Abstract
Wastewater effluents from the textile and other dye-stuff industries contain significant amounts of synthetic dyes that require treatment to prevent groundwater contamination. In research aimed at biotechnology for treatment of azo dyes, this study examined 288 strains of azo-dye degrading bacteria to identify efficient strains and determine incubation times required for decolorization. Initial enrichment cultures were carried out using a mixture of four structurally different dyes (Acid Red 88, Reactive Black 5, Direct Red 81, and Disperse Orange 3) as the sole source of C and N to isolate the bacteria from soil, activated sludge, and natural asphalt. Six strains were selected for further study based on their prolific growth and ability to rapidly decolorize the dyes individually or in mixtures. Treatment times required by the most efficient strain, AS96 (Shewanella putrefaciens) were as short as 4 h for complete decolorization of 100 mg l−1 of AR-88 and DR-81 dyes under static conditions, and 6 and 8 h, respectively, for complete decolorization of RB-5 and DO-3. To our knowledge, these bacterial strains are the most efficient azo-dye degrading bacteria that have been described and may have practical application for biological treatment of dye-polluted wastewater streams.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Azo dyes comprise a diverse group of synthetic chemicals that are widely used by the textile, leather, food, cosmetics, and paper product industries. The annual world production of azo dyes is estimated to be around one million tons (Stolz 2001; Pandey et al. 2007), and more than 2000 structurally different azo dyes are currently in use (Vijaykumar et al. 2007). The general structural characteristics of these compounds feature substituted aromatic rings that are joined by one or more azo groups (–N=N–). Azo dyes are generally recalcitrant to biodegradation due to their complex structures and xenobiotic nature and typically require an anaerobic–aerobic process to achieve complete mineralization.
Many azo dyes and their degradation intermediates are mutagenic and carcinogenic (Chung and Cerniglia 1992; Ozturk and Abdullah 2006) and contribute to the mutagenic activity of ground and surface waters that are polluted by textile effluents (Rajaguru et al. 2002; Umbuzeiro et al. 2005). Their discharge into surface water also leads to aesthetic problems, obstructing light penetration and oxygen transfer into water bodies (Slokar and Le Marechal 1998; Bae and Freeman 2007). Dye concentrations that are used for processing vats are typically around 1,000 mg l−1 (Ince and Tezcanli 1999). Depending on the class of the dyes, their loss in waste waters can range from 2% of the original concentration for basic dyes to as high as 50% for reactive dyes (O’Neill et al. 1999; Tan et al. 2000; Boer et al. 2004).
There are a variety of physicochemical methods for color removal from effluents containing dyes (Wang et al. 2004; Golab et al. 2005; Saxe et al. 2006; Alinsafi et al. 2007; Arslan-Alaton 2007). Biotechnological approaches for the remediation of contaminated wastewater are also receiving attention, but are not yet used by industry due to concern over the production of aromatic amines that are generated under anaerobic conditions (Dubrow et al. 1996). Nonetheless, there have been pilot scale investigations of anaerobic–aerobic treatment processes that are effective for complete removal and that should ultimately prove to be cost-effective and environment-friendly (see review: Dubrow et al. 1996). Activated sludge is commonly used as an inoculum to initiate degradation, and it appears that many different microorganisms can decolorize azo dyes (Khera et al. 2005; Chen 2006; Hao et al. 2007; Pandey et al. 2007). Further development of biotreatment processes will be facilitated by identifying the most effective microorganisms and ways to reduce the time that is needed to process contaminated wastewater effluents.
This study reports the isolation and identification of bacterial strains (isolated from activated sludge and soil) that were shown to be highly effective for decolorization of four azo dyes individually or in a mixture within a time span as short as 4 to 8 h. The potential application of the selected cultures was further tested by bioaugmentation of an activated sludge and was shown to enhance degradation over that achieved by the activated sludge alone. The bacterial strains that were identified should be of value for improving the treatment of azo-dye wastewater and remediation of contaminated groundwater.
