Abstract
The plant pathogenic fungus Aspergillus flavus produces several types of mycotoxins. The most well known are the carcinogenic compounds called aflatoxins. In addition, A. flavus produces cyclopiazonic acid and aflatrem mycotoxins, contributing to the toxicity of A. flavus infected crops. Cyclopiazonic acid is a specific inhibitor of calcium-dependent ATPase in the sarcoplasmic reticulum that results in altered cellular Ca++ levels. Aflatrem is a potent tremorgenic mycotoxin known to lead to neurological disorders. Previously we showed that a gene called veA controls aflatoxin and sclerotial production in A. parasiticus. In this study in A. flavus, we show that the veA homolog in A. flavus not only is necessary for the production of aflatoxins B1 and B2 and sclerotia, but also regulates the synthesis of the mycotoxins cyclopiazonic acid and aflatrem. The A. flavus ΔveA mutant was completely blocked in the production of aflatrem and showed greater than twofold decrease in cyclopiazonic acid production. The genes involved in the synthesis of cyclopiazonic acid are unknown; however, the aflatrem gene cluster has been characterized. Northern hybridization analysis showed that veA is required for expression of the A. flavus aflatrem genes atmC, atmG, and atmM. This is the first report of a regulatory gene governing the production of cyclopiazonic acid and aflatrem mycotoxins.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Among the most common toxigenic fungi affecting the food chain are Aspergillus spp. These fungi infect commodities such as corn, peanuts, cotton, tree nuts, sorghum, and other oil seeds. In particular, A. flavus is of great importance due to its impact on agriculture and human health. A. flavus produces several types of mycotoxins. The most well known are the aflatoxins (AF), with AFB1 being one of the most mutagenic and carcinogenic natural compounds known (Payne and Brown 1998; Sweeney and Dobson 1999; Trail et al. 1995). Ingestion of AF-contaminated food or feed has been associated with hepatotoxicity, teratogenicity, immunotoxicity, and even death in animals and humans (Dvorackova and Kusak 1990; Trail et al. 1995). Ingestion of AF is directly correlated with incidence of human liver cancer (Wogan et al. 1992). It has also been demonstrated that AF induces mutations in the p53 tumor suppressor gene and K-ras and H-ras protooncogenes (Denissenko et al. 1999; Lasky and Magder 1997; Riley et al. 1997; Shen and Ong 1996; Wang and Groopman 1999). Aflatoxin contamination of crops can also result in significant economic losses to both producers and processors in the USA and worldwide (Richard and Payne 2003; Wu 2004).
In addition to AF, some A. flavus strains produce other mycotoxins such as cyclopiazonic acid (CPA) and aflatrem. CPA is an indole tetramic acid (Fig. 1a) that is toxic to animals and humans. This compound is a specific inhibitor of calcium-dependent ATPase in the sarcoplasmic reticulum altering Ca++ levels, resulting in increased muscle contraction (Riley and Goeger 1992). In rats, CPA has been reported to cause lesions of the liver, kidney, pancreas, spleen, and heart (Purchase 1971). Chicken fed with rations containing 100 ppm of CPA had proventriculitis, mucosal necrosis, hepatitis, and myocardial inflammation, with a significant mortality rate (Dorner et al. 1983). The toxin accumulates in edible tissue in poultry (Norred et al. 1988; Porter et al. 1988), and it is also found in milk (in ewes) and eggs (laying hens). CPA is also produced by several Penicillium spp., including P. aurantiohriseum (P. cyclopium), where it was first described. P. camembertii, used in the production of gourmet cheese, produces CPA (Pitt et al. 1986). These findings indicate additional routes of CPA contamination in food. Aflatoxin and CPA contamination can occur together in the same food commodity (Dorner et al. 1983; Horn and Dorner 1999; Lansden and Davidson 1983; Purchase 1971; Urano et al. 1992). Both CPA and AF were isolated from ground nut meal samples related to the “turkey X disease” that caused the death of 100,000 turkey poults (Blout 1961; Goldblatt 1969), and led in large part to the initiation of AF characterization. CPA might have contributed to some of the symptoms described in this early case (Blout 1961; Cole et al. 1986).
The mycotoxin aflatrem, an indole diterpene (Fig. 1b) also produced by A. flavus, is a potent tremorgenic compound known to lead to neurological disorders (Valdes et al. 1985; Yao et al. 1988). Although the mechanism by which aflatrem causes tremors in animals is not well defined, clinical studies have shown that these symptoms are generated by alterations in the receptors and neurotransmitter release in the central and peripheral nervous system. Recently, a gene cluster containing three genes involved in aflatrem biosynthesis was identified (Zhang et al. 2004). These authors demonstrated that the genes atmG, atmC, and atmM are necessary for aflatrem biosynthesis in A. flavus.
Our previous studies in A. parasiticus showed that the veA gene is required for the production of AF intermediates as well as for the morphogenesis of resistant structures termed sclerotia (Calvo et al. 2004). The veA gene product does not present homology with any other protein of known function. Our chemical analysis showed that the synthesis of additional unknown compounds could also be affected by the veA mutation. The possible role of veA as a regulator of additional secondary metabolic pathways leading to the production of other mycotoxins in AF producers has not been investigated. In A. flavus, the genes involved in the CPA biosynthetic pathway or the regulatory mechanisms controlling CPA production are unknown. Furthermore, the regulatory mechanisms controlling expression of the aflatrem biosynthetic gene cluster have yet to be elucidated. This is the first report of a gene, veA, controlling the synthesis of the mycotoxins CPA and aflatrem. These findings add to the significance of veA as a possible genetic target to control mycotoxin contamination of food and feed commodities.
