Abstract
Quinone reductase activity of azoreductase AZR from Rhodobacter sphaeroides was reported. High homologies were found in the cofactor/substrate-binding regions of quinone reductases from different domains. 3D structure comparison revealed that AZR shared a common overall topology with mammal NAD(P)H/quinone oxidoreductase NQO1. With menadione as substrate, the optimal pH value and temperature were pH 8–9 and 50°C, respectively. Following the ping-pong kinetics, AZR transferred two electrons from NADPH to quinone substrate. It could reduce naphthoquinones and anthraquinones, such as menadione, lawsone, anthraquinone-2-sulfonate, and anthraquinone-2,6-disulfonate. However, no activity was detected with 1,4-benzoquinone. Dicoumarol competitively inhibited AZR’s quinone reductase activity with respect to NADPH, with an obtained K i value of 87.6 μM. Significantly higher survival rates were obtained in Escherichia coli YB overexpressing AZR than in the control strain when treated by heat shock and oxidative stressors such as H2O2 and menadione.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Compared to traditional physicochemical methods, microbial treatment of xenobiotic compounds, such as azo dyes and nitroaromatics, is a promising strategy. A wide range of bacteria have been reported capable of reducing azo dyes (Stolz 2001; Dos Santos et al. 2007). Enzymes that catalyze the reduction of azo groups are termed azoreductase. In the last decade, many azoreductases have been purified and characterized from different bacterial species (Chen 2006). Utilizing NAD(P)H as electron donor, they catalyze the successive two-electron-reduction of azo groups in vitro. Previously, we have cloned and characterized a gene azr (537 bp, GenBank accession number AY150311) coding for azoreductase AZR from Rhodobacter sphaeroides. AZR overexpressed in Escherichia coli catalyzes the reduction of various azo compounds with different structures (Yan et al. 2004). Further studies demonstrated that AZR adopts a flavodoxin-like fold with a three-layer α/β/α structure. AZR can also function as nitroreductase and flavin mononucleotide (FMN) reductase in vitro. It was shown that 2,4,6-trinitrotoluene was the most efficient nitro substrate and was reduced to hydroxylamino-dinitrotoluene (Liu et al. 2007). However, it has been argued that, as introduction of azo and nitro compounds into the environment is a recent anthropogenic event, enzymatic reduction of azo dyes and nitroaromatics may be secondary activities of reductases with different primary roles.
In addition to azo and nitro compounds, quinone is another kind of electrophilic compound. Quinones, characteristic of two carbonyl groups in an unsaturated six-member carbon ring, constitute an important class of ubiquitous and naturally occurring compounds, including several biologically important coenzymes, acceptors, or vitamins (Deller et al. 2008). Cellular oxidoreductases involved in the reduction of quinone compounds are termed quinone reductase. Quinones can be reduced by enzymes such as cytochrome P450 reductases via one-electron reduction to potentially harmful semiquinones. Semiquinone intermediates in turn are highly prone to react with molecular oxygen to generate superoxide radicals, which lead to oxidative stress and cell damage. On the other hand, quinones can also be competitively reduced through two-electron pathway by NAD(P)H/quinone reductases to quinols, which is believed to help minimize the oxidative stress (Deller et al. 2008; Sollner et al. 2007; Gonzalez et al. 2005).
In this paper, the quinone reductase activity of AZR was studied in detail. It was also found that the survival of E. coli strain bearing the azr gene is significantly higher than that of the control strain when treated by heat shock and oxidative stressors.
Materials and methods
Chemicals
1,4-Benzoquinone, 2-methyl-1,4-naphthoquinone (menadione), 2-hydroxy-1,4-naphthoquinone (lawsone), anthraquinone-2-sulfonate (AQS), anthraquinone-2,6-disulfonate (AQDS), paraquat, and thrombin were obtained from Sigma. Ampicillin, bovine serum albumin (BSA), and isopropyl-β-D-thiogalactopyranoside (IPTG) were purchased from TaKaRa Dalian. All other chemicals were of highest analytical grade available and used without further purification.
