Abstract
Streptomyces is an interesting host for the secretory production of recombinant proteins because of its innate capacity to secrete proteins at high level in the culture medium. In this report, we evaluated the importance of the phage-shock protein A (PspA) homologue on the protein secretion yield in Streptomyces lividans. The PspA protein is supposed to play a role in the maintenance of the proton motive force (PMF). As the PMF is an energy source for both Sec- and Tat-dependent secretion, we evaluated the influence of the PspA protein on both pathways by modulating the pspA expression. Results indicated that pspA overexpression can improve the Tat-dependent protein secretion as illustrated for the Tat-dependent xylanase C and enhanced green fluorescent protein (EGFP). The effect on Sec-dependent secretion was less pronounced and appeared to be protein dependent as evidenced by the increase in subtilisin inhibitor (Sti-1) secretion but the lack of increase in human tumour necrosis factor (hTNFα) secretion in a pspA-overexpressing strain.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Streptomyces lividans has already been shown to be a valuable host for the secretion of heterologous proteins, of both eukaryotic and prokaryotic origin (Hong et al. 2003; Lara et al. 2004; Pozidis et al. 2001; Sianidis et al. 2006). In most cases where Streptomyces was used as host for the secretion of heterologous proteins, the Sec system has been used. Nevertheless, the recently detected Tat pathway (reviewed in Berks et al. 2003) can also be used for the translocation of heterologous proteins across the cytoplasmic membrane of Streptomyces (Schaerlaekens et al. 2004). The latter pathway has some pronounced differences compared to the Sec pathway. The Sec machinery transports proteins in an unfolded state across the cytoplasmic membrane, requiring both ATP hydrolysis and the proton motive force (PMF) for energy (Economou 1999). On the other hand, the Tat pathway transports proteins that are at least partially folded before export, and this translocation route is solely dependent on the PMF (DeLisa et al. 2003). As the PMF plays a role in both secretion pathways, an S. lividans strain with an improved PMF maintenance could be advantageous for the secretion of recombinant proteins. However, little is known about the way in which the PMF is maintained and regulated in Gram-positive bacteria. In Escherichia coli, the phage-shock-protein (psp) response is presumed to play a role in PMF maintenance (reviewed in Darwin 2005).
This psp response is activated by several forms of extracytoplasmic stress that compromise the energy status of the cell. These include, but are not limited to, filamentous phage infection, high osmolarity and elevated ethanol concentrations, alkaline shock and the presence of proton ionophores such as carbonylcyanide m-chlorophenylhydrazone (CCCP) (Model et al. 1997). The complete psp system in E. coli is comprised of the pspABCD operon and its transcriptional regulator, pspF. In this system, PspA is thought to be the main effector protein, as overexpression of pspA alone can alleviate the effects caused by deletion of both pspB and pspC (Kleerebezem et al. 1996). With respect to the production of heterologous proteins, it is important to note that pspA overexpression alone was sufficient to increase almost threefold Tat-dependent secretion of the SufI, CueO and ssTorA-GFP proteins in E. coli (DeLisa et al. 2004). So far, it has been thought that homologues of the PspA protein in bacteria that did not have the complete psp system in their chromosome, such as S. lividans, were unlikely to be a part of a real psp response (Darwin 2005).
In this work, we tested the impact of the S. lividans pspA homologue on protein secretion in this strain. To evaluate the effect of pspA overexpression on protein translocation, secretion of both native and heterologous proteins routed via both the Sec and the Tat pathway were quantified in S. lividans TK24 and the pspA-overexpressing strain.