Materials and methods
Fungal strains and growth conditions
The A. flavus strain used in this study was ATCC MYA384 (a niaD −A. flavus 70S strain). This A. flavus strain produces abundant small sclerotia. ATCC MYA384 was also used as host strain for fungal transformation (see below). ATCC MYA384 and the transformant strains were inoculated on YGT medium (0.5% wt/v yeast extract, 2% wt/v glucose, and 1 ml of trace element solution per liter), unless otherwise indicated. Agar (15 g l−1) was added in the case of solid medium. Strains were maintained as glycerol stocks at −80 °C.
Identification of the A. flavus veA gene
A 3.6-kb polymerase chain reaction (PCR) product representing the A. flavus veA coding region and flanking DNA sequences was obtained by PCR amplification of A. flavus genomic DNA template using the primers FveA and RveA (Table 1) designed from the available A. parasiticus veA nucleotide sequence (Calvo et al. 2004; GenBank accession no. AY445513). The PCR product was cloned in TOPO-pCR 2.1 vector and sequenced using synthetic primers and an ABI PRISM DNA sequencing kit (Perkin Elmer Life Science). The A. flavus veA sequence was deposited in GenBank with accession no. DQ296645. This sequence information was then used for the construction of the deletion vector.
Generation of the A. flavus ΔveA mutant strain
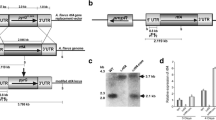
The 3.6-kb PCR product was subsequently used in the engineering of a plasmid vector for the insertional inactivation of the wild-type A. flavus ΔveA gene. The transformation vector pBS-veA-niaD was generated as follows: The 3.6-kb veA region was released from the TOPO-pCR 2.1 vector by EcoRI digestion. The veA fragment was then ligated into EcoRI digested, dephosphorylated plasmid vector pBluescript SK− (Stratagene) in which the XhoI site present in the multiple cloning site had been previously inactivated by XhoI digestion followed by a Klenow end-fill reaction. The pBS-veA vector was then digested with XhoI, which cuts at nt 65 and 638 of the veA ORF, and dephosphorylated. A 7.4-kb SalI–XhoI fragment containing the A. parasiticus niaD gene present in the vector pSL82 (Horng et al. 1990) was ligated into the XhoI digested pBS-veA, creating the plasmid pBS-veA-niaD. This construct now harbored the veA gene that was disrupted by insertion of the niaD gene region. The pBS-veA-niaD plasmid DNA was used to transform ATCC MYA384 (A. flavus 70S niaD − strain). Protoplasts of A. flavus 70S niaD − were prepared as previously described by Cary et al. (2005). Transformants in which the wild-type veA gene had been insertionally inactivated via double homologous recombination with the veA-niaD gene region (Fig. 2a) were initially identified as those colonies that did not form sclerotia during growth. Five transformants of veA-niaD (ΔveA) demonstrated this phenotype and were selected for further analysis. Three control transformants were also obtained after transformation of MYA384 with pSL82. Two different selectable markers, required for complementation, are not available in A. flavus S strains. However, the ΔveA mutants showed the same phenotype and were similar to those generated in other Aspergillus spp. with respect to aflatoxin/sterigmatocystin (ST) production and sclerotial formation (Calvo et al. 2004; Kato et al. 2003). Southern analysis confirmed the veA disruptions via recombinational inactivation (Fig. 2b).
Construction of the A. flavus veA knockout mutant. a Schematic diagram of the knockout vector pBS-veA-niaD used to generate the veA knockout mutants. The dashed lines show the results expected by replacement of the wild-type DNA with DNA containing the niaD transformant selection cassette. Direction of transcription is indicated by horizontal arrows. The lengths of expected SalI or XhoI DNA fragments upon restriction enzyme digestion of either the wild-type strain or the veA knockout transformant genomic DNA are shown under the horizontal lines. Abbreviations: Xh, XhoI; S, SalI; E, EcoRI; N, NotI; Xh/S, XhoI/SalI site inactivated by Klenow end-fill ligation. Distances depicted are not to scale. b Results of Southern hybridization. Lanes: 1, veA, SalI; 2, ΔveA, SalI; 3, veA, XhoI; 4, ΔveA, XhoI. All the transformants were examined by Southern analysis, obtaining the sample band patterns indicating gene replacement in the ΔveA strains or presence of the wild-type veA allele in the control strains. Molecular size markers are Lambda HindIII digested DNA
Physiological studies
Studies on fungal development were performed with the A. flavus pSL82 control strains (veA wild-type allele and niaD+; herein referred to as control strains) and the A. flavus ΔveA mutant strains. Plates containing 30 ml of solid YGT medium were point inoculated and incubated in the light (25 μE m−2 s−1) or in the dark at 30 °C for 7 days. Conidia were harvested with equal amounts of water and counted using a hemacytometer. Sclerotia were numerous and homogeneously distributed on the colony surface in the control strains. To evaluate sclerotial production, three cores of 1-cm diameter were collected from different areas of each replicate plate surface. Experiments were carried out with four replicates. Sclerotia found in those cores were counted under a stereo-zoom microscope (Lieder). Total sclerotial production per colony was estimated taking into account the colony diameter. Colony growth was recorded as colony diameter.
