Abstract
The engineering of Corynebacterium glutamicum is important for enhanced production of biochemicals. To construct an optimal C. glutamicum genome, a precise site-directed gene integration method was developed by using a pair of mutant lox sites, one a right element (RE) mutant lox site and the other a left element (LE) mutant lox site. Two DNA fragments, 5.7 and 10.2 kb-long, were successfully integrated into the genome. The recombination efficiency of this system compared to that obtained by single crossover by homologous recombination was 2 orders of magnitude higher. Moreover, the integrated DNA remained stably maintained on removal of Cre recombinase. The Cre/mutant lox system thus represents a potentially attractive tool for integration of foreign DNA in the course of the engineering of C. glutamicum traits.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Corynebacterium glutamicum is a nonpathogenic, high GC content Gram-positive bacterium widely used in the industrial production of amino acids, nucleic acids, and organic acids (Kinoshita 1985; Liebl 1992; Lessard et al. 1999; Malumbres et al. 1995). To improve its industrial productivity, techniques employing random mutation strategies have been preferred. Meanwhile, genome science has progressed at a rapid rate, culminating in the more than 500 completely sequenced microbial genomes at present. This constitutes an important resource for understanding cellular life. Two C. glutamicum strains have been sequenced: R (Yukawa et al. 2007) and ATCC13032 (Ikeda and Nakagawa 2003; Kalinowski et al. 2003), and strain reconstruction studies for improved industrial applications using whole genome sequences, have been initiated (Ohnishi et al. 2003).
In the C. glutamicum post-genome era, the concept of Minimum Genome Factories (MGFs), encompassing the use of recombinant strains of which metabolism has been streamlined by limiting the genome to the optimal subset required for the targeted application to maximize product formation, was described (Inui et al. 2005; Vertès et al. 2005). In efforts to improve the industrial productivity of C. glutamicum, significant progress in the development of molecular techniques, including metabolic analyses, gene manipulation, and genome engineering, has been made (Hermann 2003; Kotrba et al. 2003; Suzuki et al. 2005a; Vertès et al. 1994). However, to implement the concept of MGF(s) by the rearrangement of bacterial genomes, molecular biology tools which make multiple excisions and insertions possible are a prerequisite.
Recently, a total of 190 kb of genomic regions predicted to be nonessential for cell survival were excised by using a method dependent upon the Cre/LE- and RE-mutant lox mediated recombination systems (Suzuki et al. 2005b). Cre/LE- and RE-mutant lox is a modified DNA recombination system based on Cre/loxP. Cre/loxP is a highly efficient and simple two-component system currently recognized as a useful DNA recombination tool. As Cre recombinase can catalyze reciprocal site-specific recombination of DNA at 34 bp loxP sites, it mediates both intramolecular (deletional or inversional) and intermolecular (integrative) recombination. To overcome bidirectionality of the Cre/loxP reaction and favor integration events, several Cre/mutant lox systems have been developed (Albert et al. 1995; Araki et al. 1997; Araki et al. 2002; Lee and Saito 1998). Cre/LE- and RE-mutant lox exhibits nucleotide changes into the Cre binding site of loxP.
By using two types of mutant lox sequences, it retains recombination ability, while avoiding reversible recombination (Araki et al. 1997). Previously, we succeeded in the successive deletion of eight genomic regions of C. glutamicum by using this intramolecular recombination of Cre/LE- and RE-mutant lox (Suzuki et al 2005b). Another aspect for the realization of the concept of MGF(s) is multiple gene integration. The integration of foreign genes makes possible the addition of new metabolic pathways and enhancing of existing ones. So far, homologous recombination has been the favored method of introduction of foreign genes into a microorganism’s genome. However, as the frequency of the homologous recombination reaction is dependent upon the host cell characteristics, it is often difficult to increase the recombination efficiency artificially. For C. glutamicum, different experimental strategies to promote frequency of homologous recombination have been described (Ikeda and Katsumata 1998; Schwarzer and Puhler 1991; Vertès et al. 1993a).