Materials and methods
Strains, media and growth conditions
E. coli strain TG1 was used as host for cloning purposes while E. coli S17-1 (Simon et al. 1983) was used for conjugation of DNA from E. coli to Streptomyces. Cultures were grown at 37 °C (300 rpm) in Luria Bertani broth, supplemented with the appropriate antibiotics. S. lividans TK24 and its derivatives were precultured in 5 ml phage medium (Korn et al. 1978) supplemented with thiostrepton (10 μg/ml) or apramycin (50 μg/ml), if necessary, and grown at 27 °C with continuous shaking at 300 rpm for 48 h. After homogenization of the mycelium, the strains were inoculated in NM medium (Van Mellaert et al. 1994). For solid medium, MRYE (Anné et al. 1990) was used supplemented with thiostrepton (50 μg/ml) or apramycin (50 μg/ml), if applicable. Protoplast formation and subsequent transformation of S. lividans as well as conjugation from E. coli to S. lividans were carried out as described by Kieser et al. (2000). To test the distinct activities, precultures of S. lividans transformants grown in phage medium for 48 h were used to inoculate 50 ml NM medium and cultures were subsequently cultivated for 24–48 h.
DNA manipulations and vector constructions
For all DNA manipulations, standard techniques were followed (Sambrook et al. 1989; Kieser et al. 2000). Restriction endonucleases and DNA modifying enzymes were from Invitrogen and Roche Diagnostics. Plasmids and oligonucleotides used in this work are listed in Table 1.
To overproduce PspA in S. lividans, the pspA gene was cloned under control of the Streptomyces venezuelae subtilisin inhibitor (vsi) promoter in the Streptomyces vector pIJ486 (Ward et al. 1986). Therefore, the pspA gene was amplified by polymerase chain reaction (PCR) using the primers PspAF and PspAR. After cloning of the acquired PCR fragment in pGEM-T Easy (Promega), DNA sequences were verified, and the resulting pGEMPspA plasmid was digested with HindIII and EcoRI to allow insertion downstream of the vsi promoter into a HindIII/EcoRI-digested pBSVsi plasmid (Lammertyn 2000), resulting in pBSVsiPspA. This vector was digested with XbaI and HindIII, and the resulting pspA-expression cassette was ligated into the XbaI/HindIII-restricted pIJ486 vector, resulting in the Streptomyces vector pIJ486VPspA.
For the expression and secretion of hTNFα, XlnC and enhanced green fluorescent protein (EGFP), three pSSV05 (unpublished) derivatives were constructed. A cassette containing the cDNA of the mature hTNFα, fused in frame to the vsi signal sequence, was excised from the V-TNF plasmid (Schaerlaekens et al. 2004) as an XbaI/HindIII fragment and ligated into XbaI/HindIII-digested pSSV05 resulting in pSSV-TNF. Similarly, a cassette containing the S. lividans xlnC gene under control of the vsi promoter, was excised from pBSvsixyl (Schaerlaekens et al. 2004) as an XbaI/HindIII fragment and ligated into XbaI/HindIII-digested pSSV05 resulting in pSSV-XlnC.
For the secretory production of EGFP, the egfp gene was amplified by PCR using plasmid pIJ8668 (Sun et al. 1999) as template with the primers EGFPFor and EGFP-Rev. Upon cloning the PCR fragment in pGEM-T Easy, the resulting vector was first digested with PstI, then treated with T4 DNA polymerase to remove the 3-protruding ends, and finally digested with EcoRI. Subsequently, the egfp gene was placed under control of the vsi promoter and the xlnC signal sequence by cloning the obtained restriction fragment in NsiI/T4 DNA polymerase/EcoRI-treated pBSVX (Schaerlaekens et al. 2004). The resulting pBSVX-EGFP plasmid was digested with XbaI and EcoRI, and the resulting fragment was cloned into XbaI/EcoRI digested pSSV05, resulting in pSSVX-EGFP.
Transformation of S. lividans
To express EGFP, hTNFα and XlnC in the S. lividans TK24 wild-type and pspA-overexpressing strain, the pSSVX-EGFP, pSSV-TNF and pSSV-XlnC plasmids were, respectively, conjugated from E. coli S17-1 cells to spore suspensions of both S. lividans strains. Sti-1, which is encoded on the chromosome, was not cloned on a multicopy plasmid, as satisfactory amounts of protein could be detected when encoded from the chromosome.
