Abstract
We isolated three orange or yellow pigment-producing marine bacteria, strains 04OKA-13-27 (MBIC08261), 04OKA-17-12 (MBIC08260), and YM6-073 (MBIC06409), off the coast of Okinawa Prefecture in Japan. These strains were classified as novel species of the family Flavobacteriaceae based on their 16S rRNA gene sequence. They were cultured, and the major carotenoids produced were purified by chromatographic methods. Their structures were determined by spectral data to be (3R)-saproxanthin (strain 04OKA-13-27), (3R,2′S)-myxol (strain YM6-073), and (3R,3′R)-zeaxanthin (strains YM6-073 and 04OKA-17-12). Saproxanthin and myxol, which are monocyclic carotenoids rarely found in nature, demonstrated significant antioxidative activities against lipid peroxidation in the rat brain homogenate model and a neuro-protective effect from l-glutamate toxicity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Some species of bacteria, yeasts, and fungi, as well as algae and higher plants, synthesize a large number of carotenoids with different molecular structures (Britton et al. 2004). Several species of marine bacteria have been reported to produce dicyclic, monocyclic, or acyclic carotenoids (Goodwin 1980). The Marine Biotechnology Institute (MBI) has identified structurally novel or rare carotenoids from marine bacteria belonging to α-Proteobacteria. These carotenoids have included astaxanthin glucoside from Paracoccus sp. N81106 (re-classified from Agrobacterium aurantiacum; MBIC01143; Yokoyama et al. 1995), 2-hydroxyastaxanthin from Brevundimonas sp. SD212 (MBIC03018; Yokoyama et al. 1996b), and 4-ketonostoxanthin 3’-sulfate from Erythrobacter sp. PC6 (re-classified from Flavobacterium sp. PC-6; MBIC02351; Yokoyama et al. 1996a), these being xanthophylls derived from β-carotene. Recently, we have initiated the analysis of novel and/or rare types of carotenoids from new colored marine bacteria. The new isolates, 04OKA-13-27 (MBIC08261), 04OKA-17-12 (MBIC08260), and YM6-073 (MBIC06409), belonging to the family Flavobacteriaceae were selected for use in this study. The carotenoids produced by these isolates were purified by chromatographic methods and their structures subsequently determined by spectroscopic analyses [diode array detector (DAD) high-performance liquid chromatography (HPLC), field desorption mass spectrometry (FD-MS), proton nuclear magnetic resonance (1H-NMR), and circular dichroism (CD)].
The generation of free radicals has been suggested to play a major role in the progression of a wide range of pathological disturbances, including myocardial and cerebral ischemia (Traystman et al. 1991), atherosclerosis (Palinski et al. 1989), renal failure (Erdogan et al. 2002), inflammation (Cheeseman and Forni 1988), and rheumatoid arthritis (Bodamyali et al. 2004). Subsequent peroxidative disintegration of cells and organelle membranes has previously been implicated in various pathological processes (Mylonas and Kouretas 1999). Carotenoid pigments, which have been proven to possess strong antioxidative activities, have attracted greater attention due to their beneficial effects on human health, e.g., their potential in the prevention of such diseases as cancer and cardiovascular complaints (van den Berg et al. 2000). More than 700 carotenoids have been isolated from natural sources (Britton et al. 2004), and evaluating the pharmaceutical potential of various carotenoids with different structures could be an exciting field of medical research. However, the carotenoid species so far studied for this purpose have been restricted to a small number, including β-carotene and its derivatives, zeaxanthin, β-cryptoxanthin, canthaxanthin, and astaxanthin, and the other dicyclic carotenoids, α-carotene and lutein, and the acyclic carotenoid, lycopene. With the exception of those carotenoids that can be isolated from a species of higher plants or be chemically synthesized, it has been difficult to find natural sources for supplying sufficient amounts of carotenoids. This study shows that the monocyclic carotenoids that are very rarely found in nature, saproxanthin and myxol, are respectively produced by the marine bacteria, strains 04OKA-13-27 and YM6-073. The antioxidative activities of these isolated carotenoids are evaluated for their inhibitory activity against lipid peroxidation induced by free radicals in a rat brain homogenate (Kato et al. 1993) and for their neuro-protective effect against l-glutamate toxicity on the neuronal hybridoma cell line, N18-RE-105 (Shindo et al. 2004).