To overcome the problem of low efficiency of integration via homologous recombination, we developed a Cre/LE- and RE-mutant lox–based method described here. Integrative intermolecular recombination of Cre/LE- and RE-mutant lox was applied to DNA integration for C. glutamicum. The recombination frequency of target insertion was 2 and 4 orders of magnitude higher, respectively, than that of existing single or double crossover homologous recombination techniques (Vertès et al. 1993a). This method could be useful in creating improved cells for bioindustry.
Materials and methods
Bacterial strains and culture conditions
The bacterial strains used in this study are listed in Table 1. Escherichia coli was grown aerobically at 37°C in Luria–Bertani (LB) medium (Sambrook et al. 1989). C. glutamicum was cultivated at 33°C in complex medium containing 4% glucose (Inui et al. 2004). Antibiotics were used at the following concentrations for E. coli: kanamycin (Km) (Wako Pure Chemical, Osaka, Japan), 50 µg/ ml; chloramphenicol (Cm) (Wako), 50 µg/ ml; for C. glutamicum: kanamycin (Km), 50 µg/ ml; chloramphenicol (Cm), 5 µg/ ml. The integrants of heme biosynthesis genes were cultivated on solid complex medium plates containing 60 mM glycine (Nakalai, Kyoto, Japan).
DNA manipulations
E. coli plasmid DNA was isolated using a Qiaprep Spin kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Restriction enzymes and T4 DNA ligase (TAKARA Bio, Shiga, Japan) were used as recommended by the manufacturer. E. coli was transformed by the CaCl2 method (Sambrook et al. 1989). Transformation and integration of C. glutamicum were performed by electroporation as previously described (Vertès et al. 1993b). Purified DNA extracted from E. coli SCS110 strain with 1 µg of integrative plasmid or 50 ng of the replicative plasmid was introduced into C. glutamicum cells using GenePulsar II (Bio-Rad, Richmond, CA., USA). After electroporation, cells were incubated at 33°C for 1 h in 1 ml complex medium and plated on complex medium containing appropriate antibiotics. β-galactosidase activity was detected on complex medium plates containing 200 µg/ml X-gal (Nakalai). Preparation of C. glutamicum, E. coli and Rhodopseudomonas palustris genomic DNAs were performed as previously described (Suzuki et al. 2005c). DNA concentration was measured at 260 nm using a Beckman DU640 spectrophotometer (Beckman Coulter, CA, USA). To introduce RE mutant lox66 sequence in C. glutamicum R genome, markerless DNA integration method was performed as previously described (Inui et al. 2004). RE mutant lox66 sequence was integrated in a nonessential region for cell survival at a position 593,110 bases from dnaA gene of the genome (Suzuki et al. 2005b).
Plasmids
The plasmids used in this work are listed in Table 1. The 34 bp RE mutant lox66 site was introduced into the genome of C. glutamicum via markerless DNA integration method (Inui et al. 2004). For this purpose, markerless DNA integration plasmid pCRA420 was constructed as follows: two kinds of DNA fragments, Ins1 (~ 900 bp) and Ins2 (~ 1000 bp) were amplified by polymerase chain reaction (PCR) using C. glutamicum R genomic DNA and Ins1F/ Ins1R or Ins2F/ Ins2R primers. Ins1 was digested with EcoRI and SacI, and ligated to the same sites of pCRA725. Ins2 was digested with SphI and ligated to the SphI site of pCRA725, resulting plasmid pCRA725ins1–2. A 40-bp DNA fragment containing a RE mutant lox66 site and SacI sites was synthesized and ligated to the SacI site of pCRA725ins1–2, resulting in plasmid pCRA420.
Another mutant lox plasmid to integrate foreign DNA into the C. glutamicum genome was constructed as follows: 3.1 kb of β-galactosidase-encoding gene derived from pMC1871 was ligated into BamHI site under the lac promoter of pHSG298. Resultant plasmid was digested with XbaI and SphI and ligated with 44 base DNA fragment containing a LE mutant lox71 site, resulting in pCRA422. As a control plasmid, pCRA421 carrying 1 kb of a C. glutamicum R genomic region from 591,815 to 592,845 instead of a LE mutant lox71 site was constructed and used to integrate foreign DNA by homologous recombination. The 1-kb region is included in a larger region previously described as nonessential for cell survival (Suzuki et al. 2005a).