Materials and methods
Chemicals and culture medium
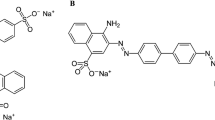
Four azo dyes, including Acid Red 88 (C.I., 75% dye content), Reactive Black 5 (C.I., 55% dye content), Direct Red 81 (C.I., 50% dye content), and Disperse Orange 3 (C.I., 90% dye content) were purchased from Sigma-Aldrich, USA. These structurally different compounds represent four classes of azo dyes (Fig. 1). All other chemicals were reagent/analytical grade. The mineral salts medium (MSM) pH 7.2, contained (g l−1) NaCl (1.0), CaCl2·2H2O (0.1), MgSO4·7H2O (0.5), KH2PO4 (1.0), and Na2HPO4 (1.0).
Isolation of efficient azo-dye decolorizing bacteria
Bacterial strains were isolated from activated sludge (Corona Wastewater Treatment Plant, CA, USA), turf grass soil (Chino loamy sand, pH 7.4, Chino Park, CA, USA) and from a natural asphalt soil mixture obtained from the Rancho La Brea tar pits in Los Angeles, CA, USA. Isolates from each inoculum source that were capable of growth on azo dyes were first enriched using MSM amended with a mixture of four dyes as the sole source of C and N. Each dye was added to the medium at 50 mg l−1 to achieve a final concentration of 200 mg l−1. This value is typical of those used in studies on treatment of azo-dye waste water effluent (Zhao and Hardin 2007). The cultures containing 200 ml of MSM with dyes in 500 ml Erlenmeyer flasks were inoculated with 10 ml volumes of activated sludge, soil, or asphalt-soil and incubated at 30°C for 5 days under static conditions. After incubation, cell suspensions from each flask were plated onto MSM agar medium and incubated at 30°C for 24 h. Microbial colonies that appeared on the agar medium were washed gently with sterile water and resuspended into the flasks containing fresh MSM broth spiked with the mixture of dyes. After a second transfer of the cell suspensions onto MSM agar plates containing 0.1% yeast extract, 100 actively growing colonies with different colony growth characters were selected from each source and were purified by streaking twice on agar medium. The purified cultures were preserved at −20°C in 15% (w/v) glycerol for subsequent study.
Bacterial isolates from the enrichment cultures were individually tested for their abilities to grow on agar plates with MSM using each dye separately as a sole source of C and N. The bacterial cells were centrifuged and washed twice with autoclaved MSM broth before cultivation on MSM agar plates containing each dye. Dyes AR-88, RB-5, and DR-81 were dissolved in distilled water and passed through 0.2 μm sterile membrane filters (Nalgene®), whereas dye DO-3 was first dissolved in small volume of ethanol (95%) and then diluted into sterilized distilled water. Further tests were carried out with megatiter plates using liquid medium supplemented with cosubstrates to enhance the bacterial growth rates. In this case, each dye medium was supplemented with 0.05% (w/v) yeast extract and 0.05% (w/v) glucose. Using 96-well plates, 1.8 ml of dye medium was added to each well and inoculated using sterile toothpicks to transfer confirmed azo-dye degrading strains from agar medium. Each plate was sealed tightly with presterile thermal adhesive sealing film and incubated at 30°C under static conditions. Uninoculated controls were included to check for abiotic decolorization. Experiments with each dye bacteria combination had six replicates to permit measurements at intervals. Decolorization was determined by taking 1.5 ml aliquots from each well and centrifuging the medium at 10,000 rpm for 15 min to remove the cells. The absorbance of the supernatants was measured at λ max of each dye (AR-88, 505 nm; RB-5, 597 nm; DR-81, 508 nm; DO-3, 443 nm) using a spectrophotometer (DU640, Beckman, USA).
On the basis of the screening in liquid medium, 24 isolates were selected and further compared in megatiter plates inoculated with uniform cell densities. The bacterial isolates were first cultured in MSM containing 0.1% yeast extract (without dye) for 24 h at 30°C with shaking at 150 rpm. The cells were harvested, and a uniform cell density (0.6 OD at 550 nm) was maintained. The mean cell counts of these cultures were between 108 and109 cfu ml−1. The cell density standardized decolorization experiment was performed using the same protocol above.
