Abstract
The expression of heterologous proteins in the cytoplasm of Escherichia coli is often accompanied by limitations resulting in uncontrollable fermentation processes, increased rates of cell lysis, and thus limited yields of target protein. To deal with these problems, reporter tools are required to improve the folding properties of recombinant protein. In this work, the well-known σ32-dependent promoters ibpAB and fxsA were linked in a tandem promoter (ibpfxs), fused with the luciferase reporter gene lucA to allow enhanced monitoring of the formation of misfolded proteins and their aggregates in E. coli cells. Overexpression of MalE31, a folding-defective variant of the maltose-binding protein, and other partially insoluble heterologous proteins showed that the lucA reporter gene was activated in the presence of these misfolded proteins. Contrary to this, the absence of damaged proteins or overexpression of mostly soluble proteins led to a reduced level of luciferase induction. Through performing expression of aggregation-prone proteins, we were able to demonstrate that the ibpfxs::lucA reporter unit is 2.5–4.5 times stronger than the single reporter units ibp::lucA and fxs::lucA. Data of misfolding studies showed that this reporter system provides an adequate tool for in vivo folding studies in E. coli from microtiter up to fermentation scales.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Escherichia coli is one of the most widely used host organisms in fermentation processes, which aim to achieve high cell densities and, thus, high yields of target proteins. One of the major limitations during expression of heterologous proteins in E. coli is the aggregation of misfolded proteins. Overexpression and aggregation of heterologous proteins often result in physiological stress, inducing a heat shock-like response (Kanemori et al. 1994; Parsell and Sauer 1989).
A major regulator of the cytoplasmic heat shock network in Escherichia coli is σ32 (RpoH), which functions as promoter-specific subunit of the RNA-polymerase holoenzyme Eσ32 (Erickson et al. 1987; Neidhardt and VanBogelen 1981; Straus et al. 1987; Yamamori and Yura 1982). Folding modulators such as the DnaK–DnaJ–GrpE or the GroEL–GroES chaperone systems belong to the σ32 heat shock regulon, encoding a group of chaperones, proteases, and other heat shock proteins (Hsps). Under normal growth conditions, such Hsps show up in low levels and build complexes with σ32. Under these conditions, σ32-dependent genes are not transcribed (Blaszczak et al. 1995; Gamer et al. 1992, 1996; Liberek et al. 1992; Liberek and Georgopoulos 1993; Straus et al. 1990; Tatsuta et al. 2000). Physiological stress such as heat shock or overproduction of recombinant proteins leads to the release of σ32 from DnaK/J-GrpE, which results in increased transcriptional levels of σ32-dependent genes (Cowing et al. 1985; Grossman et al. 1984, 1987; Landick et al. 1984).
Recently, DNA array technology has been intensively used to investigate the transcriptional regulation of the E. coli stress response induced by heat shock (Chuang et al. 1993; Richmond et al. 1999) or overproduction of recombinant proteins (Jürgen et al. 2000; Lesley et al. 2002). Not surprisingly, the majority of stress-regulated genes are associated with the σ32 heat shock regulon. The highest level of induction after heat shock shows the ibpAB operon, encoding two small Hsps (sHsps), IbpA and IbpB (Chuang et al. 1993; Richmond et al. 1999). These sHsps are involved in intracellular heat shock response, preventing aggregation of misfolded proteins and inducing refolding of stress-denatured proteins (Carrio and Villaverde 2003; Kuczyńska-Wiśnik et al. 2002; Laskowska et al. 1996; Mogk et al. 2003; Veinger et al. 1998). Other upregulated genes are, e.g., htpG and clpB (Lesley et al. 2002; Richmond et al. 1999), which are also involved in the rescue of damaged and aggregated proteins (Motohashi et al. 1999; Thomas and Baneyx 1998; Zolkiewski 1999).
In recent years, reporter genes such as the green fluorescent protein (GFP) or β-galactosidase (LacZ) have been used to examine promoter strength (Dehio et al. 1998; Lissemore et al. 2000) and transcriptional regulation of stress promoters (Bianchi and Baneyx 1999a; Vasina and Baneyx 1996). Bianchi and Baneyx (1999b) have also used the ibp promoter in fusion with LacZα to characterize antibiotics.
Other studies used protein fusions with GFP (Waldo et al. 1999) or LacZ (Wigley et al. 2001) to analyze protein folding and misfolding of overexpressed recombinant proteins. Nevertheless, many heterologous proteins often are not suitable for fusion with such reporters due to inaccessible C terminus of the target protein. The enzymatic activity of such reporters often interferes with functional assays.