Materials and methods
Survey of pigment-producing marine bacteria
Samples of algae and hard corals were collected from Majyanohama, Akajima, Kerama Islands in Okinawa Prefecture of Japan. A sediment sample was collected from Maeda–Misaki in Okinawa Prefecture of Japan.
Strain 04OKA-13-27 (MBIC08261) was isolated from the dense mats of filamentous algae, which were collected in March 2004 from within the territory of damselfish (Stegastes nigricans). The algae (1 cm3) were homogenized with a glass rod in 5 ml of sterile seawater. The homogenate was diluted ten times with sterile seawater and was used to isolate the bacterium on a 1/10 MA + Ca medium [3.74 g of 2216 marine broth (Difco, Detroit, MI, USA), 750 ml of filtered seawater, and 250 ml of distilled water with 1% CaCO3 containing 1.5% agar] after being cultivated for 3–7 days. Strain 04OKA-17-12 (MBIC08260) was isolated from hard coral (Acropora nobilis Dana 1846), which had been collected in March 2004 from 2–5 m below the surface of the sea. The coral was cut into small pieces (1 cm3), ground with a mortar and pestle, and transferred into 5 ml of sterile seawater. The bacterium was then isolated on 2216 marine broth (Difco) containing 1.5% agar after being cultivated for 3–7 days. Strain YM6-073 (MBIC06409) was isolated from the sediment sample collected in July 2003 from 0.1 m below the surface of the sea by cultivating for 30 days on an HSV medium (see “Electronic Supporting Material”).
16S rRNA gene sequence determination and analysis
Genomic DNA was extracted by using the InstaGene matrix (BioRad Laboratories, Hercules, CA, USA). PCR-mediated amplification of the 16S rRNA gene was carried out by using universal primers 27F and 1492R (Lane 1991). The PCR products were purified by Montage PCRμ96 (Millipore, Bedford, MA, USA) and sequenced by BigDye terminator cycle sequencing kit v. 3.1 (Applied Biosystems, Foster City, CA, USA). The nucleotide sequence was determined with a 3730 capillary sequencer (Applied Biosystems) after cleaning up the products of the DNA sequencing reaction with Montage SEQ96 (Millipore). The closely related sequences were searched by using the BlastN program (Altschul et al. 1997) in the DNA Data Bank of Japan (DDBJ) and Classifier in RDP-II database release 9 (Cole et al. 2005). The 16S rRNA gene sequences were aligned by using ClustalX (Thompson et al. 1997) with the related 16S rRNA gene sequences in the RDP-II database. The aligned sequence was used to construct a phylogenetic tree by the neighbor-joining method (Saitou and Nei 1987) based on genetic distances calculated from the Kimura two-parameter model (Kimura 1980) by Mega v. 3.1 (Kumar et al. 2004). The robustness of the topology was evaluated by the neighbor-joining method through 1,000 bootstrap replications (Felsenstein 1985).
Cultivation of the biological materials
Each isolated marine bacterium was cultured for 3 days in 100 ml of marine broth (Difco) in 500-ml Sakaguchi flasks at 30°C on a rotary shaker at 100 rpm. The cells grown from 2 l of the culture were collected by centrifugation, the total yield of the cells being about 20–25 g.