Aflatoxin analysis
Approximately 106 spores per milliliter were inoculated into 500-ml Erlenmeyer flasks containing 250 ml YGT media and incubated in the dark for 14 h at 30 °C with shaking at 200 rpm. Mycelia from each flask were harvested onto a sterile 87-mm nylon membrane (Schleicher & Schuell, Inc.) and placed onto the surface of a YGT agar plate and incubated at 30 °C in the light. A nylon filter was removed from the surface of a plate at 2-, 12-, 24-, or 36-h time points. The filter and macerated agar were placed in a 250-ml Erlenmeyer flask, and 100 ml sterile water and 50 ml acetone were added. The sample was gently agitated at room temperature for a minimum of 1 h. AF was extracted and analyzed by thin layer chromatography (TLC) as previously described (Cleveland et al. 1987). The residue was resuspended in acetone and spotted on a TLC plate (Si250F, J.T. Baker). The plate was then developed in toluene/ethyl acetate/formic acid (5:4:1, v/v/v), air-dried, and examined for the presence of AF under ultraviolet (UV) light.
CPA analysis
Twenty sterile vials containing 2 ml of YGT media were inoculated using a final concentration of 106 spores per milliliter. The samples were vortexed following inoculation and before they were placed in the incubator. The cultures were grown statically at 30 °C for 7 days in the light or in the dark. The experiment was carried out with five replicates. Following incubation, the cultures were harvested, homogenized, extracted with chloroform, and allowed to dry overnight. Samples were analyzed by high performance liquid chromatography (HPLC) on a Waters 600E HPLC system with Empower software in conjunction with a Waters 717 auto sampler and a Waters 486 tunable absorbance detector unit adjusted to a UV wavelength of 280 nm. The extracts were resuspended in 200 μl of mobile phase solvent mixture that consisted of n-haptane:2propanol:water:40% tetrabutylammonium hydroxide (2,560:1,120:32:8, v/v/v/v) (Sobolev et al. 1998). Twenty-five microliters was injected into a Luna 100A (Phenomenex) normal phase silica column (150×4.6 mm) of 5-μm particle size. A flow rate of 1 ml min−1 was used. The elution time of the samples was compared with pure CPA standard (Sigma) and quantified on the basis of the ratio of the peak area of CPA to that of the standard.
Aflatrem analysis
Seed cultures of 25 ml of YEPGA liquid medium (Zhang et al. 2004) containing approximately 5×106 spores per milliliter (final concentration) were incubated in 125-ml Erlenmeyer flasks at 200 rpm at 30 °C for 48 h. Two milliliters of the A. flavus seed cultures was then transferred to 250-ml Erlenmeyer flasks containing 50 ml of aflatrem production medium (Zhang et al. 2004). Cultures were incubated statically under light or dark conditions at 30 °C, and the mycelia were harvested after 7 days of incubation by filtering through Miracloth and removing the excess liquid with paper towels. One-gram aliquots were lyophilized and homogenized in 10 ml of a 2:1 mixture of chloroform–methanol and vortexed. The samples were placed at room temperature and vortexed every 10 min for 1 h. The samples were centrifuged at 2900×g at room temperature for 10 min to pellet insoluble material. The supernatants were transferred to new tubes and the solvent allowed to evaporate overnight. Five hundred microliters of acetonitrile:water (9:1) was added to each of the dried extracts and vortexed until the samples were totally resuspended. Samples were centrifuged at 150×g at room temperature for 20 min, and the supernatants were analyzed by HPLC using the Waters system described above. Aflatrem was detected at 230 and 280 nm as previously described (McMillan et al. 2003) A Luna (Phenomenex) C18 100A (46×25 cm) with a particle size of 5 μm was used for the aflatrem analysis. The mobile phase was composed of acetronitrile:water (9:1) with a flow rate of 1.0 ml min−1. Elution time and quantification of the samples were obtained by comparing with that of pure aflatrem standard (a gift provided by Merck).
Northern analysis
For Northern analysis of AF gene expression, the fungal cultures were grown as described for aflatoxin analysis above. Mycelia were carefully scraped from the nylon membrane, frozen with liquid nitrogen, and stored at −80 °C. The samples for Northern analysis of aflatrem gene expression were obtained using the same culture conditions described for aflatrem analysis above, but in this case, mycelia were harvested at 72, 96, 120, and 144 h for RNA extraction. Total RNA was isolated from fungal mycelia using Triazol reagent (Invitrogen) according to the manufacturer’s instructions. Approximately 20 μg of total RNA for each sample was separated by agarose gel electrophoresis and blotted as previously described (Sambrook et al. 1989). Agarose gel purified DNA fragments representing the A. flavus aflR, aflD (nor-1), aflM (ver-1), and aflP (omtA) gene coding regions were used to generate [α-32P]dCTP labeled probes using the RediPrime II DNA labeling system (Amersham Biosciences). The primers used to PCR amplify those fragments from A. flavus genomic DNA to generate these probes are listed in Table 1 and were based on the A. flavus AF gene cluster sequence (GenBank accession no. AY510453). The primers used to PCR fragments corresponding to the aflatrem genes atmC, atmG, and atmG are also listed in Table 1 and were based on the A. flavus aflatrem cluster sequence (GenBank accession no. AY559849).