The plasmid carrying heme biosynthesis pathway genes was constructed as follows: pHSG298 was digested with XbaI and ligated with a 38-base DNA fragment containing a LE mutant lox71 site. The resulting plasmid was designated pLE. DNA fragments containing hemA and hemB derived from R. palustris No.7 and hemC, hemD, and hemE derived from E. coli were amplified from the respective genomic DNAs by PCR by using corresponding hem forward (F) and reverse (R) primers (Table 2). The fragments were ligated to plasmid pTrc99A under the control of trc promoter. DNA fragments containing each heme gene and trc promoter were amplified by PCR with PtrcF/ PtrcR primers, digested with XbaI and SpeI, and ligated to XbaI site of pLE, resulting in plasmid pCRA423. All heme genes were constitutively expressed under trc promoter. PCR primer sequences used in this work are listed in Table 2.
Sequencing
Sequencing was performed on an ABI PRISM 3100 Genetic Analyzer (Applied Biosystems, CA, USA) using BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) according to the manufacturer’s instructions. DNA sequence data were analyzed using GENETYX WIN program (Genetyx, Tokyo, Japan).
Results
C. glutamicum R derivative for stable targeted DNA integration
Cre recombinase can catalyze reciprocal site-specific recombination at 34 bp loxP sites. Insertion of a circular DNA carrying a loxP into a loxP site on a chromosome (integrative recombination) is however, quite inefficient. Given that unimolecular reactions are kinetically favored over bimolecular reactions, the inserted DNA will often be excised (Fig. 1a). The original loxP site is composed of an asymmetric 8 bp spacer region flanked by 13 bp inverted repeats. The LE and RE mutant loxs exhibit changes in their respective peripheral 5 bp (Fig. 1c). Recombination between the LE mutant lox71 and the RE mutant lox66 produces the wild-type loxP and a LE + RE double mutant lox. Due to the weak recognition of double mutant lox by Cre, the integrated DNA is stably maintained in the genome (Fig. 1b) (Araki et al. 1997).
Illustration of Cre/loxP and Cre/LE- and RE-mutant lox system. Black regions of the triangles represent sites at which nucleotide sequence changes occurred. a Recombination between loxP sites. b Recombination between a LE mutant lox71 and a RE mutant lox66 produces a loxP and a LE + RE mutant lox. c Gray boxes indicate sites at which nucleotide sequence changes occurred
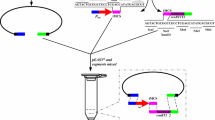
The general scheme for foreign DNA integration using Cre/LE- and RE-mutant lox system for C. glutamicum is depicted in Fig. 2. To integrate foreign DNA, first, cells carrying RE mutant lox66 sequence were constructed using a markerless DNA integration method (Inui et al. 2004). Plasmid pCRA420 was used for this purpose, and RE mutant lox sequence was integrated at a position 593,110 bases from dnaA gene of the genome (Yukawa et al. 2007). Second, the cells carrying RE mutant lox66 sequence were transformed with pCRA406, a replicative Cre recombinase expression plasmid for C. glutamicum. Transformants were selected by their growth in the presence of chloramphenicol, and the resulting cells were designated C. glutamicum CRRE.
Schematic representation of site-directed integration of foreign DNA using Cre/mutant lox system. Only RE mutant lox66 sequence was integrated into C. glutamicum R genome using conditionally lethal sacB selection marker from B. subtilis (Inui et al. 2004). Ins1 and Ins2 are short segments of C. glutamicum R genome. These segments were amplified by PCR and integrated into plasmid pCRA420 for homologous recombination using the sacB system. The cells carrying RE mutant lox66 sequence were transformed by using pCRA406 to supply Cre recombinase. Km represents Kanamycin resistance gene
Screening of transformants for acquired, pCRA406-borne chloramphenicol resistance yielded a strain designated C. glutamicum CRRE. This strain was used as a recipient of non-replicative plasmid constructions carrying the LE mutant lox71 for targeted DNA insertions. Strains in which integration events occurred were selected based on acquired kanamycin resistance (Fig 2).