Activity assays
Xylanase activity was assayed using the dinitrosalicylic acid assay as described previously (De Keersmaeker et al. 2005). Briefly, after 24 h of growth, cultures of S. lividans were centrifuged (10 min, 4,000×g, 4 °C); the obtained supernatants were diluted in the assay buffer, and the amount of reducing sugar was quantified. One unit of xylanase was defined as the amount of enzyme that produces 1 mg reducing sugar in 10 min at 60 °C from a saturated xylan solution. Values were expressed as units per mg mycelial dry weight to correct for differences in growth rate between the different S. lividans strains.
EGFP activity was assayed by measuring fluorescence of 200 μl spent medium using a Fluoroskan Ascent FL fluorophotometer (Labsystems), with the excitation filter set to 485 nm and the emission filter to 520 nm.
Subtilisin inhibitor activity was determined in the presence of subtilisin BPN’ and the substrate N-succinyl-l-Ala-l-Ala-l-Pro-l-Phe-p-nitroanilide as described by Kojima et al. (1990). Extracellular fractions of 24- or 48-h cultures were diluted in assay buffer, and the percentage of subtilisin activity was measured. One unit was defined as the amount of enzyme that inhibited 5 μg subtilisin during 10 min incubation at 25 °C.
ELISA assays
The amount of hTNFα secreted in the extracellular medium of the different S. lividans cultures was determined by enzyme linked immunosorbent assay (ELISA) (DiaMed EuroGen, Turnhout, Belgium) according to the manufacturer’s recommendations.
SDS-PAGE and Western blot analysis
To check accumulation of XlnC, and EGFP in the extracellular medium in S. lividans, Western blot analysis was performed. Proteins present in the supernatant corresponding to cultures with the same dry weight were TCA-precipitated and were subsequently separated by sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) and visualized by Western blotting and immunodetection with specific antibodies in combination with a suited alkaline phosphatase-conjugated goat anti-rabbit antibody (Sigma) and the chromogenic substrate solution NBT/BCIP (Roche Diagnostics).
Results
S. lividans pspA gene
PspA homologues are widespread in bacteria and higher organisms. In the Streptomyces coelicolor genome bank (http://www.sanger.ac.uk), we found a pspA homologue (previously SCO2168) which forms an operon with SCO2167, a gene with no homology to any known protein. With primers based on the S. coelicolor sequence, we were able to amplify the corresponding gene from S. lividans TK24. pspA was isolated as a 780-bp gene. The S. lividans and S. coelicolor pspA genes show 99.3% identity at the nucleotide level and 98.1% identity at the amino acid level, respectively.
The CLUSTALW identity and similarity scores with the E. coli pspA are 23 and 39%, respectively, but more importantly, no homologues of the pspB, pspC, pspD or pspG genes could be identified. Of these genes, pspA, B and C are, so far, assumed to be needed for a functional, minimal psp response (Darwin 2005).
The influence of pspA overexpression on the total amount of secreted protein
To overexpress pspA, S. lividans TK24 was transformed with the multicopy plasmid pIJ486VPspA, containing the pspA gene cloned behind the vsi promoter region. To compare the total amount of secreted protein, both wild-type and the pspA-overexpressing strain were grown in 50 ml minimal medium NMMP. After 24 h of growth, the amount of protein present in the culture supernatant was determined by the Bradford assay (Bio-Rad) using bovine serum albumin as the standard. The spent medium of the wild-type strain contained 27.6±0.9 μg of protein (mg dry wt)−1, whereas the pspA-overexpressing strain contained 36.1±0.8 μg of protein (mg dry wt)−1. This represents a significant increase in the total amount of secreted protein of 32%.
Furthermore, the pspA-overexpressing strain showed a similar growth curve when grown in the NM medium that was used in further secretion tests (shown in Fig. 1), indicating that the obtained results are not due to a growth difference between both strains.
Influence of pspA overexpression on Tat-dependent secretion
In a next step, we wanted to assess the specific effect of pspA overexpression on Tat-dependent secretion. To achieve this, the pspA-overexpressing and the wild-type strain were transformed with the plasmid pSSV-XlnC or pSSVX-EGFP containing, respectively, the native xylanase C gene and the “enhanced green fluorescent protein” gene under the control of the vsi promoter. Enzymatically active xylanase C can only be obtained after secretion via the Tat pathway (Faury et al. 2004), and the same applies for fluorescent, secreted EGFP (unpublished data).