Decolorization of azo dyes and their mixture in liquid media
The six most effective bacterial strains from the final screening were further examined for their decolorization potentials in 10-ml glass vials. Nine milliliters of the sterilized MSM broth containing 100 mg dye l−1 were added to 10 ml autoclaved glass vials supplemented with 0.4% yeast extract as a co-substrate. Yeast extract previously has been shown to supplement the growth of azo dye degrading bacteria (Sponza and Isik 2002). In this work, it was omitted in the enrichment where the bacteria were screened for use of azo dyes as a sole C and N source, but was included in the dye treatability study described here to determine the maximum rates of decolorization for these strains. The media were inoculated with the respective bacterial strains by adding inocula of uniform cell density (OD0.6). The medium to inoculum ratio (v/v) was 50:1 yielding approximate cell densities of 107 cfu ml−1. The vials were tightly sealed and were incubated static at 35°C. To monitor aerobic decolorization of these dyes, a similar set of treatments was incubated with loosely capped vials on a platform shaker. Uninoculated vials with MSM containing each azo dye plus yeast extract were also incubated under similar conditions to check for abiotic decolorization of each dye. Six vials were used for each strain per dye. Aliquots (1.5 ml) were taken periodically from different vials alternately to determine decolorization of the dyes. The results are presented as the average of three replications for each interval.
To check for adsorption of dyes, the cell pellets after centrifugation were resuspended in an equal volume of methanol to extract the dye (Khera et al. 2005). The suspensions were vortexed and centrifuged for 15 min at 10,000 rpm. The supernatant from each sample was read at λ max of each dye. Other experiments examined the range of concentrations that could be degraded by two selected strains (AS81 and AS96) and decolorization of a mixture of the four dyes by the six strains. The data presented in these experiments are the mean of three replications.
Decolorization of azo dyes following bioaugmention of activated sludge
Two bacterial strains (AS81 and AS96) were assessed for their ability to decolorize azo dyes after augmentation of activated sludge. Nine milliliters of the MSM supplemented with 100 mg l−1 dye and 0.4% yeast extract were added to 10 ml glass vials and inoculated by adding 180 μl of the respective bacterial culture (0.6 OD) and 180 μl of activated sludge. Treatments containing inocula of the pure cultures of bacteria, activated sludge only, and an uninoculated control were included for comparison. The experiment was performed with six replicates for each treatment. Results are presented as the average of three replications at each interval.
Bacterial identifications
The bacterial cells were collected by centrifugation, and genomic DNA was extracted using the FastDNA Spin kit following the manufacturer’s instructions. The extracted DNA was amplified to obtain near full length 16S rRNA genes by polymerase chain reaction (PCR) using primers 27f and 1492r (Operon Biotechnologies, AL). The PCR was performed with the following conditions: initial denaturation at 95°C for 5 min, 30 cycles at 94°C for 40 s, 55°C for 4 s, and 72°C for 50 s. Final elongation was at 72°C for 10 min. The amplified products were purified using a DNA purification kit (QIAquick®). Nucleotide sequences were determined by sequencing (ABI PRISM® 377 DNA Sequencer, USA). Three primers including 27f (5′-agagtttgatcmtggctcag-3′), 907r (5′-ccgtcaattcctttgagttt-3′), and 1492r (5′-agrtaccttgttacgactt-3′) were used for sequencing. The 16S rRNA gene of each strain was assembled using Pregap4 and Gap4 software (Staden-Windows Package) and compared with the 16S rRNA gene database at the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/BLAST/Blast.cgi) using the BLAST search tool.
Results
Bacterial growth on MSM plates containing azo dyes
Approximately 80% of the bacteria from the enrichment cultures were able to use all four azo dyes (AR-88, RB-5, DR-81, and DO-3) as individual growth substrates when grown on agar medium containing a mixture of the dyes and individually (data not shown). Overall, the greatest number of isolates was obtained on agar medium containing dye DR-81, while the fewest were obtained on dye DO-3.