Purification of the pigments
Pigments were extracted from the wet cell pellets with 300 ml of 90% methanol by using an ultra sonicator several times, and the methanol was removed by evaporation. Distilled water (150 ml) was then added, and the pigments were extracted twice with ethyl acetate (100 ml) to complete the pigment recovery. The ethyl acetate fraction was dried overnight over anhydrous sodium sulfate and concentrated to dryness under reduced pressure. The resulting residue was loaded into a column of silica gel 60 (20 × 200 mm, Merck, Germany), the major pigments being eluted with 2:1, v/v hexane/ethyl acetate (04OKA-13-27 and YM6-073) and 3:2, v/v hexane/ethyl acetate (3:2, v/v; 04OKA-17-12). These pigments were further purified by preparative silica gel high-performance thin layer chromatography (HPTLC; Merck) developed with dichloromethane/methanol (10:1, v/v), and finally by HPLC in a preparative μBondapak C18 column (10 × 250 mm, Waters, Milford, MS, USA), eluting with methanol in the case of the 04OKA-13-27 and YM6-073 pigments and with 95% methanol for the 04OKA-17-12 pigment at a flow rate of 3 ml/min.
Spectroscopic analysis of the carotenoids
The absorption spectra were measured with an MCPD-3600 photodiode array detector (Otsuka Electronics, Japan) attached to HPLC apparatus equipped with an analytical μBondapak C18 column (8 × 100 mm, RCM type; Waters) and eluted with methanol at a flow rate of 1.8 ml/min. The CD spectra of the carotenoids were measured at 22°C with a J-600 spectropolarimeter (Jasco, Japan) in diethyl ether/2-pentane/ethanol (5:5:2, by vol). The relative molecular masses were measured by FD-MS, using an M-2500 double-focusing gas chromatograph–mass spectrometer (Hitachi, Japan) equipped with field-desorption apparatus. The 1H-NMR spectra in CDCl3 at 18°C were measured at 400 MHz with an AMX400 NMR spectrometer (Bruker, Germany).
Inhibitory activity against lipid peroxidation in a rat brain homogenate
A rat brain homogenate was prepared according to the method of Kubo et al. (1984) with some modifications. In brief, the frozen rat brain (Wistar, 8 weeks, male) was purchased from Funakoshi (Japan). After defrosting the brain in an ice-cold 100 mM phosphate buffer at pH 7.4, 0.4 g of the brain was immediately mixed for 30 s with 15 ml of the ice-cold phosphate buffer in a Teflon homogenizer. The reaction mixture for the assay consisted of 0.2 ml of the homogenate, 0.6 ml of a 100 mM phosphate buffer, 0.1 ml of 1 mM sodium ascorbate and 0.05 ml of a carotenoid solution dissolved in methanol. The mixture was incubated at 37°C for 1 h under reciprocal agitation. Malondialdehyde (MDA) was stoichiometrically formed in the reaction mixture according to the concentration of the lipid peroxides. MDA thus formed was allowed to react with thiobarbituric acid for spectrophotomeric quantification at 532 nm. The percentage inhibition was calculated as follows: [1−(T−B)/(C−B)]×100 (%), in which T, C, and B are respectively the A532 readings of the treated carotenoid, the control (peroxidation with no reagent), and the zero-time control (no peroxidation).
Inhibitory activity against glutamate toxicity in N18-RE-105 cells
N18-RE-105 cells (mouse neuroblastoma clone N18TG-2× Fisher rat 18-day embryonic neural retina; Malouf et al. 1984) were maintained at 37°C in 25-cm2 tissue culture flasks in 90% Dulbecco’s modified Eagle’s medium containing HAT (0.1 mM hypoxanthine, 40 μM aminopterin, and 0.14 mM thymidine) and 10% fetal calf serum under a humidified atmosphere of 5% CO2 and 95% air. The cells were plated in 96-well microplates at a density of 2,000 cells per well with 100 μl of the medium. After culturing for 24 h, the medium was replaced with one containing 10 mM l-glutamate and/or reagents. The cytotoxicity was quantified after 48 h of treatment by an automated colorimetric assay based on the production of dark blue formazan crystals by living cells incubated with the tetrazolium salt, MTS. The MTS activity was measured by using a commercial kit (Promega, Madison, WI, USA). The percentage cell survival was calculated as follows: (T−C)/(B−C)×100 (%), in which T, C, and B are respectively the absorbance values at 492 nm of the carotenoid (+) plus l-glutamate (+), no carotenoid (−), and l-glutamate (+) and neither the carotenoid (−) nor l-glutamate (−), respectively. The EC50 value is the reagent concentration necessary to reduce glutamate-induced cell death by 50% (Miyamoto et al. 1989).