In this paper, we used the firefly luciferase as an indirect reporter gene under control of different σ32-dependent promoters. We showed that specific tandem promoters allow enhanced in vivo protein folding studies during cultivation from microtiter up to fermentation scale. We demonstrated that overexpression of insoluble proteins led to increased luciferase activity due to the transcriptional activation of used σ32-dependent promoters. On the contrary, overexpression of soluble proteins, including host and heterologous proteins, resulted only in a low background activity of the lucA reporter gene. In summary, our results suggest that this technology provides a tool to analyze and optimize the protein folding in vivo up to high cell density fermentations.
Materials and methods
Plasmid construction
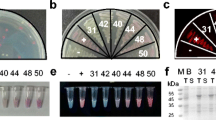
The sequences of all promoters are from E. coli MG1655 and were obtained from the E. coli database EcoCyc (http://www.ecocyc.org; Keseler et al. 2005) and coliBase (http://colibase.bham.ac.uk). Oligonucleotides, summarized in Table 1, were used to construct the ibp and fxs promoters followed by the Shine–Dalgarno sequence from the lactose operon (SDlac). The primers for all promoters were designed to insert an EcoRI site upstream of the promoter and a multicloning site (NcoI, NotI, XbaI, BamHI, HindIII) downstream of the SDlac, allowing cloning into pUC19 via EcoRI and HindIII. The luciferase reporter gene lucA from the pSP-luc+ plasmid (Promega, Mannheim, Germany) was cloned into the pUC19-promoter derivates via NcoI and XbaI. The lucA reporter units were recovered by EcoRI–BamHI digestion and ligated in the same sites of pOU61 (Larsen et al. 1984), thus obtaining the reporter plasmids. All reporter plasmids are based on pOU61, containing the ampicillin antibiotic resistance gene bla and the genetically modified R1 origin, which strongly regulates the plasmid copy number on one plasmid per cell. This results in the avoidance of accumulation of the luciferase by multicopy effects. The primers P1 and P6 (Table 1) have been used to generate the ibpfxs tandem promoter. This fragment is a SD-less ibp promoter with an additional ApoI site that allows cloning into previously generated and EcoRI-digested pfxslucA reporter plasmid. The construction of the ibpfxsT7::lucA reporter unit was achieved by a two-step polymerase chain reaction. In the first step, an assembly product was generated with the primer pair P1 and P7 (Table 1) that functions as a template, to generate the final ibpfxsT7 fusion in combination with the primer P8 (Table 1). To obtain the reporter plasmid pibpfxsT7lucA (Fig. 1), the final assembly product was digested with EcoRI and NcoI and ligated into the plasmid pibplucA, generated previously.
Reporter plasmid pibpfxsT7lucA for determination of protein misfolding-induced stress. The reporter plasmid is based on the plasmid pOU61 (Larsen et al. 1984), carrying the genetically modified R1 (par +) ori, stabilizing the plasmid at low temperature when grown in the absence of selection pressure and tightly regulating the copy number of one per cell by the repA, copA–copB system. Phage λ p R promoter and cI857 allele are required for temperature-dependent runaway replication of plasmid DNA. To determine protein misfolding, the promoters of IbpAB and FxsA were linked in a tandem and were fused with the firefly luciferase reporter gene lucA. To increase the dynamic range of detected luciferase activity, the SDT7g10 (underlined) was used to control the translation of lucA. An asterisk marks the transcription start of both promoters
Cell cultivation and protein expression
The stress-induced transcription of the luciferase reporter gene was estimated by lac-promoter mediated overexpression of different soluble model proteins. For this approach, all proteins were directly cloned downstream of the native lac promoter in the pMK3c-gfp plasmid (Gumpert et al. 2002).
For folding studies the, E. coli K-12 strain RV308 (ATCC No. 31608, Genentech, South San Francisco, CA, USA) was co-transformed with a reporter as well as with an expression plasmid. Proteins were expressed in 5-ml deep well plates at 37°C in Luria–Bertani (LB) medium containing ampicillin (100 mg/l) and kanamycin (30 mg/l). The lac-promoter mediated protein expression was induced by adding isopropylthiogalactoside (IPTG) to a final concentration of 1 mM and was performed for 4 h. To determine background activity of all reporter plasmids, E. coli RV308 was also co-transformed with an empty plasmid (pMK3c-), a derivate of the pMK3c-gfp plasmid without any target gene.