Decolorization of azo dyes in liquid media
Screening experiments assessed the potential of 288 bacterial isolates from the enrichment cultures for decolorizing four different dyes under static conditions in liquid medium. Bacteria isolated from activated sludge and soil exhibited the highest efficiency in decolorizing all of the tested dyes compared to those isolated from natural asphalt (data not shown). Although some strains isolated from asphalt were capable of decolorizing the dyes, their decolorization potential was substantially less than the bacteria isolated from activated sludge or soil (data not shown).
The six most efficient strains, which included two from soil and four from activated sludge, were identified by their 16S rRNA gene sequences and were confirmed as pure cultures (Table 1). The strains represented diverse genera including Aeromonas, Pseudomonas, Bacillus, and Shewanella from activated sludge, and Bacillus and Massillia from soil. Each of the strains matched with previously described NCBI database accessions to the species level with 98 to 99% similarities.
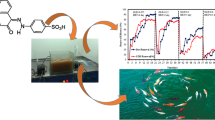
Rapid decolorization (99 and 92%) of dye AR-88 by the bacterial strain AS96 (Shewanella putrefaciens) and AS81 (Aeromonas punctata), respectively, was observed at 4 h, with complete decolorization after 6 h (Fig. 2). Strains, AS7 and S46, tentatively identified as Bacillus cereus and Bacillus thuringiensis, respectively, were slower with completely decolorization occurring at 12 h. Strains AS77 Pseudomonas nitroreducens and strain S81 Massilia timonae achieved complete decolorization of this dye in 16 h.
In the case of dye RB-5, strains AS96 and AS81 achieved 93 and 84% decolorization after 4 h, respectively, and complete decolorization in 6 and 12 h (Fig. 2). The other four strains (AS7, AS77, S46, and S81) demonstrated a lag phase in which there was little decolorization of RB-5 for the first 8 h; after which decolorization ranged from 9 to 84% at 16 h. In comparison, decolorization of RB-5 by the abiotic control was only 7% after 16 h.
Strain AS96 was the most effective for decolorization of dye DR-81 with complete decolorization occurring in 4 h (Fig. 2). The second most effective strain, AS81 achieved complete decolorization of DR-81 after 8 h. The decolorization of DR-81 by the other strains ranged from 23 to 89% after 16 h within the same detention time. Negligible (4%) decolorization of the dye occurred abiotically.
The fastest decolorization of the fourth dye DO-3 was observed after 4 h, again with strain AS96, which achieved 92% decolorization at 4 h and complete decolorization after 8 h (Fig. 2). Strains AS81 and AS7 were able to decolorize DO-3 completely after 12 and 16 h respectively. Up to 85 and 89% decolorization occurred with strains S46 and S81, respectively. Strain AS77 was not effective for decolorization of this dye.
It was observed that small amounts of two dyes (AR-88 and DO-3) were adsorbed onto the bacterial cells (<6% recovered with methanol extraction) at initial stages (within 4 h) of incubation (data not shown); however, the adsorbed dye contents were decolorized completely at later stages. The dyes (RB-5 and DR-81) were not adsorbed by the bacteria.
Incubation times required for complete decolorization of the dyes were concentration dependent over a range of 100–500 mg l−1 (Fig. 3). At 100–300 mg l−1 dye concentrations, 80 to 100% removal of dyes was observed in response to inoculation with AS96 after 8 h, while decolorization ranged from 52 to 90% at 400 mg l−1 and 19 to 79% at 500 mg l−1. Strain AS81 was relatively less efficient than AS96 and showed decolorization between 66 to 100% at dye concentrations of 100–300 mg l−1 after 8 h. At 400 and 500 mg l−1 concentrations, the maximum dye removal was 53 and 35%, respectively, after 8 h in solutions inoculated with strain AS81. Overall, both strains were capable of decolorizing RB-5 and DR-81 dyes at concentrations up to 500 mg l−1 completely after 20 h (data not shown). Decolorization of DO-3 was relatively less at high concentration than for the other three dyes.
Shaking (aeration) increased the time required for complete decolorization of 100 mg l−1 dye compared to that which was required under static conditions (Fig. 4). Under aerated conditions, the decolorization of the four dyes after 8 h incubation with AS96 was 14 to 30% less than under static conditions. Similarly, decolorization of the dyes by strain AS81 was 13 to 28% lower with aeration.