Results
Classification of the marine bacteria
The three marine bacteria, which had been collected off the coast of Okinawa Prefecture, were classified on the basis of the 16S rRNA gene sequences. The lengths of the determined sequences were 1,467 bp for strain 04OKA-13-27 (MBIC08261), 1,463 bp for strain 04OKA-17-12 (MBIC08260), and 1,465 bp for strain YM6-073 (MBIC06409). A similarity search in the databases of DDBJ and RNA Database Project II (RDPII) showed the 16S rRNA gene sequences of strains 04OKA-13-27, 04OKA-17-12, and YM6-073 to respectively be 96.5% (1,408 bp/1,459 bp) similar to Stanierella latercula ATCC 23177T, 95.5% (1,324 bp/1,386 bp) similar to Gaetbulimicrobium brevivitae strain SMK-19T, and 94.2% (1,306 bp/1,386 bp) similar to Robiginitalea biformata strain HTCC2501T. A neighbor-joining tree was constructed (Fig. 1) to deduce the phylogenetic relationship between these strains with already known species in the family Flavobacteriaceae. The result shows that the three bacterial strains should be classified as novel species of the family Flavobacteriaceae.
Phylogenetic tree based on the neighbor-joining method and using the 16S rRNA sequence to demonstrate the phylogenetic position of the carotenoid-producing strains in the family Flavobacteriaceae. Estimated distances were obtained from the Kimura 2 model parameters. Branch points supported by bootstrap resampling are indicated by solid circles (values > 90%) and open circles (values > 75%). The scale bar indicates 0.05 changes per nucleotide. EMBL/GenBank/DDBJ accession numbers are listed in parentheses
Identification of the pigment produced by bacterial strain 04OKA-13-27
The colonies of marine bacterial strain 04OKA-13-27 were red. The elution profile of HPLC for the organic solvent-soluble pigment showed one major carotenoid peak that was eluted at 4.5 min. A yield of about 0.3 mg of the purified carotenoid was obtained from 2 l of the culture after extraction and isolation by chromatographic methods.
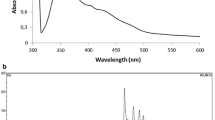
The absorption maxima of this carotenoid were at 294, 363, 445, 471, and 501 nm, and the spectral fine structure of %III/II, which is the ratio of the peak heights of the longest and the medium wavelength absorption bands from the trough between the two peaks (Takaichi and Shimada 1992), was 46 in methanol (Fig. 2, solid line). These results indicate that the carotenoid was a derivative of γ-carotene with 12 conjugated double bonds (Takaichi and Shimada 1992). The relative molecular mass (M+) was observed at 568 m/z, and the 1H-NMR spectrum (Table 1) is compatible with that of saproxanthin (Englert 1995). The CD spectrum is compatible with that of (3R)-bacteriorubixanthin (Takaichi et al. 1988), indicating a 3R configuration (Fig. 3). These data enabled the carotenoid to be identified as (3R)-saproxanthin. The International Union of Pure and Applied Chemistry–International Union of Biochemistry (IUPAC–IUB) semi-systematic name of (3R)-saproxanthin is (3R)-3′,4′-didehydro-1′,2′-dihydro-β,ψ-carotene-3,1′-diol.