Bacterial strains and culture conditions
E. coli JM109 was obtained from TaKaRa Dalian. The gene azr encoding azoreductase from R. sphaeroides AS1.1737 was cloned and inserted into plasmid pGEX 4T-1 under the control of the lac promoter (Yan et al. 2004). The recombinant plasmid was transformed into CaCl2-treated E. coli JM109 competent cell. The recombinant strain was named E. coli YB. All the strains were cultivated in Luria–Bertani (LB) media. The recombinant cells were induced by addition of 1 mM IPTG.
Sequence and structure analysis
Alignment of amino acid sequences was performed using GeneDoc (Nicholas et al. 1997). Structure comparison was conducted and viewed with Pymol (DeLano Scientific, San Carlos, CA, USA).
Enzyme assays
The purification of AZR was performed as described previously (Liu et al. 2007). Protein concentration was measured, according to the Bradford (1976) procedure, using BSA as a standard.
Quinone reductase activity was measured spectrophotometrically by monitoring NADPH disappearance at 340 nm (ɛ = 6.22 mM−1 cm−1). One-milliliter typical reaction mixtures contained 20 mM phosphate buffer (pH 7.0), 40 μM menadione, and a suitable amount of enzyme. The reaction was initiated by the addition of 100 μM NADPH. Menadione reduction was also monitored in a coupled assay (Wang and Maier 2004). Reduced menadione, menadiol, in turn reduces cytochrome c. The reaction mixtures contained 20 mM phosphate buffer (pH 7.0), 100 μM NADPH, 40 μM menadione, and 50 μM cytochrome c. Reduction of cytochrome c was monitored by the increase in absorbance at 550 nm (ɛ = 29.5 mM−1 cm−1). Optimal pH and temperature for the quinone reductase activity were determined with menadione. The optimal pH was determined with 20 mM acetate/Na-acetate buffer (pH 3–6), 20 mM Na2HPO4/NaH2PO4 buffer (pH 5–7), 20 mM Tris–HCl buffer (pH 7–10), and 20 mM glycine/NaOH buffer (pH 10–11), respectively. The optimal temperature was determined by measuring the rate of reaction at temperatures ranging from 20 to 70°C under standard assay conditions. Thermal stability of the quinone reductase activity was also studied. The enzyme was incubated in a water bath with a temperature range of 20–90°C. After 15 min incubation, the enzyme was added to an assay mixture at 30°C to study its residual activity.
For the steady-state kinetic analysis, the reaction mixtures contained 20 mM phosphate buffer (pH 7.0), a suitable amount of enzyme, and 10–50 μM menadione. The reactions were initiated by the addition of 25, 50, 75 or 100 μM NADPH.
To study the inhibition of dicoumarol on AZR’s quinone reductase activity, the initial NADPH oxidation rate was determined in the presence of various concentrations (0–100 μM) of dicoumarol. One-milliliter reaction mixtures contained 20 mM phosphate buffer (pH 7.0), 40 μM menadione, 30–100 μM NADPH, and a suitable amount of enzyme. The reaction was initiated by addition of NADPH. The dicoumarol stock solution (10 mM) was prepared in 0.1 M NaOH.
Survival of E. coli strains when treated by oxidative stressors and heat shock
Strains of E. coli JM109 and E. coli YB were grown overnight and then inoculated into fresh LB media, respectively, with an initial OD660 of 0.2. Strains were then cultivated aerobically until they reached an OD660 around 1.0. This represents around 3 × 107 cells ml−1, which is the 100% survival rate. Appropriate amounts of stressors (H2O2, 10 mM; paraquat, 0.4 mM; and menadione, 0.6 mM) were then added to cultures. Heat shock was induced by incubating cultures at 48°C. Cell survival was monitored by sampling at intervals, diluting in 20 mM phosphate buffer (pH 7.0), and plating aliquots onto LB plates to obtain viable cell counts.
All the experiments were carried out at least three times.
Results
Sequence and structure analysis
Amino acid sequence of AZR was aligned with those of other proteins possessing quinone reductase activity from Bacillus subtilis, Saccharomyces cerevisiae, E. coli, Arabidopsis thaliana, Archaeoglobus fulgidus, and Homo sapiens (Deller et al. 2006; Sollner et al. 2007; Patridge and Ferry 2006; Laskowski et al. 2002; Ross and Siegel 2004). The sequence length varies, and the overall sequence homology is not obvious. However, high similarities were found in the middle parts of their amino acid sequences. Regions of Pro72-Ser79 and Gly106-Gly111, corresponding to the two loops around the FMN group of AZR, respectively, are highly conserved (Fig. 1a). The two loops were reported to be involved in the binding of flavin cofactor and NAD(P)H, respectively (Liger et al. 2004; Liu et al. 2007).