All six strains were capable of decolorizing a mixture of the dyes in the MSM medium supplemented with 0.4% yeast extract (Fig. 5). Inoculation with strain AS96 resulted in the most rapid decolorization with complete decolorization at 4 h. In contrast, strain AS81 achieved complete decolorization of the dye mixture after 8 h. The other four strains (AS7, AS77, S46, and S81) partially decolorized the mixture, with 69 to 89% decolorization after 16 h.
Decolorization of azo dyes by the pure and mixed cultures
Strains AS81 and AS96 from the activated sludge were able to decolorize all the tested four dyes in the liquid medium after bioaugmentation into a live culture of activated sludge. The amounts of dye that were decolorized were nearly identical for bioaugmented cultures as those that were achieved using pure cultures of the bacteria (Table 2). The unamended activated sludge had little capacity to decolorize the dyes with only 14% decolorization occurring after 8 h.
Discussion
The bacteria isolated from activated sludge, soil, and asphalt exhibited variable growth on agar plates containing each of the four structurally diverse azo dyes when supplied as the sole source of C and N. The greatest number of isolates that were able to decolorize the dyes came from activated sludge and soil. None of the bacteria isolated from the asphalt showed decolorization potential equivalent to that of activated sludge and soil isolates. Our initial premise was that the complex mixture of petroleum hydrocarbons in asphalt may select for efficient degraders (Kim and Crowley 2007). Here, of the 288 isolates that were tested, the four most effective came from activated sludge, and two were obtained from soil.
The six selected bacterial strains showed the greatest ability to decolorize azo dyes in liquid medium when supplemented with 0.4% yeast extract. One of the bacterial strains (S. putrefaciens strain AS96) isolated from activated sludge was capable of completely decolorizing all four test dyes (100 mg dye l−1) in just 4–8 h of static incubation or a mixture (25 mg l−1 each) of the four dyes in 4 h. Furthermore, this bacterium was able to completely decolorize three dyes (AR-88, RB-5, and DR-81) up to a concentration of 300 mg l−1 in just 8 h (Fig. 3) and 500 mg l−1 of RB-5 and DR-81 within 20 h (data not shown). This implies that the strain AS96 (S. putrefaciens) carries an efficient enzymatic system for the cleavage of azo bonds which caused rapid decolorization of higher concentrations of different azo dyes under reduced (static) conditions. In another experiment, a related strain, Shewanella oneidensis JM6 was also tested and was shown to be effective in decolorizing these dyes (data not shown). The findings suggest that this and possibly other unidentified strains of Shewanella could potentially be useful for the treatment of wastewaters contaminated with azo dyes. The decolorization rates recorded in this study are much higher than those that have been reported previously. For comparison, other researchers have reported complete decolorization of AR-88 (20 mg l−1) in 20 h (Khera et al. 2005), Direct Blue-6 (100 mg l−1) in 72 h (Kalme et al. 2007), DR-81 in 35 h (Junnarkar et al. 2006), RB-5 (200 mg l−1) in 24 h (Lucas et al. 2006), 50% decolorization of DO-3 (200 mg l−1) in 120 h (Zhao and Hardin 2007), and 90% of Fast Acid Red GR in 12 h (Xu et al. 2005), achieved by either pure cultures or consortia. Decolorization of mixtures of azo dyes as achieved here with single strains has not been reported in the literature.