Integration of 5.7 kb DNA containing a 3.1-kb β-galactosidase gene
A model β-galactosidase-encoding gene, lacZ, of E. coli was used to confirm the feasibility of Cre/mutant lox-mediated DNA integration approach in C. glutamicum. To this end, the plasmid pCRA422 harboring 3.1 kb lacZ along with 34 bp LE mutant lox71 was prepared (Fig 3a). To compare the efficiency of this system with that of homologous recombination, via a single crossover event, the plasmid pCRA421 harboring lacZ and a 1-kb DNA region of C. glutamicum R genome was prepared (Fig 3a). Of each of pCRA421 and pCRA422, 1.0 μg was individually transformed into CRRE strain by using electroporation. After 24 h, thousands of colonies harboring pCRA422 vs 300~400 colonies harboring pCRA421 were obtained on complex medium plates (Fig. 3b). The number of resultant colonies on the plates was directly proportional to the amount of DNA used between the range of 0.01 and 1.0 μg (Table. 3).
Site-directed integration of foreign DNA in C. glutamicum R. a Structure of pCRA421 and pCRA422 plasmids. pCRA421 harbors 1 kb of the C. glutamicum R genomic region from 591,815 to 592,845 in place of the LE mutant lox71 site of pCRA422. The arrow on pCRA421 genomic region indicates the direction of the genomic sequence from 591,815 to 592,845. b 1 μg each of pCRA421 or pCRA422 was used for integration experiments. After electroporation, plates were incubated for 24 h at 33°C and photographed. PCR amplification was performed using wild-type and pCRA422 integrant. Approximately 4.2 and 2.2 kb DNA fragments were successfully amplified with primers P1/P2, or P3/P4, respectively, using the pCRA422 integrant. No fragments were observed from wild-type strain. M indicates the marker
Colony PCR using cells from the two series of plates was performed by using the P1/P2 and P3/P4 primer pairs, and approximately 4.2 and 2.2 kb DNA fragments were amplified from the integrants obtained upon transformation with pCRA422, but not from wild type (Fig. 3b). Finally, PCR products amplified with P1/P2 or P3/P4 primers were isolated and sequenced. The Ins1 and Ins2 sequences were flanked by loxP and LE + RE mutant lox, respectively, confirmative of site-specific integration (data not shown). Ninety-six colonies of pCRA422 were transferred to a new complex medium plate containing kanamycin and X-gal, and all of them were blue in color, demonstrating expression of β-galactosidase activity, which is naturally absent from C. glutamicum R (data not shown).
To remove the Cre recombinase-expressing plasmid, integrants were cultivated for 24 h in complex medium without chloramphenicol, and plated. Approximately 2–5% of resultant cells showed chloramphenicol sensitivity due to the loss of pCRA406. One of them was used in subsequent cultivations. After 100 generations, an aliquot of the culture was spread on a plate, and all colonies were confirmed to be still kanamycin resistant and possess β-galactosidase activity, indicating stability of integrated DNA.
Integration of 10.2 kb DNA containing five heme biosynthesis genes
To further demonstrate the potential of Cre/mutant lox integration strategy for manipulation of C. glutamicum R, a 7 kb DNA element harboring the heme biosynthesis pathway genes was integrated into the genome of this bacterium. To achieve this, the hemA and hemB of R. palustris No. 7 and hemE and hemCD of E. coli were each fused to E. coli trc promoter, which is operative also in C. glutamicum. The pathway was subsequently assembled in lox71-containing vector, giving rise to plasmid pCRA423 (Fig. 4a). Transformation with 0.01 μg of pCRA423 by standard electroporation procedures yielded 177–495 kanamycin resistant colonies. The expected chromosomal localization of integrated pCRA423 (Fig. 4a) was confirmed by amplifying specific 4.8 and 5.1 kb DNA fragments from several colonies by PCR with primer pairs P1/P5 and P3/P6 (Fig. 3b). To demonstrate the activity of the engineered heme biosynthesis pathway, selected integrants were plated on complex medium supplemented with 60 mM glycine as a 5-aminolevulinate precursor. After 24 h incubation at 33°C, the pCRA423 integrant cells turned red, which provided evidence of efficient production of porphyrins in C. glutamicum R.