Identification of the pigments produced by bacterial strain YM6-073
The colonies of marine bacterial strain YM6-073 were red. The elution profile of HPLC for the pigments showed two major carotenoid peaks eluted at 3.6 and 4.2 min. The polar and less polar carotenoids were separated from each other by chromatographic methods to respectively yield about 0.3 and 0.5 mg of the purified compounds from 2 l of the culture.
The absorption maxima of the polar carotenoid were at 294, 365, 445, 472, 489, and 502 nm, and the spectral fine structure of %III/II was 58 in methanol (Fig. 2). This absorption spectrum is compatible with that of saproxanthin from strain 04OKA-13-27. The relative molecular mass (M+) was observed at 584 m/z. The 1H-NMR spectrum (Table 1) is compatible with that of myxol (Yokoyama and Miki 1995; Takaichi et al. 2001), and the CD spectrum is also compatible with that of (3R,2′S)-myxol (Takaichi et al. 2001). The polar carotenoid was therefore identified to be (3R,2′S)-myxol (Fig. 3). The IUPAC–IUB semi-systematic name is (3R,2′S)-3′,4′-didehydro-1′,2′-dihydro-β,ψ-carotene-3,1′,2′-triol.
The absorption maxima of the less polar carotenoid were 276, (345), (425), 449, 466, and 475 nm, and the spectral fine structure of %III/II were 25 in methanol (Fig. 2, broken line). These results indicate that the carotenoid was a derivative of β-carotene (Takaichi and Shimada 1992). The relative molecular mass (M+) was observed at 568 m/z. The 1H-NMR spectrum (Table 1) is compatible with that of zeaxanthin, and the CD spectrum is also compatible with that of (3R,3′R)-zeaxanthin (Takaichi et al. 1990). The structure of the carotenoid was therefore identified to be that of (3R,3′R)-zeaxanthin (Fig. 3). The IUPAC–IUB semi-systematic name is (3R,3′R)-β,β-carotene-3,3′-diol.
Identification of the pigment produced by bacterial strain 04OKA-17-12
The colonies of marine bacterial strain 04OKA-17-12 were yellow. The elution profile of HPLC for the pigments showed one major carotenoid peak that was eluted at 4.2 min. The yield of the purified carotenoid was about 0.3 mg from 2 l of the culture.
The absorption spectrum (Fig. 2, broken line) and the retention time on HPLC of the carotenoid are compatible with those of zeaxanthin from strain YM6-073. The relative molecular mass (M+) was also observed at 568 m/z, and the 1H-NMR spectrum (Table 1) and CD spectrum are compatible with those of (3R,3′R)-zeaxanthin. The carotenoid was therefore identified to be (3R,3′R)-zeaxanthin (Fig. 3).
Antioxidative activities of the carotenoids
The hydroxyl radicals (OH) produced by Fenton’s reaction (Walling 1975) in the presence of Fe2+ have been suggested to initiate lipid peroxidation and lead to subsequent disintegration of the cell membrane. The rat brain homogenate model has been accepted as being adequate to reflect this lipid peroxidation damage (Kato et al. 1993). The inhibitory activities against lipid peroxidation in the rat brain homogenate by saproxanthin, myxol, and zeaxanthin were evaluated (Fig. 4). The IC50 values were 2.1, 6.2, and 13.5 μM, respectively (10.9 μM for β-carotene), indicating these carotenoids to be more active than flunarizine (IC50 of 55.0 μM), which is a brain protective calcium antagonist with free radical-scavenging activity (Kubo et al. 1984). These carotenoids can therefore be expected to be useful for alleviating tissue damage resulting from the generation of free radicals and subsequent peroxidative disintegration of the cell membrane.