Initial decolorization of azo dyes is known to involve a reductive process and is thus facilitated by anaerobic, static culture conditions (Chang and Lin 2001; Junnarkar et al. 2006; Kalme et al. 2007). This was confirmed here for all of the isolates that were tested. The study also demonstrated that the selected bacterial strains were capable of removing the color of azo dyes from solid agar medium in test tubes or on agar plates (data not shown), which suggest the accumulation of redox active enzymes or biochemical substances that were released into the medium during growth of the bacterial cells. This has been observed previously (Barragan et al. 2007) and suggest that it may be possible to develop encapsulated cell methods with agarose or other carriers to contain the bacterial cells in flow through reactors. The bacterial cells were also effective in short-term experiments after bioaugmentation into activated sludge. This suggests it may be possible to augment activated sludge or wastewater treatment ponds with live cultures of effective bacteria to enhance azo dye degradation. Long-term experiments will be required to determine longevity of the bioaugmented strains over time. Nonetheless, the ability to efficiently utilize the azo dyes as a carbon and nitrogen source provides a niche that should favor long-term survival of the bacteria in wastewater streams that contain sufficient levels of azo dyes to support bacterial growth. Further studies will examine metabolite production and development of anaerobic aerobic treatment methods to achieve complete mineralization.
Conclusions
The present study reports accelerated in vitro decolorization of four structurally different azo dyes by the highly efficient bacterial cultures both in liquid and solid media. Some strains completely decolorized individual and mixture of dyes in liquid medium in a very short time span of 4 h and were effective when inoculated into activated sludge. To our knowledge, these are among the most effective strains that have been described in the literature and our results suggest that these bacterial species have potential application for bioremediation of dye-polluted waters/sludge or could be used in bioreactors to treat wastewater streams. Work is in progress to identify the biodegradation products of these dyes formed by the selected bacterial strains and to identify the enzyme(s) responsible for rapid decolorization of the dyes.
References
Alinsafi A, Evenou F, Abdulkarim EM, Pons MN, Zahraa O, Benhammou A, Yaacoubi A, Nejmeddine A (2007) Treatment of textile industry wastewater by supported photocatalysis. Dyes Pigm 74:439–445
Arslan-Alaton I (2007) Degradation of a commercial textile biocide with advanced oxidation processes and ozone. J Env Manage 82:145–154
Bae JS, Freeman HS (2007) Aquatic toxicity evaluation of new direct dyes to the Daphnia magna. Dyes Pigm 73:81–85
Barragan BE, Costa C, Marquez MC (2007) Biodegradation of azo dyes by bacteria inoculated on solid media. Dyes Pigm 75:73–81
Boer CG, Obici L, Souza CG, Peralta RM (2004) Decolourization of synthetic dyes by solid state cultures of Lentinula (Lentinus) edodes producing manganese peroxidase as the main lignolytic enzyme. Bioresour Technol 94:107–112
Chang JS, Lin YC (2001) Decolorization kinetics of recombinant E. coli strain harboring azo dye decolorization determinants for Rhodococcus sp. Biotech Lett 23:631–636
Chen BY (2006) Biologically-feasible screening strategy for optimal decolorization strain to diazo Evercion Red H-E7B. J Chinese Inst Chem Eng 37:117–124
Chung KT, Cerniglia CE (1992) Mutagenicity of azo dyes: structure–activity relationships. Mut Res 277:201–220
Dubrow SF, Boardman GD, Michelsen DL (1996) Chemical pretreatment and aerobic–anaerobic degradation of textile dye wastewater. In: Reife A and Freeman HS (eds) Environmental chemistry of dyes and pigments. Wiley Interscience. pp 75–102
Golab V, Vinder A, Simonic M (2005) Efficiency of the coagulation/flocculation method for the treatment of dye bath effluent. Dyes Pigm 67:93–97