Site-directed integration of pCRA423. a Structure of pCRA423 plasmid and the resultant integrant. White arrows and triangles indicate heme genes and trc promoters, respectively. Black arrows represent PCR primers. b PCR amplification results of pCRA423 integrant. Colony-PCR was performed and approximately 4.8 and 5.1 kb DNA fragments were successfully amplified with primers P1/P5, or P3/P6, respectively, using the pCRA423 integrant. M indicates the marker. c Phenotype of pCRA423 integrants. The cells were plated on a complex medium plate containing 60 mM glycine. The integrants of pCRA423 were red in color, while those of pCRA421 remained yellow
Discussion
DNA integration techniques in genome are basic genetic tools to express desirable genes stably in bacteria. However, C. glutamicum has been shown to possess restriction systems that severely affect the introduction of foreign DNA into its genome (Vertès et al. 1993b; Schäfer et al. 1997). Due to the development and utilization of efficient DNA transfer techniques, different experimental strategies to integrate a defined DNA efficiently in C. glutamicum genome have continued to emerge. Initially, DNA molecules were transferred from E. coli into C. glutamicum using mobilizable E. coli vectors and conjugation and integrated in the C. glutamicum genome by homologous recombination, with transformation frequencies between 10−5 and 10−6 per incoming donor, resulting in 102–3 integrants from 108 cells of each strain used (Schwarzer and Puhler 1991). Later, a more convenient method using non-replicative and non-methylated vectors improved the integration efficiency to 102 integrants per microgram DNA by electrotransformation and subsequent homologous recombination (Vertès et al. 1993a). However, as these methods depend on homologous recombination, it remained difficult to increase the recombination efficiency artificially. More recently, highly efficient site-specific integration (5 × 103 integrants per microgram DNA) using ϕ16 integrase derived from temperate corynephage ϕ16 has been reported (Moreau et al. 1999). Methods based on recombinase might therefore be useful to integrate DNA fragments within the C. glutamicum genome.
The Cre/loxP system comprised of Cre/LE- and RE-mutant lox system, utilizes the action of Cre recombinase. It is an efficient and simple recombination system which does not require any host-specific system. The LE and RE mutant lox has many potential uses because mono directional recombination is possible. We earlier confirmed that it can work well in C. glutamicum (Suzuki et al. 2005b). To overcome limitations of recombination efficiency of existing methods, Cre/LE- and RE-mutant lox system was exploited. By using this system, integration efficiency of foreign DNAs was 4 orders of magnitude higher than that of homologous recombination of double crossover described by Vertès et al (1993a).
We showed the feasibility of Cre/mutant lox strategy by integrating 5.7 kb pCRA422 harboring the lacZ gene of E. coli, which provided C. glutamicum R with functional β-galactosidase activity. To further demonstrate the potential of the designed system, we implemented the more complex heme biosynthesis pathway into C. glutamicum R by chromosome integration of 10.2 kb plasmid pCRA423. The 7 kb heme region encodes five porphyrin biosynthesis genes, hemA, hemB, hemC, hemD, and hemE. hemA encodes 5-aminolevulinate synthase (ALA-S), which synthesizes 5-aminolevulinate (ALA) from the condensation of glycine with succinyl-coA (Beale 1996; Kwon et al. 2003). C. glutamicum is predicted not to have this pathway for ALA synthesis. hemB–hemE encode enzymes downstream of ALA synthesis in the porphyrin biosynthesis pathway. Normally C. glutamicum dose not produce detectable amounts of porphyrin in medium, but by the integration of the 10.2-kb DNA fragment in the genome, integrants expressed red color, indicating the production of porphyrins. In E. coli, overexpression of hemA gene from R. spheroides resulted in the production of ALA and in the appearance of red-colored colonies containing tetrapyrroles (van der Werf and Zeikus 1996). These results suggest that the integration and overexpression of a heterologous hemA in C. glutamicum R could elevate the intracellular ALA level and of four additional heterologous hemB–hemE enhance the production of porphyrins.