Cell cultivation during scale-up fermentation
For monitoring of cytoplasmic protein misfolding in scale-up processes, we used a 500-ml fermentation system (Infors, Bottmingen, Switzerland), which allows continuous sampling during fermentation via bypass (Fig. 2). To perform folding studies in bioreactors three separate fermentations were carried out: (1) wild-type GFP, (2) UV-optimized GFP (GFPuv; Crameri et al. 1996), and (3) a control fermentation to determine background level activity of the tandem reporter unit ibpfxsT7::lucA. The fermentations were performed at 28°C in LB medium, containing appropriate antibiotics. The fermentation cultures were inoculated with the respective precultures to a start at an optical density at 550 nm (OD550 nm) between 1.0 and 2.0. Reaching an OD550 nm in the range of 16–18, the protein expression was induced by adding IPTG to a final concentration of 1 mM. To increase cell density, addition of a glucose-containing feeding solution (50% w/v in fivefold concentrated LB) was simultaneously started using a constant inflow of 10 ml/h. Samples were taken continuously from a 1:2 prediluted fermenter sample in 0.9% NaCl. An OD controller consisting of a flow-through photometer (Jenway, Princeton, NJ, USA), computer-controlled pumps (Ismatec GmbH, Wertheim-Mondfeld, Germany), and the software Dasylab (GBMmbH, Mönchengladbach, Germany), was used to determine the actual OD550 nm of the predilution and to further dilute the sample to a constant OD550 nm of 1.0. To determine luciferase activity, the diluted sample was mixed at a ratio of 1:1 with luciferin assay buffer (25 mM tricine, 15 mM MgCl2, 5 mM ATP, 7 mM beta-mercaptoethanol, 0.5 mg bovine serum albumin per milliliter, 13 mM d-luciferin Na-salt, pH 7.8) by using another pump with constant speed and then was injected into the online flow-through luminometer LEO (Wallac GmbH, Freiburg, Germany).
Luciferase assay
For luciferase assay, 200 μl of expression cultures were added to 100 μl luciferase assay buffer in microtiter plates. The light emission was measured for 15 min using the luminescence-reader FluostarOptima (BMG Labtech, Offenburg, Germany). The luminescence value in the saturation phase was taken for data analysis. To correlate the signal with a defined amount of cells, luminescence values were standardized to the light scattering of the sample at 550 nm. All values of luminescence were termed as relative luminescence units (RLU).
Fluorescence assay for determination of soluble and active GFP was performed in 96-well microtiter plates using the fluorescence reader FluostarOptima. The GFP activity was assayed in 200 μl of diluted fermenter sample with an OD550 nm of 1.0 and was followed at 365 nm (Ex) and 510 nm (Em).
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis solubility analysis
To examine the folding properties and expression levels of selected model proteins, samples from folding studies at 37°C, described previously, were used. Samples with a defined amount of cells (1 ml of OD550 nm = 10.0) were collected, the cells were harvested by centrifugation, and the bacterial pellet was resuspended in lysis buffer (50 mM Tris–HCl, pH 8.0, 1 mM EDTA, 0.1 mg/ml lysozyme). The cells were lyzed for 10 min on ice by gently inverting them, and the lysates were cleared by centrifugation for 10 min at 20,000 × g at 4°C. Subsequently, aliquots were separated by reducing sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE; Invitrogen, Karlsruhe, Germany), and bands were visualized using Coomassie brilliant blue staining. The accumulated MalE31 was determined by correlation of the band intensity with a calibration curve, which was obtained from purified MalE on SDS-PAGE. For this purpose, the software Phoretix 1D (version 3.0, Phoretix International) was used.
The accession number of sequences of promoters from E. coli MG1655 is U00096 (Blattner et al. 1997).
Results
Design of specific reporter plasmids for detection of cytoplasmic protein misfolding
Five different reporter plasmids were constructed by fusion of well-known stress-inducible promoters of the σ32-regulon to the firefly luciferase reporter gene lucA to detect protein misfolding in vivo. In addition to the stringently regulated ibpAB promoter, we used the promoter of the inner membrane protein FxsA of E. coli (Wang et al. 1999). The promoter is significantly upregulated during overproduction of different misfolding-prone proteins (Lesley et al. 2002). To eliminate factors that influence the comparability between these promoters, the native Shine–Dalgarno (SD) sequences have been replaced by the SD sequence of the lactose operon (SDlac). To enhance the misfolding-induced signal transduction, we designed a reporter construct containing two independent σ32-recognition sites performed by coupling two stress-dependent promoters in tandem. In an alternative approach, the ibp::lucA reporter unit and the ibpfxs tandem promoter were placed under control of the stronger SDT7 to increase the dynamic range of detected luciferase.