Sequence and structure analysis of AZR. a Multiple sequence alignment of amino acid sequences of quinone reductases from Rhodobacter sphaeroides (AZR, GenBank accession number AAN17400), Bacillus subtilis (YhdA, GenBank accession number CAB12762), Saccharomyces cerevisiae (Lot6p, GenBank accession number 1T0IA), Arabidopsis thaliana (FQR1, GenBank accession number NP200261), Escherichia coli (WrbAEc, GeneBank accession number AAA24759), Archaeoglobus fulgidus (WrbAAf, GenBank accession number NP069179), and Homo sapiens (NQO1, GenBank accession number P15559). b Alignment of 3D structures of AZR and NQO1. Ribbon representation of AZR and NQO1 are shown in red and green, respectively. The flavin cofactors are shown as stick models
As shown in Fig. 1b, although the overall sequences of AZR and NQO1 demonstrate low homology, monomers of the two proteins share a common α/β topology (rmsd of 9.5 Å over 467 atoms). They both adopt a typical flavodoxin-like fold, consisting of a central five-stranded parallel β sheet surrounded by α helices on both sides. In addition, different flavin cofactors (FMN for AZR while flavin adenine dinucleotide for NQO1) are bound in a similar manner, with the si-face partly exposed to the solvent and the re-face buried.
Characterization of quinone reductase
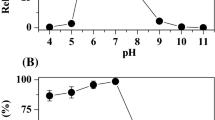
The quinone reductase had maximal activity at pH 8–9. Maximum quinone reductase activity was observed at 50°C. Thermal stability studies demonstrated that, although the enzyme was relatively stable up to 50°C, it lost activity rapidly at temperatures above 50°C.
Using NADPH as electron donors, several naturally occurring and artificial quinone substrates were studied for AZR’s quinone reductase activity. The enzyme could use menadione, lawsone, AQS, and AQDS as electron acceptors. No activity was detected with 1,4-benzoquione. According to the k cat/K m value, menadione was the best substrate for AZR among the quinones examined (Table 1).
The quinone reductase activity of AZR was determined using a coupled menadione-cytochrome c reduction assay that measures an increase in absorbance at 550 nm due to cytochrome c reduction. Using this assay, AZR showed an activity of 10.9 ± 1.3 μmol of cytochrome c reduced per minute per milligram of protein.
Using menadione as electron acceptor at various fixed concentrations of NADPH, the reaction mechanism of AZR quinone reductase was investigated by steady-state kinetic analysis. The obtained parallel lines in double reciprocal plots (Fig. 2) indicated that AZR reduces quinones with bi–bi ping-pong kinetics.
The effects of dicoumarol on AZR’s quinone reductase activity were studied. As shown in Fig. 3, dicoumarol was a competitive inhibitor of NADPH. The inhibition constant (K i) for NADPH was 87.6 μM.
Enhancing survival of E. coli by expression of AZR
It was reported that quinone reductase might be involved in combating oxidative stress (Wang and Maier 2004; Gonzalez et al. 2005). Using stressors such as H2O2, menadione, and paraquat, the viabilities of E. coli JM109 and the recombinant E. coli YB overexpressing AZR were compared. The effect of heat shock, which may increase oxidative stress (Kim et al. 2005), was also studied.
When E. coli strains were treated with 10 mM H2O2, a protective effect of AZR was observed from the beginning. The survival rate of E. coli YB was over 30% higher than that of the control strain. When treated with 0.4 mM paraquat, similar enhanced resistance was obtained. The recombinant strain showed around 20–30% higher survival rates. When menadione was used as stressors, in contrast to the decreasing survival rate of E. coli JM109, the survival rate of E. coli YB increased in the first 3 h. While the survival rate of E. coli YB was almost four times higher than that at the beginning, nearly no viable count of the control strain was obtained at 4 h. A protection against heat stress by AZR was also observed from the beginning. The recombinant strain showed over 20% higher survival rate than the control one in 1 h (Fig. 4).