Results
veA regulates morphogenesis in A. flavus
Evaluation of the ΔveA morphological phenotype revealed a slight increase in vegetative growth (estimated as colony growth) compared with the control strains, particularly in the dark (Fig. 3a, data not shown). Cultures exposed to light showed higher variation among control strains (approximately ±14 mm).
In addition, all the A. flavus ΔveA mutants were unable to form sclerotia under conditions where the control strains abundantly produced these resistant structures (Figs. 3b and 4). Although conidiation varied among the transformants analyzed, a general decrease in aerial mycelia was observed in all the ΔveA mutants compared with the control strains (Fig. 4 and data not shown).
veA controls AF production in A. flavus at transcriptional level
We examined AF production in both ΔveA and control A. flavus strains in a time course experiment in which the mycelia were shifted to solid medium after growing in liquid culture for 14 h (Fig. 5a). Our results indicate that ΔveA was unable to produce either AFB1 or AFB2, whereas both aflatoxins were clearly detected in the veA control at 12, 24, and 36 h after the shift to solid medium.
TLC and Northern hybridization of ΔveA mutant and control strain for AF production and AF gene expression, respectively. a TLC analysis of AF production in the A. flavus ΔveA mutant and control strain. Metabolites were extracted from cultures grown on YGT agar medium in the light and harvested at 2, 12, 24, and 36 h. Abbreviation: Std., AF standard. The experiment was repeated, with similar results. Results were also confirmed by HPLC (data not shown). b Northern hybridization of total RNA from the ΔveA mutant and control strain using 32P-labeled AF gene probes. The ethidium-bromide-stained 16S rRNA region of each gel is shown to indicate RNA loading
The effect of veA deletion on AF biosynthesis was further analyzed by Northern analysis (Fig. 5b). In the control strain, transcripts of the AF-specific regulatory transcription factor gene aflR (Woloshuk et al. 1995; Yabe and Nakajima 2004) were detected 2 h after the shift, and transcripts corresponding to the enzymatic genes aflD, aflM, and aflP (Yabe and Nakajima 2004; Yu et al. 2004) were detected mainly at 2 and 12 h after the shift, indicating cluster activation. At 24 h, the accumulation of these transcripts decreased, and at 36 h, they were not detected. In the ΔveA, aflR, aflD, aflM, and aflP transcripts were not detected at any time point analyzed. These results were confirmed by quantitative reverse transciptase (qRT)-PCR (data not shown). The experiment was repeated twice, obtaining similar results.
CPA production is controlled by veA
Our TLC studies (Fig. 5a) indicated that the biosynthesis of additional metabolites is also affected by the veA deletion. For this reason, we explored the possible role of veA as a global regulator controlling other mycotoxins produced by A. flavus besides AF. Our CPA HPLC analysis revealed that the production of this A. flavus mycotoxin is also veA-dependent (Fig. 6). The ΔveA mutant presented a reduction in CPA production of 48% in the light and 66% in dark culture conditions with respect to the control strain. An additional compound was consistently detected eluting at 3.1 min (Fig. 6b). Remarkably, this unknown compound was more abundant in the ΔveA strain than in the control strain (approximately fivefold higher). The results were consistent in all replicates.
Effects of the veA mutation on CPA production in A flavus. a HPLC quantification of CPA in the ΔveA mutant and control samples obtained from cultures grown in YGT liquid medium in light or dark conditions for 7 days. The experiment was carried out with five replicates and repeated twice, with similar results. b Overlaid chromatograms showing the production of CPA in one of the samples analyzed from the A.flavus ΔveA mutant and control strain. Total time corresponding to the void volume at 1 ml min−1 flow rate was 1 min. The CPA standard elution time coincided with the peaks indicated by arrows in all the replicates
veA is necessary for aflatrem production in A. flavus
Results from our HPLC analysis indicate that the veA deletion completely prevented the synthesis of aflatrem under conditions that allowed production of this mycotoxin in the A. flavus control strain (Fig. 7). The amount of aflatrem in the control strain was higher in the light than in the cultures in the dark (approximately 3.6-fold higher) in all the samples analyzed. Besides the peak corresponding to aflatrem (the elution time coinciding with pure aflatrem standard elution time), we detected other peaks corresponding to unknown compounds of lower abundance. Some of those compounds were only observed in the control strain (Fig. 7b). Other compounds were detected at higher levels in the ΔveA mutant.