Expression of soluble and insoluble protein variants for evaluation of reporter plasmids
To determine how the described reporter plasmids are suited to visualize misfolding-induced stress, we used a set of three protein pairs. Each set consists of two closely related proteins with different folding properties: (1) the well-known and fully soluble maltose-binding protein (MBP) MalE and the folding-defective MalE31 derivate (Betton and Hofnung 1996); (2) the mainly insoluble wild-type GFP and the UV-optimized GFP (GFPuv), which is optimized for bacterial expression in E. coli (Crameri et al. 1996); and (3) two forms of the protease from the tobacco etch virus (TEV protease). One was N-terminally fused to the folding-mediator MBP (soluble) and the other one to glutathione S-transferase (insoluble; Kapust and Waugh 1999; Fig. 4b).
Expression of misfolded proteins induced σ32-dependent reporter gene activation
Lac-promoter mediated expression of the insoluble mutant of the maltose-binding protein (MBP) MalE31 has been carried out as an example to investigate whether σ32-dependent reporter gene activity correlates with the accumulation of misfolded and insoluble proteins in E. coli. Misfolding was examined by measuring the luciferase activity, controlled by the stress-inducible promoters ibp, fxs and the tandem promoter ibpfxs in a time frame of overexpression of MalE31 (Fig. 3). To demonstrate that the courses of luminescence correlate with the expression of insoluble MalE31, its accumulation is shown in the same time frame by SDS-PAGE analysis (Fig. 3a). This protein gel was also taken to quantify the amount of MalE31 using Phoretix 1D. Contrary to the wild-type MalE protein, which is mainly expressed in the soluble form (folding properties shown in Fig. 4b), lac-promoter directed expression of the folding defective mutant MalE31 resulted in an approximate 20-fold increased yield of misfolded MalE31 molecules (Fig. 3b). As shown in Fig. 3c,d, the accumulation of insoluble MalE31 resulted in a significant 5- or 4.5-fold increase of luciferase activity over background level, utilizing the ibp::lucA and fxs::lucA reporter units. Contrary to these results, the tandem promoter ibpfxs showed a 16-fold increase of luciferase activity during accumulation of MalE31 (Fig. 3e). As indicated by luminescence assays, all progress curves of luminescence saturate in a time range of approximately 2.5–4 h. These findings correlate well with expression of MalE31 that is also saturated. Contrary to these results, a background activity at the range of only 30 ± 15 RLU (black bars) was observed using cells harboring the control plasmid pMK3c-.
Stimulation of the σ32-dependent lucA reporter gene after overexpression of the folding-defective maltose-binding protein MalE31. The accumulation of MalE31 in the soluble (S) or insoluble (I) protein fraction at 37°C was determined by SDS-PAGE analysis and is shown in a. The quantification of MalE31 was performed through comparing protein bands on SDS-PAGE, using an MBP calibration curve and the software Phoretix 1D (b). The course of the luciferase activity was measured during lac-promoter mediated overexpression of MalE31 (white bars) and in the controls, carrying an empty plasmid (black bars) by utilization of the ibp::lucA (c), fxs::lucA (d), and ibpfxs::lucA (e) reporter fusions and was standardized to a cell density at 550 nm of 1.0. The represented data are from three independent experiments (average±SD)
The presence of aggregation-prone proteins effects the induction of the lucA reporter gene. The activation of all reporter units was measured after induction of protein expression (white bars) and without expression (black bars) and is shown in a. Samples carrying the reporter plasmid, co-transformed with an empty plasmid, were used to determine the basal luciferase activity (control). The luciferase activities (standardized to an OD550 nm of 1.0) were examined in 200-μl aliquots, taken 4 h after induction at 37°C. The luminescence during overexpression of all selected model proteins was determined by utilizing the designed reporter units (order per group, from left): ibp::lucA, fxs::lucA, ibpT7::lucA, ibpfx::lucA, and ibpfxsT7::lucA. Equivalent volumes of these samples were used to correlate the luminescence levels with the expression level and the folding property of each model protein. The soluble (S) and insoluble (I) protein fractions were separated as described previously and were fractionated by 4–14% SDS-PAGE (b). An arrow indicates the corresponding band of each model protein. Used protein standard (M): SeeBlue® Plus2 (Invitrogen). Represented data are from three independent experiments (average±SD)
Effect of the stress dependent promoters on the luciferase level, when activated by σ32
Results illustrated in Fig. 4a showed that the expression of all insoluble variants of the model proteins led to the highest level of luminescence in cells, harboring the pibplucA reporter plasmid, whereas cells co-transformed with the fxs::lucA reporter unit showed only a reduced transcriptional activity of lucA under the same conditions. With an approximate fivefold increase, the highest luciferase activity observed as a misfolding of MalE31 was determined with the reporter plasmid pibplucA. On the contrary, expression of the soluble MalE variant led to a 1.2-fold increase of luciferase activity over the background level.