The improvement of C. glutamicum genome is important for enhanced production of biochemicals. The Cre/mutant lox system should be useful for various gene integrations and should greatly contribute in creating a C. glutamicum minimum genome factory (MGF).
References
Albert H, Dale EC, Lee E, Ow DW (1995) Site-specific integration of DNA into wild-type and mutant lox sites placed in the plant genome. Plant J 7:649–659
Araki K, Araki M, Yamamura K (1997) Targeted integration of DNA using mutant lox sites in embryonic stem cells. Nucleic Acids Res 25:868–872
Araki K, Araki M, Yamamura K (2002) Site-directed integration of the cre gene mediated by Cre recombinase using a combination of mutant lox sites. Nucleic Acids Res 30:e103
Beale SI (1996) Biosynthesis of hemes. In: Neidhard, FC (eds) Escherichia coli and Salmonella typhimurium. ASM, Washington, p 731–748
Fujii T, Nakazawa A, Sumi N, Tani H, Ando A, Yabuki M (1983) Utilization of alcohols by Rhodopseudomonas sp. No. 7 isolated from n-propanol-enrichment cultures. Agric Biol Chem 47:2747–2753
Hermann T (2003) Industrial production of amino acids by coryneform bacteria. J Biotechnol 104:155–172
Ikeda M, Katsumata R (1998) A novel system with positive selection for the chromosomal integration of replicative plasmid DNA in Corynebacterium glutamicum. Microbiology 144:1863–1868
Ikeda M, Nakagawa S (2003) The Corynebacterium glutamicum genome: features and impacts on biotechnological processes. Appl Microbiol Biotechnol 62:99–109
Inui M, Kawaguchi H, Murakami S, Vertès AA, Yukawa H (2004) Metabolic engineering of Corynebacterium glutamicum for fuel ethanol production under oxygen-deprivation conditions. J Mol Microbiol Biotechnol 8:243–254
Inui M, Tsuge Y, Suzuki N, Vertès AA, Yukawa H (2005) Isolation and characterization of a native composite transposon, Tn14751, carrying 17.4 kilobases of Corynebacterium glutamicum chromosomal DNA. Appl Environ Microbiol 71:407–416
Kalinowski J, Bathe B, Bartels D, Bischoff N, Bott M, Burkovski A, Dusch N, Eggeling L, Eikmanns BJ, Gaigalat L, Goesmann A, Hartmann M, Huthmacher K, Kramer R, Linke B, McHardy AC, Meyer F, Mockel B, Pfefferle W, Puhler A, Rey DA, Ruckert C, Rupp O, Sahm H, Wendisch VF, Wiegrabe I, Tauch A (2003) The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of . l-aspartate-derived amino acids and vitamins. J Biotechnol 104:5–25
Kinoshita S (1985) Glutamic acid bacteria. In: Demain AL, Solomon NA (eds) Biology of industrial microorganisms. Cummings, London, pp 115–146
Kotrba P, Inui M, Yukawa H (2003) A single V317A or V317M substitution in Enzyme II of a newly identified beta-glucoside phosphotransferase and utilization system of Corynebacterium glutamicum R extends its specificity towards cellobiose. Microbiology 149:1569–1580
Kwon SJ, de Boer AL, Petri R, Schmidt-Dannert C (2003) High-level of porphyrins in metabolically engineered Escherichia coli: systematic extension of a pathway assembled from overexpressed genes involved in heme biosynthesis. Appl Environ Microbiol 69:4875–4883
Lee G, Saito I (1998) Role of nucleotide sequences of loxP spacer region in Cre-mediated recombination. Gene 216:55–65
Lessard PA, Guillouet S, Willis LB, Sinskey AJ (1999) Corynebacteria, Brevibacteria. In: Flickinger, MC, Drew, SW (eds) Encyclopedia of bioprocess technology: fermentation, biocatalysis and bioseparation. Wiley, New York, pp 729–740