Effects of the veA mutation on aflatrem production in A flavus. a HPLC quantification of aflatrem in ΔveA and control cultures grown in aflatrem-inducing medium (Zhang et al. 2004). The experiment was carried out with three replicates and repeated twice, with similar results. b Overlaid chromatograms showing the aflatrem analysis results of one sample obtained from the A. flavus ΔveA mutant and control strain. Total time corresponding to the void volume at 1 ml min−1 flow rate was 1.7 min. The aflatrem standard elution time coincided with the peaks indicated by arrows in all the replicates
veA is a positive regulator for the activation of the aflatrem gene cluster
The discovery of the aflatrem gene cluster (Zhang et al. 2004) allowed us to further study the possible role of veA in the transcriptional regulation of enzymatic genes responsible for the synthesis of this tremorgenic mycotoxin. We examined the expression of atmC, atmG, and atmM in a time course experiment (Fig. 8). Transcripts of these three genes were detected in the control strain but not in the ΔveA mutant. Our Northern analysis indicated that accumulation of atmC, atmG, and atmM transcripts in the control strain occurred earlier in the light than in the dark (96 and 120 h, respectively, after culture shift to aflatrem producing medium) (Fig. 8). The experiment was repeated twice, obtaining similar results.
Effect of veA mutation on the transcription of the aflatrem gene cluster. Northern blots were hybridized with atmC, atmM, and atmG probes. Approximately 20 μg of total RNA from the ΔveA mutant and control samples was used for the RNA blot analysis. RNAs were extracted from mycelial samples during a time course experiment (72-, 96-, 120-, and 144-h time points). The ethidium-bromide-stained 16S rRNA region of each gel is shown to indicate RNA loading
Discussion
Effect of the veA mutation in A. flavus development and aflatoxin production, and comparison with previous findings
Previous studies support that mycotoxin production in Aspergillus is associated with morphogenesis (Calvo et al. 2002). Specifically, AF/ST production in Aspergillus spp. has been closely linked to asexual development (conidiation). Most studies of genetic regulation of fungal development have been performed in the model system A. nidulans and have focused on conidiation (Adams et al. 1998). A. nidulans also develops resistant structures called cleistothecia where sexual spores (ascospores) are produced. A. flavus and A. parasiticus form resistant structures called sclerotia, which are similar to cleistothecia but are unable to form spores. One role of cleistothecia and sclerotia is to survive environmental extremes (Coley-Smith and Cooke 1971; Malloch and Cain 1972; Wicklow 1987). Interestingly, evidence presented by Geiser et al. (1996) indicates that asexual Aspergilli are often derived from meiotic lineages and that sclerotia might be vestigial cleistothecia that lost spore-producing capacity. We hypothesize that conserved signaling pathways could control both cleistothecial and sclerotial development. Molecular studies on regulation of fungal sexual development or formation of resistant structures are still limited, and only a few genes involved have been identified, mainly in A. nidulans. One of these genes is called velvet, or veA. In previous studies in A. nidulans, we showed that veA not only regulates morphogenesis but also is necessary for the production of ST (Kato et al. 2003), the penultimate precursor in the AF biosynthetic pathway. We also found a veA homolog in A. parasiticus and generated a veA deletion mutant unable to produce AF intermediates (Calvo et al. 2004) or sclerotia. Our TLC analysis indicated that A. nidulans and A. parasiticus mutants present a different profile compared with the wild-type controls with respect to other metabolites besides ST and AF intermediates, suggesting that the veA gene could have a broader effect, perhaps over additional metabolic pathways. We found further evidence analyzing penicillin (PN) production in A. nidulans (Kato et al. 2003), where the deletion of veA resulted in alteration of the PN gene expression and concomitant reduction in PN production. In the current study, we investigated the possibility that additional metabolites, specifically mycotoxins, are also regulated by veA in A. flavus, the predominant Aspergillus species responsible for AF contamination of crops in the USA and worldwide. In addition, we tested if the requirement for veA in AF production and normal morphogenesis observed in A. parasiticus also holds true in A. flavus.
The morphological analysis of the A. flavus ΔveA mutant revealed a slight increase in vegetative growth compared with the control strain, mainly in the dark (Fig. 3). The results obtained from cultures in the dark differ from those observed in the A. parasiticus and A. nidulans ΔveA mutants (Calvo et al. 2004; Kato et al. 2003), where deletion of veA caused a reduction in colony growth, particularly in the A. parasiticus ΔveA mutant (only a slight reduction was observed in the A. nidulans mutant).
A reduction in conidiation was previously shown in veA deletion mutants in A. parasiticus (Calvo et al. 2004) and in A. fumigatus (Krappmann et al. 2005). In contrast, an increase in conidiation that was medium-dependent was observed in the A. nidulans veA mutant (Kato et al. 2003; Kim et al. 2002). In addition, differences in conidiation levels among A. parasiticus and A. flavus have commonly been observed even in natural isolates, e.g., between L and S A. flavus strains (unpublished observations). The differences in genetic background that determine the variation in conidiation levels in these isolates are unknown. Our results showed variation in conidiation, particularly among the control transformants. However, all the control strains presented more aerial mycelia than the veA mutants (Fig. 4, data not shown), suggesting that deletion of veA has an effect on vegetative growth in A. flavus.