Hao JJ, Song FQ, Huang F, Yang CL, Zhang ZJ, Zheng Y, Tian XJ (2007) Production of laccase by a newly isolated deuteromycete fungus Pestalotiopsis sp. and its decolorization of azo dye. J Industrial Microbiol Biotechnol 34:233–240
Ince H, Tezcanli G (1999) Treatability of textile dye-bath effluents by advanced oxidation: preparation for reuse. Water Sci Technol 40:183–190
Junnarkar N, Murty DS, Bhatt NS, Madamwar D (2006) Decolorization of diazo dye Direct Red 81 by a novel bacterial consortium. World J Microbiol Biotech 22:163–168
Kalme SD, Parshetti GK, Jadhav SU, Govindwar SP (2007) Biodegradation of benzidine based dye Direct Blue-6 by Pseudomonas desmolyticum NCIM 2112. Bioresor Tech 98:1405–1410
Khera MS, Saini HS, Sharma DK, Chadha BS, Chimni SS (2005) Comparative studies on potential of consortium and constituent pure bacterial isolates to decolorize azo dyes. Water Res 39:5135–5141
Kim JS, Crowley DE (2007) Microbial diversity in natural asphalts of the Rancho La Brea tar pits. Appl Env Microbiol 73:4579–4591
Lucas MS, Amaral C, Sampaio A, Peres JA, Dias AA (2006) Biodegradation of the diazo dye Reactive Black 5 by a wild isolate of Candida oleophila. Enzyme Microbiol Tech 39:51–55
O’Neill C, Hawkes FR, Hawkes DL, Lourenco ND, Pinheiro HM, Delee W (1999) Color in textile effluents sources, measurement, discharge consents and simulation: a review. J Chem Technol Biotechnol 74:1009–1018
Ozturk A, Abdullah MI (2006) Toxicological effect of indole and its azo dye derivatives on some microorganisms under aerobic conditions. Sci Total Env 358:137–142
Pandey A, Singh P, Iyengar L (2007) Bacterial decolorization and degradation of azo dyes. Int Biodeter Biodegr 59:73–84
Rajaguru P, Vidya L, Baskarasethupathi B, Kumar PA, Palanivel M, Kalaiselvi K (2002) Genotoxicity evaluation of polluted ground water in human peripheral blood lymphocytes using the comet assay. Mut Res 517:29–37
Saxe JP, Lubenow BL, Chiu PC, Huang CP, Cha DK (2006) Enhanced biodegradation of azo dyes using an integrated elemental iron-activated sludge system: II. Effects of physical–chemical parameters. Water Env Res 78:26–30
Slokar YM, Le Marechal AM (1998) Methods of decoloration of textile wastewater. Dyes Pigm 37:335–356
Sponza DT, Isik M (2002) Decolorization and azo dye degradation by anaerobic/aerobic sequential process. Enzyme Microb Technol. 31:102–110
Stackebrandt E, Goebel BM (1994) A place for DNA-DNA reassociation and 16 S ribosomal-RNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol 44:846–849
Stolz A (2001) Basic and applied aspects in the microbial degradation of azo dyes. Appl Microbiol Biotechnol 56:69–80
Tan NCG, Borger A, Slender P, Svitelskaya AV, Lettinga G, Field JA (2000) Degradation of azo dye Mordant Yellow 10 in a sequential anaerobic and bioaugmented aerobic bioreactor. Water Sci Technol 42:337–344
Umbuzeiro GA, Freeman H, Warren SH, Oliveira DP, Terao Y, Watanabe T, Claxton LD (2005) The contribution of azo dyes to the mutagenic activity of the Cristais River. Chemosphere 60:55–64
Vijaykumar MH, Vaishampayan PA, Shouche YS, Karegoudar TB (2007) Decolourization of naphthalene-containing sulfonated azo dyes by Kerstersia sp strain VKY1. Enzyme Microbial Technol 40:204–211
Wang A, Qu J, Liu H, Ge J (2004) Degradation of azo dye Acid Red 14 in aqueous solution by electrokinetic and electrooxidation process. Chemosphere 55:1189–1196
Xu M, Guo J, Cen Y, Zhong X, Cao W, Sun G (2005) Shewanella decolorationis sp. nov., a dye decolorizing bacterium isolated from activated sludge of a waste-water treatment plant. Int J Syst Evol Microbiol 55:363–368
Zhao X, Hardin IR (2007) HPLC and spectrophotometric analysis of biodegradation of azo dyes by Pleurotus ostreatus. Dyes Pigm 73:322–325
Acknowledgements
Financial support for this study was provided by the Higher Education Commission, Pakistan. We thank Dr. Jason Cantera for assistance in 16S rRNA gene analysis.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Khalid, A., Arshad, M. & Crowley, D.E. Accelerated decolorization of structurally different azo dyes by newly isolated bacterial strains. Appl Microbiol Biotechnol 78, 361–369 (2008). https://doi.org/10.1007/s00253-007-1302-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-007-1302-4