Figure 2 shows the effect of the pspA coexpression on the xylanase activity, measured using the dinitrosalicylic acid assay, of S. lividans overexpressing xlnC over a period of 42 h of growth. When grown in the absence of xylan, as is the case here, the wild-type S. lividans does not produce detectable levels of the native XlnA, B and C proteins (Mondou et al. 1986; Vats-Mehta et al. 1990). Similarly, no xylanase activity could be detected in the S. lividans [pIJ486PspA] strain, meaning that the native xlnA, B and C genes were not responsible for the observed xylanase activity in the xlnC-overexpression strain. In contrast, in the XlnC-overproducing strain, the amount of XlnC measured in the supernatant was 4.8 U. When pspA was concomitantly overexpressed, values up to 18.5 units xylanase (mg dry wt)−1 could be obtained, an almost fourfold increase in xylanase activity when compared to the XlnC-overproducing strain.
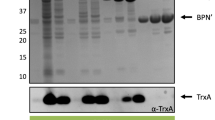
a Effect of pspA coexpression on the xylanase activity found in spent medium of S. lividans strains expressing the xlnC gene and grown for 18, 24 and 42 h: S. lividans [pIJ486PspA+pSSV-XlnC] (light gray bars), S. lividans [pSSV-XlnC] (dark gray bars), S. lividans [pIJ486PspA] (dark bars). b Western blotting with anti-XlnC antibodies on the extracellular fraction of S. lividans TK24 [pSSV-XlnC] (lane 2) and S. lividans TK24 [pIJ486PspA+pSSV-XlnC](lane 3). Cultures were grown for 24 h (left blot) or 42 h (right blot) in 50 ml NM medium. In each lane, the amount of supernatant corresponding to 4 mg of dry weight was loaded. Lane 1 shows a molecular weight marker
In parallel, the effect of PspA overproduction on heterologous protein secretion via the Tat pathway was evaluated. Figure 3 shows that in the PspA-overproducing strain, secretion of active EGFP was significantly enhanced after 24 h of growth. The relative medium fluorescence increased from 4.13 to 15.89 RFU (mg dry wt)−1, an almost fourfold increase. This corresponds with an increase in active EGFP yield of 6.9 to 20 mg EGFP/l. From the data above, it is clear that overproduction of the S. lividans PspA protein has a dramatically positive effect on the secretion of the tested Tat-dependent proteins.
a Effect of pspA coexpression on EGFP secretion in S. lividans strains grown for 24 h in NM medium. The amount of secreted EGFP was determined by measuring fluorescence intensities, corrected for mycelial dry weight, of spent medium from S. lividans [pIJ486], S. lividans [pSSVX-EGFP) and from S. lividans [pIJ486PspA+pSSVX-EGFP]. b Western blotting with anti-GFP antibodies on the extracellular fraction of S. lividans [pIJ486PspA] (lane 2), S. lividans TK24 [pSSV-EGFP] (lane 3) and S. lividans TK24 [pIJ486PspA+pSSV-EGFP] (lane 4). Cultures were grown for 24 h in 50 ml NM medium. In each lane, the amount of supernatant corresponding to 0.1 mg of dry weight was loaded. Lane 1 shows a molecular weight marker
Influence of pspA overexpression on Sec-dependent secretion
To test if PspA overproduction also positively affected the Sec-dependent secretion, we evaluated the secretion of the S. lividans subtilisin inhibitor 1 (Sti-1) in a PspA-overproducing strain by introducing pIJ486PspA in the wild type. The Sti-1 protein is already efficiently secreted from the wild-type S. lividans. Secretion of Sti-1 was assayed both for the wild-type and the PspA-overproduction strain grown in NM medium for 18–24 h. Subtilisin inhibitor activity in the supernatant of 18-h cultures ranged from 15.7±0.3 to 18.9±2.0 U (mg dry wt)−1 for the wild type and the pspA strain, respectively (Fig. 4), indicating a small but statistically significant increase in subtilisin inhibitor activity of 20%. Similar values were obtained after 24 h of growth, confirming the positive effect of pspA overexpression on Sec-dependent secretion of native proteins in S. lividans.