At this time, the veA mode of action is unknown and the gene product does not present homology with any known protein to help in predicting its functionality. Further research on the veA mechanism is needed to understand how veA regulates fungal growth and development. Despite some differences in the role of veA in growth in different Aspergillus species, the effect of the veA mutation in A. flavus on sclerotial production was consistent among the transformants, with previous findings in A. parasiticus (Calvo et al. 2004), and with the blockage of cleistothecial production in A. nidulans (Kato et al. 2003). The mutation in A. flavus veA completely prevented the formation of sclerotia in all five deletion mutants under conditions that allowed the production of these structures in all control strains tested, indicating that veA is a necessary gene for the production of these resistant structures (Figs. 3b and 4). This is relevant since the role of sclerotia is to survive environmental extremes, even during long periods (Coley-Smith and Cooke 1971; Malloch and Cain 1972; Wicklow 1987). When the conditions are again favorable, fungal sclerotia give rise to new mycelia and the infection process is reestablished. These results also strengthen the hypothesis that sclerotia and cleistothecia might have a common origin (Geiser et al. 1996). In addition, in A. flavus, wild-type sclerotia were produced in both light and dark, whereas in A. parasiticus and in A. nidulans, sclerotial/cleistothecial production takes place mainly in the dark (Calvo et al. 2004; Kato et al. 2003; Yager 1992), suggesting that the light-dependent regulation of the formation of these structures in A. flavus, A. parasiticus, and A. nidulans is not identical.
Besides regulating morphological differentiation, veA also regulates secondary metabolism in A. flavus. Our TLC results demonstrated that in A. flavus, the deletion of veA eliminated the production of AF, whereas AFB1 and AFB2 were present in strains with the veA wild-type allele (Fig. 5a). This result was confirmed in all the transformants and also by HPLC analysis (data not shown). Furthermore, the expression of aflR (encoding a specific transcription factor that simultaneously activates the AF gene cluster) and AF biosynthetic genes aflD, aflM, and aflP was not detected in the ΔveA mutant by Northern hybridization analysis (Fig. 5b). The blockage of AF biosynthesis in the A. flavus ΔveA mutant is consistent with previous finding on AF regulation in A. parasiticus (Calvo et al. 2004) and ST production in A. nidulans (Kato et al. 2003), suggesting that the mechanism through which veA regulates AF/ST production is conserved among Aspergillus spp. Furthermore, based on our results that the veA mutation causes alteration in morphogenesis as well as in AF production, it is possible that the VeA protein could be part of a signaling pathway controlling both processes. Further studies on this matter are under way.
A. flavus veA controls CPA and aflatrem biosynthesis
We also examined if veA is involved in regulating the production of other toxins in A. flavus, specifically CPA and aflatrem. CPA is a specific inhibitor of calcium-dependent ATPase and induces alterations in ion transport across cell membranes (Riley and Goeger 1992). The results of our chemical analysis showed that the ΔveA mutation results in a decrease in CPA production in either light or dark conditions (Fig. 6). In this analysis, we also detected a metabolite that was produced in much higher levels in ΔveA compared with the control. These results indicate that veA not only can act as a positive regulator in the biosynthesis of a variety of metabolites in A. flavus, but it could also have a role as a negative regulator in the synthesis of other compounds. It is also possible that some of the precursors utilized in the metabolic routes blocked by the veA mutation accumulate or are available in higher abundance for other routes, thus enhancing the production of other metabolites.
Another mycotoxin produced by A. flavus is the potent tremorgen aflatrem (Zhang et al. 2004). Our HPLC studies revealed that deletion of veA results in a complete blockage of aflatrem production, indicating that veA is also required for the biosynthesis of this mycotoxin (Fig. 7). In addition to the peak corresponding to aflatrem, we also observed a number of other peaks in extracts of both strains that correspond to other metabolites. These metabolites are found in differing abundance in the ΔveA mutant and in the control strain. In some cases, their levels are higher in the control strain with respect to the ΔveA mutant or vice versa, supporting the hypothesis that veA has a differential effect on the production on a variety of A. flavus metabolites.
veA regulates aflatrem production at the transcriptional level
The enzymatic genes necessary for aflatrem production have been previously described (Zhang et al. 2004). The aflatrem genes atmC, atmG, and atmM are clustered. Until this study, no regulatory gene(s) controlling the aflatrem cluster activation had been described. Our time course Northern analysis revealed that veA is necessary for the activation of the aflatrem biosynthetic genes atmC, atmG, and atmM (Fig. 8). We also noticed that illumination had an effect, inducing earlier expression of the aflatrem genes (Fig. 8) and, consequently, higher aflatrem production in light cultures (Fig. 7). Further research is needed to elucidate the influence of additional environmental factors on aflatrem gene expression and subsequent production of this potent tremorgenic compound.
This study represents the first report of a gene that regulates the biosynthesis of CPA and aflatrem mycotoxins. The fact that veA regulates CPA and aflatrem biosynthesis in addition to AF suggests the involvement of veA in a global regulatory mechanism controlling secondary metabolism in A. flavus. Our previous findings of veA-mediated regulation of both ST and penicillin production in A. nidulans further support this hypothesis (Kato et al. 2003). Another global regulator gene, laeA, has been described to control the expression of several gene clusters involved in secondary metabolism in Aspergillus spp. (Bok and Keller 2004; Bok et al. 2006; Keller et al. 2005). LaeA is postulated to be involved in the conversion of heterochromatin to euchromatin (Bok and Keller 2004; Keller et al. 2005). We are currently studying the possible connection between veA and laeA. The fact that veA controls the production of AF, CPA, and aflatrem adds to the importance of veA as a potential target to control Aspergillus mycotoxin contamination. In addition, inhibiting the activity of veA could also affect A. flavus survivability since veA is required for sclerotial formation, thus making veA a desirable candidate to target in control strategies.