An enhanced induction of the reporter gene could be observed under the condition that transcription of lucA was directed by the ibpfxs promoter (ibp, fxs were linked in tandem). If insoluble proteins occur, this tandem promoter showed on average a 2.5- to 4.5-fold increased luciferase activity in comparison to the single promoter units ibp::lucA and the fxs::lucA. As a consequence, expression of, e.g., MalE31, led to a 16-fold increase of luciferase activity, whereas the ibp::lucA and fxs::lucA reporter units showed a 5- or 4.5-fold increased luciferase level.
An improvement of this lucA-based reporter system was achieved after replacement of the common lacZ SD sequence by the stronger SDT7. A two- to fourfold increased range of luciferase activity was observed after expression of all misfolding-prone proteins (Fig. 4a, samples 3 and 5 of each group). As a result of the SD substitution, the background activity also increased by approximately 10%. Nevertheless, utilization of the ibpfxsT7::lucA tandem reporter unit resulted in the highest detected luciferase level, caused by overexpression of all used misfolding-prone model proteins. Low luminescence levels, detected during overexpression of mainly soluble model proteins, are not caused by low expression levels but are a result of its folding properties (Fig. 4b).
To demonstrate whether this reporter system is independent of E. coli strain and temperature, subsequent experiments were performed in E. coli K-12 strains W3110 and MG1655 and at 22 and 30°C. By performing misfolding studies in these strains, equal ratios of luminescence levels were detected as described previously (data not shown). Lowering of the temperature during cultivation resulted in decreased luciferase levels. Reduced levels of luciferase activity were caused by reduced expression levels for the model proteins GFP and improved solubility for MalE31 (data not shown).
Monitoring of folding stress during cultivation in bioreactors
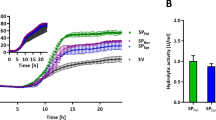
Folding studies with wild-type GFP during scale-up fermentation processes led to an approximate sixfold increase of the luciferase activity during expression. Under the same conditions, expression of GFPuv resulted only in a twofold increase of cytoplasmic luciferase activity and in a fivefold increase of fluorescence activity, which represents the soluble and functional part of GFP (Fig. 5a). In contrary to the GFPuv expression, accumulation of wild-type GFP resulted in an increased rate of cell lysis after 2 h post induction (data not shown). Likewise to folding studies performed in microtiter plates, the luminescence activity also runs into saturation 2.5–4 h after induction of GFP. To determine the background level of lucA, an additional fermentation culture with cells carrying an empty plasmid was performed under the same conditions. In performing this control fermentation, only a minimal increase of luciferase activation could be observed (Fig. 5b).
Online monitoring of folding stress during fermentation of GFP variants. The wild-type GFP (filled symbols) and the UV-optimized GFP (open symbols) were expressed at 28°C. The luminescence (circle) and the fluorescence (square) were measured every 30 min up to 4 h after induction and are represented in a. Both activities were standardized to an OD550 nm of 1.0. In b, the luminescence (circle) and fluorescence (square) are shown, also determined in the same time frame for the control fermentation
Discussion
To improve folding properties of target proteins in E. coli, fusion with well-known folding-promoting tags or variations of expression parameters were commonly used. In addition to these approaches, the protein itself can be improved either by molecular modeling or directed evolution.
Reporter systems are required to validate whether these strategies are sufficient for optimizing the folding properties of target proteins. Recently, fusions with GFP (Waldo et al. 1999) or LacZ (Wigley et al. 2001) have been applied as direct approaches to analyze expression and folding of recombinant proteins. In spite of the widely spread application of these reporters, many heterologous proteins are inappropriate for fusion or the enzymatic activity of these reporters interferes with functional assays.
In this paper, we describe an improved reporter system allowing estimation of the current folding status of overexpressed proteins in E. coli. It is well-established that overexpression of nonnative, aggregation-prone proteins in E. coli results in a heat shock-like response (Kanemori et al. 1994; Parsell and Sauer 1989), which is triggered by members of the σ32-regulon. Former studies showed that the heat shock response, induced by several stress factors, is a fast cellular response with a significant but transient increase of the mRNA level of stress-dependent genes (Bianchi and Baneyx 1999a; Jürgen et al. 2000). To demonstrate that this mechanism can be applied to online in vivo folding studies of heterologous proteins, we constructed five reporter plasmids by fusing different σ32-dependent promoters to the firefly luciferase gene lucA. Stress-inducible promoters were chosen depending on the rate of induction after heat shock response (Jürgen et al. 2000; Lesley et al. 2002).