Liebl W (1992) The genus Corynebacterium—nonmedical. In: Balowa, A, Truper, HG, Dworkin, M, Harder, W, Schleifer, H (eds) The prokaryotes 2nd ed. Springer, New York, pp 1157–1171
Malumbres, M, Mateos LM, Martin JF (1995) Microorganisms for amino acid production: Escherichia coli and corynebacteria. In: Hui, YH, Kachatourians, GG (eds) Food biotechnology microorganisms, vol 2. VCH, New York, pp 423–469
Moreau S, Blanco C, Trautwetter A (1999) Site-specific integration of corynephage phi16: construction of an integration vector. Microbiology 145:539–548
Ohnishi J, Hayashi M, Mitsuhashi S, Ikeda M (2003) Efficient 40 degrees C fermentation of l-lysine by a new Corynebacterium glutamicum mutant developed by genome breeding. Appl Microbiol Biotechnol 62:69–75
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning, a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory, New York
Schäfer A, Tauch A, Droste N, Pühler A, Kalinowski J (1997) The Corynebacterium glutamicum cglIM gene encoding a 5-cytosine methyltransferase enzyme confers a specific DNA methylation pattern in an McrBC-deficient Escherichia coli strain. Gene 203:95–101
Schwarzer A, Puhler A (1991) Manipulation of Corynebacterium glutamicum by gene disruption and replacement. Biotechnology (N Y) 9:84–87
Suzuki N, Okayama S, Nonaka H, Tsuge Y, Inui M, Yukawa H (2005a) Large-scale engineering of the Corynebacterium glutamicum genome. Appl Environ Microbiol 71:3369–3372
Suzuki N, Nonaka H, Tsuge Y, Inui M, Yukawa H (2005b) New multiple-deletion method for the Corynebacterium glutamicum genome, using a mutant lox sequence. Appl Environ Microbiol 71:8472–8480
Suzuki N, Tsuge Y, Inui M, Yukawa H (2005c) Cre/loxP mediated deletion system for large genome rearrangements in Corynebacterium glutamicum. Appl Microbiol Biotechnol. 67:225–233
Yukawa H, Omumasaba CA, Nonaka H, Kos P, Okai N, Suzuki N, Suda M, Tsuge Y, Watanabe J, Ikeda Y, Vertes AA, Inui M (2007) Comparative analysis of the Corynebacterium glutamicum group and complete genome sequence of strain R. Microbiology 153:1042–1058
van der Werf MJ, Zeikus JG (1996) 5-Aminolevulinate production by Escherichia coli containing the Rhodobacter sphaeroides hemA gene. Appl Environ Microbiol 62:3560–3566
Vertès AA, Hatakeyama K, Inui M, Kobayashi M, Kurusu Y, Yukawa H (1993a) Replacement recombination in Coryneform bacteria: high efficiency integration requirement for non-methylated plasmid DNA. Biosci. Biotech. Biochem. 57:2036–2038
Vertès AA, Inui M, Kobayashi M, Kurusu Y, Yukawa H (1993b) Presence of mrr- and mcr-like restriction systems in coryneform bacteria. Res Microbiol 144:181–185
Vertès AA, Asai Y, Inui M, Kobayashi M, Kurusu Y, Yukawa H (1994) Transposon mutagenesis of coryneform bacteria. Mol Gen Genet 245:397–405
Vertès AA, Inui M, Yukawa H (2005) Manipulating corynebacteria, from individual genes to chromosomes. Appl Environ Microbiol 71:7633–7642
Acknowledgments
We wish to thank Dr. C. Omumasaba (internal) for critical reading of the manuscript. We are also grateful to Ms. S. Minakuchi and Mr. S. Okayama for technical support. This study was funded by New Energy and Industrial Technology Development Organization (NEDO).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Suzuki, N., Inui, M. & Yukawa, H. Site-directed integration system using a combination of mutant lox sites for Corynebacterium glutamicum . Appl Microbiol Biotechnol 77, 871–878 (2007). https://doi.org/10.1007/s00253-007-1215-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-007-1215-2