References
Adams TH, Wieser JK, Yu JH (1998) Asexual sporulation in Aspergillus nidulans. Microbiol Mol Biol Rev 62:35–54
Blout WP (1961) Turkey disease. Turkeys 9:52, 55–58, 61, 77
Bok JW, Keller NP (2004) LaeA, a regulator of secondary metabolism I Aspergillus. Eukaryot Cell 3:527–535
Bok JW, Hossmeister D, Maggio-Hall LA, Murillo R, Glasner JD, Keller NP (2006) Genomic mining for Aspergillus natural products. Chem Biol 1:31–37
Calvo AM, Wilson RA, Bok JW, Keller NP (2002) Relationship between secondary metabolism and fungal development. Microbiol Mol Biol Rev 66:447–459
Calvo AM, Bok J, Brooks W, Keller NP (2004) veA is required for toxin and sclerotial production in Aspergillus parasiticus. Appl Environ Microbiol 70:4733–4739
Cary JW, Ehrlich KC, Bland JM, Montalbano BG (2005) The aflatoxin biosynthesis cluster gene, aflX, encodes an oxidoreductase involved in conversion of vesicolorin A to demethylsterigmatocystin. Appl Environ Microbiol 72:1096–1101
Cleveland TE, Bhatnagar D, Foell CJ, McCormick SP (1987) Conversion of a new metabolite to aflatoxin B2 by Aspergillus parasiticus. Appl Environ Microbiol 53:2804–2807
Cole RJ, Hill RA, Blankenship PD, Sanders TH (1986) Color mutants of Aspergillus flavus and Aspergillus parasiticus in a study of preharvest invasion of peanuts. Appl Environ Microbiol 52:1128–1131
Coley-Smith JR, Cooke RC (1971) Survival and germination of fungal sclerotia. Annu Rev Phytopathol 9:65–92
Denissenko MF, Cahill J, Koudriakova TB, Gerber N, Pfeifer GP (1999) Quantitation and mapping of aflatoxin i DNA damage in genomic DNA using aflatoxin B1-8,9-epoxide and microsomal activation systems. Mutat Res 425:205–211
Dorner JW, Cole RJ, Lomax LG, Gosser HS, Diener UL (1983) Cyclopiazonic acid production by Aspergillus flavus and its effects on broiler chickens. Appl Environ Microbiol 46:698–703
Dvorackova I, Kusak V (1990) Hepatocellular carcinoma (a 28-year necropsy review). J Environ Pathol Toxicol Oncol 10:220–224
Geiser DM, Timberlake WE, Arnold ML (1996) Loss of meiosis in Aspergillus. Mol Biol Evol 13:809–817
Goldblatt L (1969) Aflatoxin: scientific background, control, and implications. Academic, New York, NY
Horn BW, Dorner JW (1999) Regional differences in production of aflatoxin B1 and cyclopiazonic acid by soil isolates of Aspergillus flavus along a transect within the United States. Appl Environ Microbiol 4:1444–1449
Horng JS, Chang PK, Pestka JJ, Linz JE (1990) Development of a homologous transformation system for Aspergillus parasiticus with the gene encoding nitrate reductase. Mol Gen Genet 224:294–296
Kato N, Brooks W, Calvo AM (2003) The expression of sterigmatocystin and penicillin genes in Aspergillus nidulans is controlled by veA, a gene required for sexual development. Eukaryot Cell 2:1178–1186
Keller NP, Turner G, Bennet JW (2005) Fungal secondary metabolism—from biochemistry to genomics. Nat Rev Microbiol 12:937–947
Kim H, Han K, Kim K, Han D, Jahng K, Chae K (2002) The veA gene activates sexual development in Aspergillus nidulans. Fungal Genet Biol 37:72–80
Krappmann S, Bayram O, Braus GH (2005) Deletion and allelic exchange of Aspergillus fumigatus veA locus via a novel recyclable marker module. Eukaryot cell 4:1298–1307
Lansden JA, Davidson JI (1983) Occurrence of cyclopiazonic acid in peanuts. Appl Environ Microbiol 45:766–769
Lasky T, Magder L (1997) Hepatocellular carcinoma p53 G>T transversions at codon 249: the fingerprint of aflatoxin exposure? Environ Health Perspect 105:392–397
Malloch D, Cain RF (1972) The Trichocomataceae: ascomycetes with Aspergillus, Paecilomyces and Penicillium imperfect states. Can J Bot 50:2613–2628
McMillan LK, Carr RL, Young CA, Astin JW, Lowe RGT, Parker EJ, Jameson GB, Finch SC, Miles CO, McManus OB, Chmalhofer WA, Garcia L, Kaczorowski GJ, Goetz MA, Tkacz JS, Scott B (2003) Molecular analysis of two cytochrome P450 monooxygenase genes required for paxilline biosynthesis in Penicillium paxilli and effects of paxilline intermediates on mammalian maxi-K ion channels. Mol Genet Genomics 270:9–23
Norred WP, Porter JK, Doner JW, Cole RJ (1988) Occurrence of the mycotoxin cyclopiazonic acid in meat after oral administration to chickens. J Agric Food Chem 36:113–116