To show the application of this system, extensive folding studies were carried out using different model proteins, including utilization of different fusion partners and expression regimes.
Results summarized in Fig. 3 demonstrate that the heat shock-like response induced by expression of the folding defective MalE31 (Betton and Hofnung 1996) protein significantly activated the transcription of lucA. Comparing the luminescence activity of these samples with those of control samples or with samples overexpressing the soluble MalE variant (Fig. 4), we propose that the increase of luminescence is caused by proneness of MalE31 to accumulate in the insoluble protein fraction. By achieving these results, we were able to show that the activation of lucA was not influenced by factors like transcriptional or translational stress. The luciferase activity of all reporter units showed saturation 2.5 h after induction of MalE31 expression. These observed plateaus of luminescence correlate with the expression of MalE31, which also showed saturation.
Contrary to the fxs::lucA and ibp::lucA reporter units, the tandem reporter unit ibpfxs::lucA showed an enhanced luciferase activity during overexpression of various insoluble model proteins.
On the basis of the results of the MalE31 overexpression, we investigated a set of further model proteins to verify the function of our reporter units. These studies were done with related protein pairs with known folding properties. Two GFP versions were used to determine the soluble amount of GFP by direct fluorescence assay and its insoluble counterpart by indirect misfolding study using luminescence assay. All applied insoluble proteins showed significantly increased luciferase activity, depending on the used promoter. On the contrary, samples from expression of the soluble counterpart only showed background levels of luciferase activity. Measured basal level of luminescence from sample accumulated soluble proteins was caused by its soluble properties and was not affected by low expression levels (Fig. 4b). This indicates that the luciferase activity is merely a result of the accumulation of misfolded cytoplasmic proteins and not an effect of the induction of the protein itself.
To reach a significant increase of the dynamic range of the luciferase signal during expression of aggregation-prone proteins, we constructed the synthetic tandem promoter ibpfxs. This tandem promoter led to a higher luciferase signal of the induced samples compared with their single promoters. Furthermore, we demonstrated that the substitution of SDlac sequence with the stronger SDT7 sequence resulted in an enhanced luciferase activity. Unfortunately, we simultaneously obtained an increase of the basal activity of the luciferase. This indicates that the utilization of a stronger SD sequence increases the range of luminescence in the presence of misfolded proteins.
To prevent protein misfolding and protein aggregation, a huge number of alternative approaches have been developed. A useful approach is the exploitation of the ability of diverse proteins like the MBP and the glutathione S-transferase (GST) to enhance the solubility of their fusion partners (Kapust and Waugh 1999; Sachdev and Chirgwin 1998). Following this, the lucA-based reporter system was applied to demonstrate how such tags promote the folding of target protein. For this purpose, the catalytic domain of the mature TEV protease was fused to MBP and GST.
Lac-promoter mediated expression of both forms therefore resulted in different levels of luciferase activity, which was verified by simultaneous estimation of soluble and insoluble protein ratios (Fig. 4).
In conclusion, in this study we used the native stress response of E. coli as an approach to detect folding properties of different recombinant proteins and their tendency to accumulate during overexpression. We demonstrated that fusion of well-known stress-dependent promoters, especially the designed tandem promoter to the luciferase reporter gene lucA, allows efficient determination of protein misfolding or misfolding-induced stress.
In addition, we showed that this approach allows kinetic studies of protein folding in vivo during fermentation processes. In contrast to described reporter systems in previous studies, our luciferase-based reporter system works independently of reporter protein-target protein fusions. This indirect reporter system based on the firefly luciferase, which is nontoxic for E. coli, allows in vivo determination of protein misfolding simply through light emission detection.