It is widely accepted that l-glutamate, which acts as a neurotransmitter in most areas of the central nervous system, induces delayed neuronal cell death after an ischemic assault (Malouf et al. 1984; Choi 1990). Thus, substances that inhibit l-glutamate toxicity can be expected to prevent or amelionate the brain damage caused by brain ischemia. The toxicity of l-glutamate in N18-RE-105 cells has reportedly been mediated by the inhibition of cysteine uptake, which consequently suppresses synthesis of the intracellular reducing agent, glutathione (Murphy et al. 1990). Thus, such antioxidants as α-tocopherol (EC50 of 57 nM) suppress the toxic effects of l-glutamate in N18-RE-105 cells by scavenging the oxygen radicals in place of glutathione. Saproxanthin, myxol, and zeaxanthin were tested for their inhibitory activities against l-glutamate toxicity in N18-RE-105 cells by the evaluation system described by Murphy et al. (1990; Fig. 5). The respective EC50 values were 3.1, 8.1, and >500 μM (β-carotene >100 μM). These results show that saproxanthin and myxol possessed potent neuro-protective activity and may be useful for use against cerebral ischemic disease.
Discussion
(3R)-saproxanthin has only previously been found from Saprospira grandis (Saprospiraceae; Aasen and Liaaen-Jensen 1966). Hence, marine bacterial strain 04OKA-13-27 is the second species to produce saproxanthin. (3R,2′S)-Myxol has only previously been found in marine bacterial strain P99-3 (MBIC03313) belonging to the family Flavobacteriaceae (Fig. 1; Yokoyama and Miki 1995) and in cyanobacterium Anabaena variabilis ATCC 29413 (Takaichi et al. 2006). Hence, marine bacterial strain YM6-073 is the third species to produce myxol. On the other hand, myxol 2′-glycosides are widely distributed in cyanobacteria such as myxol 2′-dimethyl-fucoside from Synechocystis sp. PCC 6803 (Takaichi et al. 2001). It is interesting that myxol has only been found in marine bacteria belonging to the family Flavobacteriaceae, apart from in cyanobacteria. If 2′-hydroxylase works on saproxanthin, this carotenoid is capable of being converted into myxol (Fig. 3). The saproxanthin-producing bacterial strain 04OKA-13-27, which phylogenetically belongs to a cluster different from a cluster containing strain P99-3 (Fig. 1), seems not to possess a gene encoding 2′-hydroxylase that strains P99-3 and YM6-073 should possess. Phylogenetic position of strain YM6-073 is distant from those of strains P99-3 and 04OKA-13-27 (Fig. 1). Thus, monocyclic carotenoids such as saproxanthin and myxol that contain γ-carotene skeleton may widely be present in bacteria belonging to the family Flavobacteriaceae. The crtYm gene coding for lycopene β-monocyclase, which converts lycopene into γ-carotene, has already been isolated from bacterial strain P99-3 (Teramoto et al. 2003).
The hydroxyl groups of saproxanthin and myxol are thought to influence the orientation of carotenoids in the membrane (Woodall et al. 1997). They are very likely to be anchored in the head-group region of the phospholipids that form the bilayer. In respect of the orientation in the membrane, the hydroxyl group(s) at the acyclic terminal seem very likely to play the major role, compared with the hydroxyl group at β-end group (Albrecht et al. 2000). They have also reported that carotenoids possessing hydroxyl groups at both acyclic terminals or with no hydroxyl groups were less antioxidative. Such integration of saproxanthin or myxol should lead to the reinforcement and stabilization of biological membranes, which may decrease their permeability for oxygen (Subczynski et al. 1991) and may enhance protection against radical-induced peroxidation. The superior antioxidative activities of saproxanthin and myxol that were demonstrated in the two bioassays used, when compared with those of zeaxanthin and β-carotene, seem logical for the foregoing reason.
In conclusion, we isolated three novel marine bacteria belonging to the family Flavobacteriaceae. Two rare carotenoids, (3R)-saproxanthin and (3R,2′S)-myxol, were identified from these strains and proved to possess strong antioxidative activities. These rare carotenoids need to be evaluated for their potential as development materials for pharmaceuticals or foods preventing such diseases as cancer and cardiovascular complains. The result of the present work also suggests that analyses of carotenoids in marine bacteria belonging to Flavobacteriaceae would be promising for finding diverse rare carotenoids.