Survival of E. coli treated with oxidative stressors or heat shock. Cells were treated with 10 mM H2O2 (a), 0.4 mM paraquat (b), 0.6 mM menadione (c), and heat shock (d). Samples were taken at intervals, diluted and plated on LB solid media to monitor cell viability. Filled circles and squares represent cells of recombinant and control E. coli strains, respectively
Discussion
Bacterial decolorization of azo dyes has been widely studied in recent years. Many NAD(P)H-dependent cytoplasmic azoreductases catalyzing the reduction of azo bond have been characterized and classified into two families (Chen 2006). However, these enzymes were shown to be unimportant in vivo, as their reductase activities were only significant when using cell extracts and not using intact cells. It was speculated that intracellular azoreductase might not be involved in bacterial decolorization (Russ et al. 2000; Blümel et al. 2002). In addition, actually, there exist very few naturally occurring azo compounds. Most azo compounds are introduced into the environment anthropogenically. The primary role of azoreductase protein remains unknown and controversial.
AZR was first identified as an NADH-dependent azoreductase (Yan et al. 2004). Its amino acid sequence shows 97% identity to that of azoreductase of Bacillus sp. OY1-2, which is representative of a flavin-dependent azoreductase family. Further studies showed the nitroreductase activity of AZR for the reduction of nitroaromatics such as nitrofurazone and trinitrotoluene (Liu et al. 2007). However, these nitro compounds are also xenobiotics produced by human activity and may not be physiological substrate of AZR.
It was revealed that AZR’s amino acid sequences that involved in the binding of substrates and cofactors, namely, the signature sequence and the glycine-rich region, are relatively conserved among those of other quinone reductases from domains of Bacteria, Archaea, and Eucarya. Structure alignment further demonstrated that AZR and NQO1, a well-studied mammal quinone reductase, showed similar typical flavodoxin-like fold. Lot6p of S. cerevisiae was firstly reported to be a ferric reductase. Recent study showed its quinone reductase activity. It displays a flavodoxin-like five-stranded α/β structure, which is similar with that of AZR (Liu et al. 2007). WrbA of E. coli has similar sandwich structure. However, it has a conserved and unique insertion forming an additional α/β unit after strand β4 (Patridge and Ferry 2006; Andrade et al. 2007). Thus, all these quinone reductases have a common flavodoxin-like α/β core.
The structure similarities of AZR with other quinone reductases led us to investigate its ability to catalyze the reduction of quinone substrates. AZR could reduce several naphthoquinone and anthraquinone compounds, among which menadione was shown to be the best substrate. No activity was detected when 1,4-benzoquinone was used as substrate. However, the study on Lot6p showed that 1,4-benzoquinone was a good substrate with the highest turnover number, and 1,4-anthraquinone was not reduced at all (Sollner et al. 2007). Thus, there are some differences in substrate recognition of the two quinone reductases possessing similar overall structures. Further studies are needed to elucidate these differences. It should also be noted that, by comparison of k cat/K m, quinones are much better substrates of AZR than azo and nitro compounds studied before (Liu et al. 2007). It was shown that AZR could catalyze the reduction of menadione via two-electron mechanism. NQO1 was shown to reduce quinone to quinol by a compulsory two-electron transfer (Iyanagi 1987), which is different from the deleterious one-electron pathway.
Similar to the results of its azoreductase and nitroreductase activities (Yan et al. 2004; Liu et al. 2007), AZR reduces quinones with bi–bi ping-pong kinetics. The FMN cofactor of AZR is firstly reduced to FMNH2 by NADPH, and then, two electrons are transferred from FMNH2 to the quinone substrates. This is also in agreement with previous observations for some other quinone reductases, such as NQO1, Lot6p, and ChrR (Bianchet et al. 2004; Sollner et al. 2007; Gonzalez et al. 2005).