Finally, to assess the effect of pspA overexpression on a Sec-routed heterologous protein, human tumour necrosis factor alpha (hTNFα) was tested. To this end, both the wild-type and the pspA-overexpressing strain were transformed with plasmid pSSV-TNF, encoding a Sec-dependent substrate consisting of the Vsi signal peptide fused in frame to the mature hTNFα. Cultures of both strains were grown for 24 h at which point the amount of secreted hTNFα was quantified by ELISA. Extracellular values of 0.72±0.15 μg hTNFα (mg dry wt)−1 for the wild type and 1.05±0.27 μg hTNFα (mg dry wt)−1 for the PspA-overproducing strain were obtained. Although the PspA-overproducing strain appears to secrete more hTNFα (mg dry wt)−1, this difference is statistically not significant, and we can conclude that pspA overexpression does not significantly increase the secreted hTNFα yield in S. lividans. Taken together, pspA overexpression increases the secretion of all tested proteins, but the effect on Tat-dependent protein translocation is more pronounced than on the Sec-routed secretion.
Discussion
Based on the S. coelicolor genome, the S. lividanspspA gene could be isolated and identified. Remarkably, the known genomic sequence of the S. coelicolor, which is closely related to S. lividans (Bentley et al. 2002), revealed only a homologue for pspA but none for the pspB or C genes, thought to be needed for a functional psp response (Darwin 2005). The PspB and PspC proteins are presumed to be the sensors of extracytoplasmic stress (Adams et al. 2003), and depending on what stress condition is used, a different protein is needed to get a psp response. The response to most inducing stimuli, such as hyper-osmotic shock and 10% ethanol shock is stimulated by, but not entirely dependent on, PspB and C (Model et al. 1997). Some stress conditions, however, rely on only one of the pspB/C genes. In Yersinia enterocolitica, for instance, only the pspB gene is absolutely necessary to induce a psp response upon overproduction of the YsaC secretin (Maxson and Darwin 2006). Finally some responses, like the induction of PspA by heat shock in E. coli, do not require PspB or PspC at all (Weiner et al. 1991). The fact that pspA homologues are present in the vast majority of the sequenced bacterial chromosomes and that most of them are not adjacent to any other psp genes, indicates that PspA, as such, may play an important role in bacteria, even when PspB and C are not present.
The work presented here clearly shows that overproduction of PspA can positively influence both the Tat- and the Sec-dependent protein secretion in S. lividans although the effect on the Tat pathway is much more pronounced. As at least one of the roles of PspA appears to be the maintenance of the PMF (Kleerebezem et al. 1996), the more dramatic effect on the Tat secretion may be explained by the difference in energy requirement of both pathways. The Sec pathway requires ATP as its main energy source (Economou 1999), but the PMF lowers the level of ATP required for the translocation of proteins in E. coli (Shiozuka et al. 1990) and can increase the translocation rate of certain proteins (Yamada et al. 1989). On the other hand, the Tat pathway is solely dependent on the PMF (Berks et al. 2003).
It is clear from the obtained results that pspA overexpression increases the Tat-dependent protein secretion in S. lividans at least threefold, which is similar to the results in E. coli, where pspA overexpression resulted in a threefold increase of Tat-dependent secretion of both native and heterologous proteins (DeLisa et al. 2004). The Tat pathway has recently gained much attention, thanks to its ability to secrete proteins in a folded state, which can make it a valuable alternative for these proteins that could otherwise not be translocated in an active conformation via the Sec pathway, e.g., EGFP (Santini et al. 2001) and tPA (Kim et al. 2005). This makes the S. lividans pspA-overexpressing strain an interesting alternative host for recombinant protein production using the Tat pathway, as evidenced here by a yield of up to 20 mg/l EGFP, a protein that is notoriously difficult to secrete.
