libertellenones A–D, marinamides A and B, and emericellamides A and B (Fig. 1). How did these natural products remain undetected for so long? Are the genes not expressed under standard growth conditions? Are external signals required to turn the genes on? Is it simply that chemical detection methods are not powerful enough? Certainly not in every situation, as Fenical’s group was able to detect trace quantities of the emericellamides in pure culture (Oh et al. 2007), but not the libertellenones (Oh et al. 2005). In any case, the emericellamides would not have been isolated without the structure determination that resulted from increased yields in mixed culture.
Another encouraging result from early natural product-directed mixed fermentation studies was the isolation of new lipoaminopeptide analogues, where the core structures were provided by Acremonium, and the nitrogen termini by M. rosea (Degenkolb et al. 2002). While biological activities were not reported, the parent leucinostatins inhibit bacteria and fungi. Thus, co-cultivation of microbes producing related products is another way to increase the chemical diversity of microbial secondary metabolites (Degenkolb et al. 2002).
Competition for limiting natural resources is believed to be the selective force that promotes biosynthesis of biologically active compounds. It is intriguing that in the majority of reports summarized here, microbes from highly competitive environments, e.g., plant and animal surfaces, were responsible for enhanced or previously undescribed syntheses of bioactive molecules. Other fascinating questions for researchers entering this field to consider include: What is the optimum timing of inoculation of the cultivation partners and of extraction? What, exactly, is responsible for inducing synthesis of the bioactive constituent? Inducing factors varied dramatically in the reports summarized here, from chitinase (Shin et al. 1998) to viable cells (Oh et al. 2005).
Microbes have already played a pivotal role in human medicine. More thorough exploitation of the metabolic potential of microbes via mixed fermentation is proving to be an effective way to augment natural product libraries. With countless possible microbe combinations and increasingly sophisticated chemical isolation and structure determination methods, the potential for mixed fermentation in natural product drug discovery seems quite promising.
References
Banerjee S, Wang Z, Mohammad M, Sarkar FH, Mohammad RM (2008) Efficacy of selected natural products as therapeutic agents against cancer. J Nat Prod 71:492–496
Bode HB, Bethe B, Höfs R, Zeeck A (2002) Big effects from small changes: possible ways to explore nature’s chemical diversity. ChemBioChem 3:619–627
Burgess JG, Jordan EM, Bregu M, Mearns-Spragg A, Boyd KG (1999) Microbial antagonism: a neglected avenue of natural products research. J Biotechnol 70:27–32
Cueto M, Jensen PR, Kauffman C, Fenical W, Lobkovsky E, Clardy J (2001) Pestalone, a new antibiotic produced by a marine fungus in response to bacterial challenge. J Nat Prod 64:1444–1446
Degenkolb T, Heinze S, Schlegel B, Strobel G, Gräfe U (2002) Formation of new lipoaminopeptides, acremostatins A, B, and C, by co-cultivation of Acremonium sp. Tbp-5 and Mycogone rosea DSM 12973. Biosci Biotechnol Biochem 66:883–886
Knight V, Sanglier JJ, DiTullio D, Braccili S, Bonner P, Waters J, Hughes D, Zhang L (2003) Diversifying microbial natural products for drug discovery. Appl Microbiol Biotechnol 62:446–458
Kurosawa K, Ghiviriga I, Sambandan TG, Lessard PA, Barbara JE, Rha C, Sinskey AJ (2008) Rhodostreptomycins, antibiotics biosynthesized following horizontal gene transfer from Streptomyces padanus to Rhodococcus fascians. J Am Chem Soc 130:1126–1127
Long RA, Azam F (2001) Antagonistic interactions among marine pelagic bacteria. Appl Environ Microbiol 67:4975–4983
Mearns-Spragg A, Bregu M, Boyd KG, Burgess JG (1998) Cross-species induction and enhancement of antimicrobial activity produced by epibiotic bacteria from marine algae and invertebrates, after exposure to terrestrial bacteria. Lett Appl Microbiol 27:142–146
Newman DJ, Cragg GM (2007) Natural products as sources of new drugs over the last 25 years. J Nat Prod 70:461–477