Dicoumarol, a vitamin K antagonist, was shown to be an efficient inhibitor of NQO1 and Lot6p (K i = 2 and 410 nM, respectively). The inhibition is competitive with respect to NAD(P)H (Li et al. 1995; Prestera et al. 1992; Chen et al. 1999; Sollner et al. 2007). Our results demonstrated that the inhibition of dicoumarol for AZR is competitive with respect to NADPH. However, with a K i value of 87.6 μM, dicoumarol is not an efficient inhibitor of AZR. The cofactors of AZR and NQO1 are bound in a similar manner. Recently, the crystal structure of NQO1 in complex with dicoumarol has been reported. It was shown that dicoumarol stacks parallel to the isoalloxazine ring of the flavin cofactor and forms a wall of the catalytic pocket (Asher et al. 2006).
Quinone reductase is suggested to be involved in quinone detoxification and oxidative stress resistance. It exercises its antioxidant effects directly by quinol-mediated quenching of reactive oxygen species and indirectly by diverting quinones away from one-electron reducers (Gonzalez et al. 2005). It was proposed that NQO1 serve as a cellular control device against quinone toxicity (Lind et al. 1982). Further studies indicated that NQO1 also plays a role in directly supporting the overall antioxidant functions of the cell (Beyer 1994). Lot6p was also shown to play a role in quinone detoxification and important for managing oxidative stress caused by quinones (Sollner et al. 2007). Similar protective roles were also reported for prokaryotic quinone reductases. The physiological role of WrbA of E. coli can be seen in the oxidative stress response. It is upregulated by various stressors, such as acids, salts, H2O2, and diauxie (Patridge and Ferry 2006). ChrR of Pseudomonas putida and MdaB of Helicobacter pylori and Helicobacter hepaticus were also reported to be involved in oxidative stress resistance (Gonzalez et al. 2005; Wang and Maier 2004; Hong et al. 2008).
To evaluate whether the expression of AZR could afford protection from oxidative species participating in bacterial killing, the control and the recombinant E. coli strains were treated with heat shock or several stressors, such as H2O2, paraquat, and menadione, respectively. The results clearly showed that the survival rates of the recombinant strain were higher than those of the control one. Quinone metabolism within a cell has a direct effect on the cell’s ability to deal with oxidative stress (Soballe and Poole 1999). Quinols have been shown to lower the levels of superoxide ions in E. coli cell membrane (Soballe and Poole 2000). When using 0.6 mM menadione as stressor, the survival rate of the recombinant strain even increased in the first 3 h. This is in accordance with the fact that menadione is an effective substrate of AZR. However, when more than 3 mM menadione was used, no increase of the survival rate of E. coli YB was observed (data not shown). Our results indicated that the expressed AZR catalyzes the two-electron reduction of quinones to quinols, a mechanism protecting the recombinant strain from stress mediated damage. The survival rate seemed to show in two-step decay kinetics. This may be due to the transcription of other E. coli stress–response proteins after exposure to oxidative stress. For instance, NfsA and WrbA, both of which possess quinone reductase activities and are upregulated in response to various oxidative stressors, may help cells deal with oxidative stress (Zenno et al. 1996; Paterson et al. 2002; Patridge and Ferry 2006).
In addition to H2O2, which is generated as a by-product of aerobic respiration, there are various oxidative burdens from external sources including some pollutants. An immediate burst of H2O2 is released by plant cells coming into contact with bacteria (Baker et al. 1991). Heavy metals are suggested to induce oxidative stress in cells (Ercal et al. 2001). Bioremediation of chromate through enzymatic reduction, which is a promising process, is unavoidably associated with H2O2 generation (Gonzalez et al. 2005). Nitroaromatics such as 2,4,6-trinitrotoluene are thought to induce oxidative stress in cell by enhancing superoxide and hydrogen peroxide production (Cenas et al. 2001). It was suggested that, due to its higher ability of overexpressing specific enzyme, gene-engineered microorganism may be more suited to environments where the pollutant is found at a high concentration (Garbisu and Alkorta 1999; Cases and de Lorenze, 2005). Our results demonstrated that enhancing activities of AZR azo/nitro/quinone reductase may not merely increase the bioremediation effects; it may also promote the ability of remediating cells to cope with oxidative stress. Further studies on modification of the recombinant strain with suicide system and its bio-safety assessment are underway for possible applications.