References
Aasen AJ, Liaaen-Jensen S (1966) The carotenoids of flexibacteria: II. A new xanthophyll from Saprospira grandis. Acta Chem Scand 20:811–819
Albrecht M, Takaichi S, Steiger S, Wang Z-Y, Sandmann G (2000) Novel hydroxycarotenoids with improved antioxidative properties produced by gene combination in Escherichia coli. Nat Biotechnol 18:843–846
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Bodamyali T, Kanczler JM, Millar TM, Stevens CR, Blake DR (2004) Free radicals in rheumatoid arthritis: mediators and modulators. Oxidative Stress and Disease 10:591–610
Britton G, Liaaen-Jensen S, Pfander H (2004) Carotenoids Handbook, Birkhäuser Verlag, Basel
Cheeseman KH, Forni LG (1988) An investigation of the novel anti-inflammatory agents ONO-3144 and MK-447. Studies on their potential antioxidant activity. Biochem Pharmacol 37:4225–4233
Choi WD (1990) Cerebral hypoxia: some new approaches and unanswered questions. J Neurosci 10:2493–2501
Cole JR, Chai B, Farris RJ, Wang Q, Kulam SA, McGarrell DM, Garrity GM, Iedje JM (2005) The ribosomal database project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res 33:D294–D296
Englert G (1995) NMR spectroscopy. In: Britton G, Liaaen-Jensen S, Pfander H (eds) Carotenoids, vol. 1B. Spectroscopy. Birkhäuser, Basel, pp 147–260
Erdogan C, Unlucerci Y, Turkmen A, Kuru A, Cetin O, Bekpinar S (2002) The evaluation of oxidative stress in patients with chronic renal failure. Clin Chim Acta 322:157–161
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Goodwin TW (1980) The Biochemistry of the carotenoids, 2nd edn, vol. 1. Plant. Chapman and Hall, London, pp 291–319
Kato S, Shindo K, Kawai H, Odagawa A, Matsuoka M, Mochizuki J (1993) Pyrrolostatin, a novel lipid peroxidation inhibitor from Streptomyces chrestomyceticus. J Antibiot 46:892–899
Kimura M (1980) A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Kubo K, Yoshitake I, Kumada Y, Shuto K, Nakamizo N (1984) Radical scavenging action of flunarizine in rat brain in vitro. Arch Int Pharmacodyn 272:283–295
Kumar S, Tamura K, Nei M (2004) MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. Wiley, New York, pp 115–175
Malouf AT, Schnaar RL, Coyle JT (1984) Characterization of a glutamic acid neurotransmitter binding site on neuroblastoma hybrid cells. J Biol Chem 259:12756–12762
Miyamoto M, Murphy TH, Schnaar RL, Coyle JT (1989) Antioxidants protect against glutamate-induced cytotoxicity in a neuronal cell line. J Pharmacol Exp 250:1132–1140
Murphy HT, Schnaar LR, Coyle TJ (1990) Immature cortical neurons are uniquely sensitive to glutamate toxicity by inhibition of cysteine uptake. FASEB J 4:1624–1633
Mylonas C, Kouretas D (1999) Lipid peroxidation and tissue damage. In Vivo 13:295–309
Palinski W, Rosenfeld ME, Yla HS, Gurtner GC, Socher SS, Butler SW, Parthasarathy S, Carew TE, Steinberg D, Witztum JL (1989) Low density lipoprotein undergoes oxidative modification in vivo. Proc Natl Acad Sci USA 86:1372–1376
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Shindo K, Kimura M, Iga M (2004) Potent antioxidant activity of cacalol, a sesquiterpene contained in Cacalia delphiniifolia et Zucc. Biosci Biotechnol Biochem 68:1393–1394
Subczynski WK, Markowska E, Sielewiesiuk J (1991) Effect of polar carotenoids on the oxygen diffusion-concentration product in lipid bilayers. An EPR label study. Biochim Biophys Acta 1068:68–72