Oh DC, Jensen PR, Kauffman CA, Fenical W (2005) Libertellenones A-D: induction of cytotoxic diterpenoid biosynthesis by marine microbial competition. Bioorg Med Chem 13:5267–5273
Oh DC, Kauffman CA, Jensen PR, Fenical W (2007) Induced production of emericellamides A and B from the marine-derived fungus Emericella sp. in competing co-culture. J Nat Prod 70:515–520
Ōmura S, Ikeda H, Ishikawa J, Hanamoto A, Takahashi C, Shinose M, Takahashi Y, Horikawa H, Nakazawa H, Osonoe T, Kikuchi H, Shiba T, Sakaki Y, Hattori M (2001) Genome sequence of an industrial microorganism Streptomyces avermitilis: deducing the ability of producing secondary metabolites. Proc Natl Acad Sci U S A 98:12215–12220
Payne GA, Nierman WC, Wortman JR, Pritchard BL, Brown D, Dean RA, Bhatnagar D, Cleveland TE, Machida M, Yu J (2006) Whole genome comparison of Aspergillus flavus and A. oryzae. Med Mycol 44:S9–S11
Pel HJ et al (2007) Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat Biotech 25:221–231
Salmon I, Bull AT (1984) Mixed-culture fermentations in industrial microbiology. Curr Perspect Microb Ecol; Proc of the Third Intl Symp on Microb Ecol (Michigan State University, 7–12 Aug., 1983):656–662
Schneider P, Misiek M, Hoffmeister D (2008) In vivo and in vitro production options for fungal secondary metabolites. Mol Pharmcol 5:234–242
Shin CS, Kim HJ, Kim MJ, Ju JY (1998) Morphological change and enhanced pigment production of Monascus when co-cultured with Saccharomyces cerevisiae or Aspergillus oryzae. Biotechnol Bioeng 59:576–581
Slattery M, Rajbhandari I, Wesson K (2001) Competition-mediated antibiotic induction in the marine bacterium Streptomyces tenjimariensis. Microb Ecol 41:90–96
Sonnenbichler J, Dietrich J, Peipp H (1994) Secondary fungal metabolites and their biological activities, V. Investigations concerning the induction of biosynthesis of toxic secondary metabolites in basidiomycetes. Biol Chem 375:71–79
Suh JH, Shin CS (2000a) Analysis of the morphologic changes of Monascus sp. J101 cells cocultured with Saccharomyces cerevisiae. FEMS Microbiol Lett 193:143–147
Suh JH, Shin CS (2000b) Physiological analysis on novel coculture of Monascus sp. J101 with Saccharomyces cerevisiae. FEMS Microbiol Lett 190:241–245
Udwary DW, Zeigler L, Asolkar RN, Singan V, Lapidus A, Fenical W, Jensen PR, Moore BS (2007) Genome sequencing reveals complex secondary metabolome in the marine actinomycete Salinispora tropica. Proc Natl Acad Sci U S A 104:10376–10381
Ueda K, Kawai S, Ogawa HO, Kiyama A, Kubota T, Kawanobe H, Beppu T (2000) Wide distribution of interspecific stimulatory events on antibiotic production and sporulation among Streptomyces species. J Antibiot 53:979–982
Wiener P (1996) Experimental studies on the ecological role of antibiotic production in bacteria. Evol Ecol 10:405–421
Yakovleva EP, Sokolova EN (1978) Dissociation of a Candida tropicalis culture and its capacity to stimulate levorin synthesis when cultured together with Actinomyces levoris. Antibiotiki 23:199–203
Zhang L (2005) Integrated approaches for discovering novel drugs from microbial natural products. In: Zhang L, Demain AL (eds) Natural products: drug discovery and therapeutic medicine. Humana, Totowa NJ, pp 33–55
Zhu F, Lin Y (2006) Marinamide, a novel alkaloid and its methyl ester produced by the application of mixed fermentation technique to two mangrove endophytic fungi from the South China Sea. Chin Sci Bull 51:1426–1430
Zhu F, Lin Y, Ding J, Wang X, Huang L (2007) Secondary metabolites of two marine-derived mangrove endophytic fungi (strain nos. 1924# and 3893#) by mixed fermentation. Chem Ind Forest Prod 27:8–10
Acknowledgments
Financial support was provided by grants R01 CA90441-01-05, 2R56 CA090441-06A1, and 5R01 CA090441-07 from the Division of Cancer Treatment, Diagnosis and Centers, National Cancer Institute, DHHS, and the Arizona Biomedical Research Commission. Many thanks to Dr. F. Hogan for preparing Fig. 1.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Pettit, R.K. Mixed fermentation for natural product drug discovery. Appl Microbiol Biotechnol 83, 19–25 (2009). https://doi.org/10.1007/s00253-009-1916-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-009-1916-9