References
Betton J, Hofnung M (1996) Folding of a mutant maltose-binding protein of Escherichia coli which forms inclusion bodies. J Biol Chem 271:8046–8052
Bianchi AA, Baneyx F (1999a) Hyperosmotic shock induces the sigma32 and sigmaE stress regulons of Escherichia coli. Mol Microbiol 34:1029–1038
Bianchi AA, Baneyx F (1999b) Stress responses as a tool to detect and characterize the mode of action of antibacterial agents. Appl Environ Microbiol 65:5023–5027
Blaszczak A, Zylicz M, Georgopoulos C, Liberek K (1995) Both ambient temperature and the DnaK chaperone machine modulate the heat shock response in Escherichia coli by regulating the switch between sigma 70 and sigma 32 factors assembled with RNA polymerase. EMBO J 14:5085–5093
Blattner FR, Plunkett G 3rd, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y (1997) The complete genome sequence of Escherichia coli K-12. Science 277:1453–1474
Carrio MM, Villaverde A (2003) Role of molecular chaperones in inclusion body formation. FEBS Lett 537:215–521
Chuang SE, Burland V, Plunkett 3rd G, Daniels DL, Blattner FR (1993) Sequence analysis of four new heat-shock genes constituting the hslTS/ibpAB and hslVU operons in Escherichia coli. Gene 134:1–6
Cowing DW, Bardwell JC, Craig EA, Woolford C, Hendrix RW, Gross CA (1985) Consensus sequence for Escherichia coli heat shock gene promoters. Proc Natl Acad Sci USA 82:2679–2683
Crameri A, Whitehorn EA, Tate E, Stemmer WP (1996) Improved green fluorescent protein by molecular evolution using DNA shuffling. Nat Biotechnol 14:315–319
Dehio M, Knorre A, Lanz C, Dehio C (1998) Construction of versatile high-level expression vectors for Bartonella henselae and the use of green fluorescent protein as a new expression marker. Gene 215:223–229
Erickson JW, Vaughn V, Walter WA, Neidhardt CF, Gross CA (1987) Regulation of the promoters and transcripts of rpoH, the Escherichia coli heat shock regulatory gene. Genes Dev 1:419–432
Gamer J, Bujard H, Bukau B (1992) Physical interaction between heat shock proteins DnaK, DnaJ, and GrpE and the bacterial heat shock transcription factor sigma 32. Cell 69:833–842
Gamer J, Multhaup G, Tomoyasu T, McCarty JS, Rüdiger S, Schönfeld HJ, Schirra C, Bujard H, Bukau B (1996) A cycle of binding and release of the DnaK, DnaJ and GrpE chaperones regulates activity of the Escherichia coli heat shock transcription factor sigma32. EMBO J 15:607–617
Grossman AD, Erickson JW, Gross CA (1984) The htpR gene product of E. coli is a sigma factor for heat-shock promoters. Cell 38:383–390
Grossman AD, Straus DB, Walter WA, Gross CA (1987) Sigma 32 synthesis can regulate the synthesis of heat shock proteins in Escherichia coli. Genes Dev 1:179–184
Gumpert J, Hoischen C, Kujau M (2002) Protein expression in L-form bacteria. In: Weiner MP, Lu Q (eds) Gene cloning and expression technologies. BioTechniques Press, Eaton Publishing, Westborough, MA, pp 213–226
Jürgen B, Lin HY, Riemschneider S, Scharf C, Neubauer P, Schmid R, Hecker M, Schweder T (2000) Monitoring of genes that respond to overproduction of an insoluble recombinant protein in Escherichia coli glucose-limited fed-batch fermentations. Biotechnol Bioeng 70:217–224
Kanemori M, Mori H, Yura T (1994) Induction of heat shock proteins by abnormal proteins results from stabilization and not increased synthesis of sigma 32 in Escherichia coli. J Bacteriol 176:5648–5653
Kapust RB, Waugh DS (1999) Escherichia coli maltose-binding protein is uncommonly effective at promoting the solubility of polypeptides to which it is fused. Protein Sci 8:1668–1674
Keseler IM, Collado-Vides J, Gama-Castro S, Ingraham J, Paley S, Paulsen IT, Peralta-Gil M, Karp PD (2005) EcoCyc: a comprehensive database resource for Escherichia coli. Nucleic Acids Res 33:334–337
Kuczyńska-Wiśnik D, Kedzierska S, Matuszewska E, Lund P, Taylor A, Lipinska B, Laskowska E (2002) The Escherichia coli small heat-shock proteins IbpA and IbpB prevent the aggregation of endogenous proteins denatured in vivo during extreme heat shock. Microbiology 148:1757–1765
Landick R, Vaughn V, Lau ET, VanBogelen RA, Erickson JW, Neidhardt FC (1984) Nucleotide sequence of the heat shock regulatory gene of E. coli suggests its protein product may be a transcription factor. Cell 38:175–182
Larsen JE, Gerdes K, Light J, Molin S (1984) Low-copy-number plasmid-cloning vectors amplifiable by derepression of an inserted foreign promoter. Gene 28:45–54
Laskowska E, Wawrzynow A, Taylor A (1996) IbpA and IbpB, the new heat-shock proteins, bind to endogenous Escherichia coli proteins aggregated intracellularly by heat shock. Biochimie 78:117–122