The effect on the Sec-dependent secretion is less pronounced and will probably vary from protein to protein. This is in correspondence with the observation made by Yamada et al. (1989) that the dissipation of the PMF has a differential effect on different secretory proteins. For some proteins, this dissipation resulted in a large decrease in translocation, whereas others were less influenced. This means that the S. lividans pspA-overexpressing strain might be a valuable host for the secretion of some, but not all, proteins via the Sec pathway. Whether or not overexpression of pspA can protect S. lividans against other conditions that might induce membrane-related stress is a question that we are currently investigating. Finally, this work also supports the possibility of increasing protein secretion in a wide range of hosts, all containing a pspA homologue but not a complete psp system.
References
Adams H, Teertstra W, Demmers J, Boesten R, Tommassen J (2003) Interactions between phage-shock proteins in Escherichia coli. J Bacteriol 85:1174–1180
Anné J, Van Mellaert L, Eyssen H (1990) Optimum conditions for efficient transformation of Streptomyces venezuelae protoplasts. Appl Microbiol Biotechnol 32:431–435
Bentley SD, Chater KF, Cerdeno-Tarraga AM (2002) Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141–147
Berks BC, Palmer T, Sargent F (2003) The Tat protein translocation pathway and its role in microbial physiology. Adv Microb Physiol 47:187–254
Darwin AJ (2005) The phage-shock-protein response. Mol Microbiol 57:621–628
De Keersmaeker S, Van Mellaert L, Lammertyn E, Vrancken K, Anné J, Geukens N (2005) Functional analysis of TatA and TatB in Streptomyces lividans. Biochem Biophys Res Commun 335:973–982
DeLisa MP, Tullman D, Georgiou G (2003) Folding quality control in the export of proteins by the bacterial twin-arginine translocation pathway. Proc Natl Acad Sci USA 100:7467–7473
DeLisa MP, Lee P, Palmer T, Georgiou G (2004) Phage shock protein PspA of Escherichia coli relieves saturation of protein export via the Tat pathway. J Bacteriol 186:366–373
Economou A (1999) Following the leader: bacterial protein export through the Sec pathway. Trends Microbiol 7:315–320
Faury D, Saidane S, Li H, Morosoli R (2004) Secretion of active xylanase C from Streptomyces lividans is exclusively mediated by the Tat protein export system. Biochem Biophys Acta 1699:155–162
Hong B, Wu B, Li Y (2003) Production of C-terminal amidated recombinant salmon calcitonin in Streptomyces lividans. Appl Biochem Biotechnol 110:113–123
Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA (2000) Practical Streptomyces genetics. John Innes Foundation, Norwich
Kim JY, Fogarty EA, Lu FJ, Zhu H, Wheelock GD, Henderson LA, DeLisa MP (2005) Twin-arginine translocation of active human tissue plasminogen activator in Escherichia coli. Appl Environ Microbiol 71:8451–8459
Kleerebezem M, Crielaard W, Tommassen J (1996) Involvement of stress protein PspA (phage shock protein A) of Escherichia coli in maintenance of the protonmotive force under stress conditions. EMBO J 15:162–171
Kojima S, Obata S, Kumagai I, Miura K (1990) Alteration of the specificity of the Streptomyces subtilisin inhibitor by gene engineering. Biotechnology (NY) 8:449–452
Korn F, Weingärtner B, Kutzner HJ (1978) Genetics of the actinomycetales. In: Freechsen E, Tarnak I, Thumin JH (eds) A study of twenty actinophages: morphology, serological relationship and host range. Fisher G., Stuttgart, pp 251–270
Lara M, Servin-Gonzalez L, Singh M, Moreno C, Cohen I, Nimtz M, Espitia C (2004) Expression, secretion, and glycosylation of the 45- and 47-kDa glycoprotein of Mycobacterium tuberculosis in Streptomyces lividans. Appl Environ Microbiol 70:679–685
Lammertyn E (2000) doctoral thesis, Katholieke Universiteit Leuven, pp 148