In summary, we have shown the quinone reductase activity of AZR, which was previously reported as azoreductase and nitroreductase in vitro. The expression of AZR in E. coli enhances its survival under oxidative stress conditions. It was demonstrated that quinones are better substrates compared to azo and nitro compounds. As synthetic azo and nitro compounds are not cognate substrates in an original physiological context, the finding of this new activity of AZR may shed some light on its primary role. Further biochemical and physiological studies of AZR are in process and will be reported later.
References
Andrade SLA, Patridge EV, Ferry JG, Einsle O (2007) Crystal structure of the NADH:quinone oxidoreductase WrbA from Escherichia coli. J Bacteriol 189:9101–9107
Asher G, Dym O, Tsvetkov P, Adler J, Shaul Y (2006) The crystal structure of NAD(P)H quinone oxidoreductase 1 in complex with its potent inhibitor dicoumarol. Biochemistry 45:6372–6378
Baker CJ, O’Neill NR, Keppler LD, Orlandi EW (1991) Early responses during plant-bacteria interactions in tobacco cell suspensions. Phytopathology 81:1504–1507
Beyer RE (1994) The relative essentiality of the antioxidative function of coenzyme Q-the interactive role of DT-diaphorase. Mol Aspects Med 15(suppl.):117–129
Bianchet MA, Faig M, Amzel LM (2004) Structure and mechanism of NAD(P)H:quinone acceptor oxidoreductases (NQO). Methods Enzymol 382:144–174
Blümel S, Knackmuss HJ, Stolz A (2002) Molecular cloning and characterization of the gene coding for the aerobic azoreductase from Xenophilus azovorans KF46F. Appl Environ Microbiol 68:3948–3955
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Cases I, de Lorenze V (2005) Genetically modified organisms for the environment: stories of success and failure and what we have learned from them. Int Microbiol 8:213–222
Cenas N, Nemeikaite-Ceniene A, Sergediene E, Nivinskas H, Anusevicius Z, Sarlauskas J (2001) Quantitative structure-activity relationships in enzymatic single-electron reduction of nitroaromatic explosives: implications for their cytotoxicity. Biochim Biophys Acta 1528:31–38
Chen H (2006) Recent advances in azo dye degrading enzyme research. Curr Protein Pept Sci 7:101–111
Chen S, Wu K, Zhang D, Sherman M, Knox R, Yang CS (1999) Molecular characterization of binding of substrates and inhibitors to DT-diaphorase: combined approach involving site-directed mutagenesis, inhibitor-binding analysis, and computer modeling. Mol Pharmacol 56:272–278
Deller S, Sollner S, Trenker-El-Toukhy R, Jelesarov I, Gubitz GM, Macheroux P (2006) Characterization of a thermostable NADPH:FMN oxidoreductase from the mesophilic bacterium Bacillus subtilis. Biochemistry 45:7083–7091
Deller S, Macheroux P, Sollner S (2008) Flavin-dependent quinone reductases. Cell Mol Life Sci 65:141–160
Dos Santos AB, Cervantes FJ, Van Lier JB (2007) Review paper on current technologies for decolourisation of textile wastewaters: pespectives for anaerobic biotechnology. Biores Technol 98:2369–2385
Ercal N, Gurer-Orhan H, Aykin-Burns N (2001) Toxic metals and oxidative stress part I: mechanisms involved in metal-induced oxidative damage. Curr Top Med Chem 1:529–539
Garbisu C, Alkorta I (1999) Utilization of genetically engineered microorganisms (GEMs) for bioremediation. J Chem Technol Biotechnol 74:599–606
Gonzalez CF, Ackerley DF, Lynch SV, Matin A (2005) ChrR, a soluble quinone reductase of Pseudomonas putida that defends against H2O2. J Biol Chem 280:22590–22595
Hong Y, Wang G, Maier RJ (2008) The NADPH quinone reductase MdaB confers oxidative stress resistance to Helicobacter hepaticus. Microb Pathog 44:169–174
Iyanagi T (1987) On the mechanisms of one- and two-electron transfer by flavin enzymes. Chemica Scripta 27A:31–36