Lesley SA, Graziano J, Cho CY, Knuth MW, Klock HE (2002) Gene expression response to misfolded protein as a screen for soluble recombinant protein. Protein Eng 15:153–160
Liberek K, Georgopoulos C (1993) Autoregulation of the Escherichia coli heat shock response by the DnaK and DnaJ heat shock proteins. Proc Natl Acad Sci USA 90:11019–11023
Liberek K, Galitski TP, Zylicz M, Georgopoulos C (1992) The DnaK chaperone modulates the heat shock response of Escherichia coli by binding to the sigma 32 transcription factor. Proc Natl Acad Sci USA 89:3516–3520
Lissemore JL, Jankowski JT, Thomas CB, Mascotti DP, deHaseth PL (2000) Green fluorescent protein as a quantitative reporter of relative promoter activity in E. coli. Biotechniques 28:82–84, 86, 88–89
Mogk A, Schlieker C, Friedrich KL, Schönfeld HJ, Vierling E, Bukau B (2003) Refolding of substrates bound to small Hsps relies on a disaggregation reaction mediated most efficiently by ClpB/DnaK. J Biol Chem 278:31033–31042
Motohashi K, Watanabe Y, Yohda M, Yoshida M (1999) Heat-inactivated proteins are rescued by the DnaK J-GrpE set and ClpB chaperones. Proc Natl Acad Sci USA 96:7184–7189
Neidhardt FC, VanBogelen RA (1981) Positive regulatory gene for temperature-controlled proteins in Escherichia coli. Biochem Biophys Res Commun 100:894–900
Parsell DA, Sauer RT (1989) Induction of a heat shock-like response by unfolded protein in Escherichia coli: dependence on protein level not protein degradation. Genes Dev 3:1226–1232
Richmond CS, Glasner JD, Mau R, Jin H, Blattner FR (1999) Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Res 27:3821–3835
Sachdev D, Chirgwin JM (1998) Solubility of proteins isolated from inclusion bodies is enhanced by fusion to maltose-binding protein or thioredoxin. Protein Expr Purif 12:122–132
Straus DB, Walter WA, Gross CA (1987) The heat shock response of E. coli is regulated by changes in the concentration of sigma 32. Nature 329:348–351
Straus D, Walter W, Gross CA (1990) DnaK, DnaJ, and GrpE heat shock proteins negatively regulate heat shock gene expression by controlling the synthesis and stability of sigma 32. Genes Dev 4:2202–2209
Tatsuta T, Joob DM, Calendar R, Akiyama Y, Ogura T (2000) Evidence for an active role of the DnaK chaperone system in the degradation of sigma(32). FEBS Lett 478:271–275
Thomas JG, Baneyx F (1998) Roles of the Escherichia coli small heat shock proteins IbpA and IbpB in thermal stress management: comparison with ClpA, ClpB, and HtpG in vivo. J Bacteriol 180:5165–5172
Vasina JA, Baneyx F (1996) Recombinant protein expression at low temperatures under the transcriptional control of the major Escherichia coli cold shock promoter cspA. Appl Environ Microbiol 62:1444–1447
Veinger L, Diamant S, Buchner J, Goloubinoff P (1998) The small heat-shock protein IbpB from Escherichia coli stabilizes stress-denatured proteins for subsequent refolding by a multichaperone network. J Biol Chem 273:11032–11037
Waldo GS, Standish BM, Berendzen J, Terwilliger TC (1999) Rapid protein-folding assay using green fluorescent protein. Nat Biotechnol 17:691–695
Wang WF, Margolin W, Molineux IJ (1999) Increased synthesis of an Escherichia coli membrane protein suppresses F exclusion of bacteriophage T7. J Mol Biol 292:501–512
Wigley WC, Stidham RD, Smith NM, Hunt JF, Thomas PJ (2001) Protein solubility and folding monitored in vivo by structural complementation of a genetic marker protein. Nat Biotechnol 19:131–136
Yamamori T, Yura T (1982) Genetic control of heat-shock protein synthesis and its bearing on growth and thermal resistance in Escherichia coli K-12. Proc Natl Acad Sci USA 79:860–864
Zolkiewski M (1999) ClpB cooperates with DnaK, DnaJ, and GrpE in suppressing protein aggregation A novel multi-chaperone system from Escherichia coli. J Biol Chem 274:28083–28086
Acknowledgements
Thanks to Gisela Sudermann for excellent technical support. Special thanks to Giles Johnson and Friedrich Kring for critical reading of the manuscript. This work was funded by Merck KGaA Darmstadt, Germany.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kraft, M., Knüpfer, U., Wenderoth, R. et al. An online monitoring system based on a synthetic sigma32-dependent tandem promoter for visualization of insoluble proteins in the cytoplasm of Escherichia coli . Appl Microbiol Biotechnol 75, 397–406 (2007). https://doi.org/10.1007/s00253-006-0815-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-006-0815-6