Maxson ME, Darwin AJ (2006) PspB and PspC of Yersinia enterocolitica are dual function proteins: regulators and effectors of the phage-shock-protein response. Mol Microbiol 59:1610–1623
Model P, Jovanovic G, Dworkin J (1997) The Escherichia coli phage-shock-protein (psp) operon. Mol Microbiol 24:255–261
Mondou F, Shareck F, Morosoli R, Kluepfel D (1986) Cloning of the xylanase gene of Streptomyces lividans. Gene 49:323–329
Pozidis C, Lammertyn E, Politou AS, Anné J, Tsiftsoglou AS, Sianidis G, Economou A (2001) Protein secretion biotechnology using Streptomyces lividans: large-scale production of functional trimeric tumor necrosis factor alpha. Biotechnol Bioeng 72:611–619
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor, New York
Santini CL, Bernadac A, Zhang M, Chanal A, Ize B, Blanco C, Wu LF (2001) Translocation of jellyfish green fluorescent protein via the Tat system of Escherichia coli and change of its periplasmic localization in response to osmotic up-shock. J Biol Chem 276:8159–8164
Schaerlaekens K, Lammertyn E, Geukens N, De Keersmaeker S, Anné J, Van Mellaert L (2004) Comparison of the Sec and Tat secretion pathways for heterologous protein production by Streptomyces lividans. J Biotechnol 112:279–288
Sianidis G, Pozidis C, Becker F, Vrancken K, Sjoeholm C, Karamanou S, Takamiya-Wik M, Van Mellaert L, Schaefer T, Anné J, Economou A (2006) Functional large-scale production of a novel Jonesia sp. xyloglucanase by heterologous secretion from Streptomyces lividans. J Biotechnol 121:498–507
Simon R, Priefer U, Pühler A (1983) A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Biotechnology 1:784–791
Shiozuka K, Tani K, Mizushima S, Tokuda H (1990) The proton motive force lowers the level of ATP required for the in vitro translocation of a secretory protein in Escherichia coli. J Biol Chem 265:18843–18847
Sun J, Kelemen GH, Fernandez-Abalos JM, Bibb MJ (1999) Green fluorescent protein as a reporter for spatial and temporal gene expression in Streptomyces coelicolor A3(2). Microbiology 145:2221–2227
Van Mellaert L, Dillen C, Proost P, Sablon E, DeLeys R, Van Broekhoven A, Heremans H, Van Damme J, Eyssen H, Anné J (1994) Efficient secretion of biologically active mouse tumor necrosis factor alpha by Streptomyces lividans. Gene 150:153–158
Vats-Mehta S, Bouvrette P, Shareck F, Morosoli R, Kluepfel D (1990) Cloning of a second xylanase-encoding gene of Streptomyces lividans 66. Gene 86:119–122
Ward JM, Janssen GR, Kieser T, Buttner MJ, Bibb MJ (1986) Construction and characterisation of a series of multi-copy promoter-probe plasmid vectors for Streptomyces using the aminoglycoside phosphotransferase gene from Tn5 as indicator. Mol Gen Genet 203:468–478
Weiner L, Brissette JL, Model P (1991) Stress-induced expression of the Escherichia coli phage shock protein operon is dependent on σ54 and modulated by positive and negative feedback mechanisms. Genes Dev 5:1912–1923
Yamada H, Tokuda H, Mizushima S (1989) Proton motive force-dependent and -independent protein translocation revealed by an efficient in vitro assay system of Escherichia coli. J Biol Chem 264:1723–1728
Acknowledgements
KV and SDK are research fellows of IWT (Vlaams Instituut voor de bevordering van het Wetenschappelijk-Technologisch onderzoek in de industrie). This research was supported by grants G.0389.05. from Fonds voor Wetenschappelijk Onderzoek Vlaanderen (FWO) and OT/00/37 from K.U.Leuven and QLK3-CT-2002-02056 from the European Union.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Vrancken, K., De Keersmaeker, S., Geukens, N. et al. pspA overexpression in Streptomyces lividans improves both Sec- and Tat-dependent protein secretion. Appl Microbiol Biotechnol 73, 1150–1157 (2007). https://doi.org/10.1007/s00253-006-0571-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-006-0571-7