Payne GA, Brown MP (1998) Genetics and physiology of aflatoxin biosynthesis. Annu Rev Phytopathol 36:329–362
Pitt JI, Cruickshank RH, Leistner L (1986) Penicillium commune, P. camembertii, the origin of white cheese mould, and the production of cyclopiazonic acid. Food Microbiol 3:363–371
Porter JK, Norred WP, Cole RJ, Dorner JW (1988) Neurochemical effects of cyclopiazonic acid in chickens. Proc Soc Exp Biol Med 187:335–340
Purchase IFH (1971) The acute toxicity of the mycotoxin cyclopiazonic acid to rats. Toxicol Appl Pharmacol 18:114–123
Richard JL, Payne GA (2003) Mycotoxins: risks in plant, animal and human health. CAST Report 139. Council for Agricultural Science and Technology, p 199
Riley RT, Goeger DE (1992) Cyclopiazonic acid: speculations on its function in fungi. In: Bhatnagar D, Lillehoj EB, Arora DK (eds) Handbook of applied mycology. Mycotoxins in ecological systems. Marcel Dekker, New York, NY, pp 5:385–402
Riley J, Mandel HG, Sinha S, Judah DJ, Neal GE (1997) In vitro activation of the human Harvey-ras proto-oncogene by aflatoxin B1. Carcinogenesis 18:905–910
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
Shen HM, Ong CN (1996) Mutations of the p53 tumor suppressor gene and ras oncogenes in aflatoxin hepatocarcinogenesis. Mutat Res 366:23–44
Sobolev VS, Horn BW, Dorner JW, Cole RJ (1998) Liquid chromatographic determination of major secondary metabolites produced by Aspergillus species from section Flavi. J AOAC Int 81:57–60
Sweeney MJ, Dobson AD (1999) Molecular biology of mycotoxin biosynthesis. FEMS Microbiol Lett 175:149–163
Trail F, Mahanti N, Linz JE (1995) Molecular biology of aflatoxin biosynthesis. Microbiology 141:755–765
Urano T, Trucksess MW, Beaver RW, Wilson DM, Dorner JW, Dowell FE (1992) Co-occurrence of cyclopiazonic acid and aflatoxins in corn and peanuts. J AOAC Int 75:838–841
Valdes JJ, Cameron JE, Cole RJ (1985) Aflatrem: a tremorgenic mycotoxin with acute neurotoxic effects. Environ Health Perspect 62:459–463
Wang JS, Groopman JD (1999) DNA damage by mycotoxins. Mutat Res 424:167–181
Wicklow DT (1987) Survival of Aspergillus flavus sclerotia in soil. Trans Br Mycol Soc 89:131–134
Wogan GN, Househam KC, Hundt HK (1992) Aflatoxins as risk factors for hepatocellular carcinoma in humans. Aflatoxin exposure and its relationship to kwashiorkor in African children. Cancer Res 52:2114–2118
Woloshuk CP, Yousibova GL, Rollins JA, Bhatnagar D, Payne GA (1995) Molecular characterization of the afl-1 locus in Aspergillus flavus. Appl Environ Microbiol 61:3019–3023
Wu F (2004) Mycotoxin risk assessment for the purpose of setting international regulatory standards. Environ Sci Technol 38:4049–4055
Yabe K, Nakajima H (2004) Enzyme reactions and genes in aflatoxin biosynthesis. Appl Microbiol Biotechnol 64:745–755
Yager LN (1992) Early developmental events during sexual and asexual sporulation in Aspergillus nidulans. Biotechnology 23:19–41
Yao I, Peter AB, Baur R, Sigel E (1988) The tremorigen aflatrem is a positive allosteric modulator of γ-aminobutyric acid A receptor channel expressed in Xenopus oocytes. Mol Pharmacol 35:319–323
Yu J, Chang PK, Ehrlich KC, Cary JW, Bhatnagar D, Cleveland TE, Payne GA, Linz JE, Woloshuk CP, Bennett JW (2004) Clustered pathways genes in aflatoxin biosynthesis. Appl Environ Microbiol 70:1253–1262
Zhang S, Monahan JB, Tkacz JS, Berry S (2004) Indole diterpene gene cluster from Aspergillus flavus. Appl Environ Microbiol 70:6875–6883
Acknowledgements
A.M.C. and J.W.C. thank Jan Tkacz and Mike Goetz from Merck Co. for providing the aflatrem standard. A.M.C. gratefully acknowledges John Mitchell for his suggestions for the chemical analysis and Barry Scott for his useful comments on aflatrem-inducing cultures. J.W.C. gratefully acknowledges Pam Harris and Jolie Bonanno for their technical support.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Duran, R.M., Cary, J.W. & Calvo, A.M. Production of cyclopiazonic acid, aflatrem, and aflatoxin by Aspergillus flavus is regulated by veA, a gene necessary for sclerotial formation. Appl Microbiol Biotechnol 73, 1158–1168 (2007). https://doi.org/10.1007/s00253-006-0581-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-006-0581-5