Takaichi S, Shimada K (1992) Characterization of carotenoids in photosynthetic bacteria. Methods Enzymol 213:374–385
Takaichi S, Shimada K, Ishidsu J (1988) Monocyclic cross-conjugated carotenal from an aerobic photosynthetic bacterium, Erythrobacter longus. Phytochemistry 27:3605–3609
Takaichi S, Shimada K, Ishidsu J (1990) Carotenoids from the aerobic photosynthetic bacterium, Erythrobacter longus: β-carotene and its hydroxyl derivatives. Arch Microbiol 153:118–122
Takaichi S, Maoka T, Masamoto K (2001) Myxoxanthophyll in Synechocystis sp. PCC 6803 is myxol 2′-dimethyl-fucoside, (3R,2′S)-myxol 2′-(2,4-di-O-methyl-α-l-fucoside), not rhamnoside. Plant Cell Physiol 42:756–762
Takaichi S, Mochimaru M, Maoka T (2006) Presence of free myxol and 4-hydroxymyxol and absence of myxol glycosides in Anabaena variabilis ATCC 29413, and proposal of biosynthetic pathway of carotenoids. Plant Cell Physiol 47:211–216
Teramoto M, Takaichi S, Inomata Y, Ikenaga H, Misawa N (2003) Structural and functional analysis of a lycopene β-monocyclase gene isolated from a unique marine bacterium that produces myxol. FEBS Lett 545:120–126
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24:4876–4882
Traystman RJ, Kirsch JR, Koehler RC (1991) Oxygen radical mechanisims of brain injury following ischemia and reperfusion. J Appl Physiol 71:1185–1195
van den Berg H, Faulks R, Granado HF, Hirschberg J, Olmedilla B, Sandmann G, Southon S, Stahl W (2000) The potential for the improvement of carotenoid levels in foods and the likely systemic effects. J Sci Food Agric 80:880–912
Walling C (1975) Fenton’s reagent revised. Acc Chem Res 8:125–131
Woodall AA, Britton G, Jackson MJ (1997) Carotenoids and protection of phospholipids in solution or in liposomes against oxidation by peroxyl radicals: Relationship between carotenoid structure and protective ability. Biochim Biophys Acta 1336:575–586
Yokoyama A, Miki W (1995) Isolation of myxol from a marine bacterium Falvobacterium sp. associated with a marine sponge. Fish Sci 61:684–686
Yokoyama A, Adachi K, Shizuri Y (1995) New carothenoid glycosides, astaxanthin glucoside and adonixanthin glucoside, isolated from the astaxanthin-producing marine bacterium, Agrobacterium aurantiacum. J Nat Prod 58:1929–1933
Yokoyama A, Izumida H, Shizuri Y (1996a) New carotenoid sulfates isolated from a marine bacterium. Biosci Biotech Biochem 60:1877–1878
Yokoyama A, Miki W, Izumida H, Shizuri Y (1996b) New trihydroxy-keto-carotenoids isolated from an astaxanthin-producing marine bacterium. Biosci Biotech Biochem 60:200–203
Acknowledgement
The hard coral used was identified by Hiroki Taniguchi of Akajima Marine Science Laboratory, to whom we are grateful. This work was supported in part by the Biotechnology and Medical Technology Development Department of the New Energy and Industrial Technology Development Organization of Japan (NEDO).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Shindo, K., Kikuta, K., Suzuki, A. et al. Rare carotenoids, (3R)-saproxanthin and (3R,2′S)-myxol, isolated from novel marine bacteria (Flavobacteriaceae) and their antioxidative activities. Appl Microbiol Biotechnol 74, 1350–1357 (2007). https://doi.org/10.1007/s00253-006-0774-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-006-0774-y