Kim HJ, Kang BS, Park JW (2005) Cellular defense against heat shock-induced oxidative damage by mitochondrial NADP+-dependent isocitrate dehydrogenase. Free Radic Res 39:441–448
Laskowski MJ, Dreher KA, Gehring MA, Abel S, Gensler AL, Sussex IM (2002) FQR1, a novel primary auxin-response gene, encodes a flavin mononucleotide-binding quinone reductase. Plant Physiol 128:578–590
Li R, Bianchet MA, Talalay P, Amzel LM (1995) The three-dimensional structure of NAD(P)H:quinone reductase, a flavoprotein involved in cancer chemoprotection and chemotherapy: mechanism of the two-electron reduction. Proc Natl Acad Sci U S A 92:8846–8850
Liger D, Graille M, Zhou CZ, Leulliot N, Quevillon-Cheruel S, Blondeau K, Janin J, van Tilbeurgh H (2004) Crystal structure and functional characterization of yeast YLR011wp, an enzyme with NAD(P)H-FMN and ferric iron reductase activities. J Biol Chem 279:34890–34897
Lind C, Hochstein P, Ernster L (1982) DT-diaphorase as a quinone reductase: a cellular control device against semiquinone and superoxide radical formation. Arch Biochem Biophys 216:178–185
Liu G, Zhou J, Lv H, Xiang X, Wang J, Zhou M, Qv Y (2007) Azoreductase from Rhodobacter sphaeroides AS1.1737 is a flavodoxin that also functions as nitroreductase and flavin mononucleotide reductase. Appl Microbiol Biotechnol 76:1271–1279
Nicholas KB, Nicholas HB, Jr, Deerfield DW (1997) GeneDoc: analysis and visualization of genetic variation. EMBnet News 4:1–4
Paterson ES, Boucher SE, Lambert IB (2002) Regulation of the nfsA gene in Escherichia coli by SoxS. J Bacteriol 184:51–58
Patridge EV, Ferry JG (2006) WrbA from Escherichia coli and Archaeoglobus fulgidus is an NAD(P)H:quinone oxidoreductase. J Bacteriol 188:3498–3506
Prestera T, Prochaska HJ, Talalay P (1992) Inhibition of NAD(P)H:(quinone-acceptor) oxidoreductase by cibacron blue and related anthraquinone dyes: a structure-activity study. Biochemistry 31:824–833
Ross D, Siegel D (2004) NAD(P)H:quinone oxidoreductase 1 (NQO1, DT-diaphorase), functions and pharmacogenetics. Methods Enzymol 382:115–144
Russ R, Rau J, Stolz A (2000) The function of cytoplasmic flavin reductases in the reduction of azo dyes by bacteria. Appl Environ Microbiol 66:1429–1434
Soballe B, Poole RK (1999) Microbial ubiquinones: multiple roles in respiration, gene regulation and oxidative stress management. Microbiology 145:1817–1830
Soballe B, Poole RK (2000) Ubiquinone limits oxidative stress in Escherichia coli. Microbiology 146:787–796
Sollner S, Nebauer R, Ehammer H, Prem A, Deller S, Palfey BA, Daum G, Macheroux P (2007) Lot6p from Saccharomyces cerevisiae is a FMN-dependent reductase with a potential role in quinone detoxification. FEBS J 274:1328–1339
Stolz A (2001) Basic and applied aspects in the microbial degradation of azo dyes. Appl Microbiol Biotechnol 56:69–80
Wang G, Maier RJ (2004) An NADPH quinone reductase of Helicobacter pylori plays an important role in oxidative stress resistance and host colonization. Infect Immun 72:1391–1396
Yan B, Zhou J, Wang J, Du C, Hou H, Song Z, Bao Y (2004) Expression and characteristics of the gene encoding azoreductase from Rhodobacter sphaeroides AS1.1737. FEMS Microbiol Lett 236:129–136
Zenno S, Koike H, Kumar AN, Jayaraman R, Tanokura M, Saigo K (1996) Biochemical characterization of NfsA, the Escherichia coli major nitroreductase exhibiting a high amino acid sequence homology to Frp, a Vibrio harveyi flavin oxidoreductase. J Bacteriol 178:4508–4514
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Liu, G., Zhou, J., Jin, R. et al. Enhancing survival of Escherichia coli by expression of azoreductase AZR possessing quinone reductase activity. Appl Microbiol Biotechnol 80, 409–416 (2008). https://doi.org/10.1007/s00253-008-1555-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-008-1555-6