Abstract
Toluene-o-xylene monooxygenase (ToMO) from Pseudomonas stutzeri OX1 has been shown to degrade all chlorinated ethenes individually and as mixtures. Here, DNA shuffling of the alpha hydroxylase fragment of ToMO (TouA) and saturation mutagenesis of the TouA active site residues I100, Q141, T201, F205, and E214 were used to enhance the degradation of chlorinated aliphatics. The ToMO mutants were identified using a chloride ion screen and then were further examined by gas chromatography. Escherichia coli TG1/pBS(Kan)ToMO expressing TouA saturation mutagenesis variant I100Q was identified that has 2.8-fold better trichloroethylene (TCE) degradation activity (apparent V max of 1.77 nmol min−1 mg−1 protein−1 vs 0.63 nmol min−1 mg−1 protein−1). Another variant, E214G/D312N/M399V, has 2.5-fold better cis-1,2-dichloroethylene (cis-DCE) degradation activity (apparent V max of 8.4 nmol min−1 mg−1 protein−1 vs 3.3 nmol min−1 mg−1 protein−1). Additionally, the hydroxylation regiospecificity of o-xylene and naphthalene were altered significantly for ToMO variants A107T/E214A, T201G, and T201S. Variant T201S produced 2.0-fold more 2,3-dimethylphenol (2,3-DMP) from o-xylene than the wild-type ToMO, whereas variant A107T/E214A had 6.0-fold altered regiospecificity for 2,3-DMP formation. Variant A107T/E214A also produced 3.0-fold more 2-naphthol from naphthalene than the wild-type ToMO, whereas the regiospecificity of variant T201S was altered to synthesize 3.0-fold less 2-naphthol, so that it made almost exclusively 1-naphthol (96%). Variant T201G was more regiospecific than variants A107T/E214A and T201S and produced 100% 3,4-DMP from o-xylene and >99% 1-naphthol from naphthalene. Hence, ToMO activity was enhanced for the degradation of TCE and cis-DCE and for the regiospecific hydroxylation of o-xylene and naphthalene through DNA shuffling and saturation mutagenesis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chlorinated aliphatics are a group of health-threatening pollutants. For example, tetrachloroethylene (PCE) is one of 14 volatile organic compounds on the United States Environmental Protection Agency (US EPA) Priority Pollutant List (Carter and Jewell 1993). Vinyl chloride (VC) is a known human carcinogen (McCarty 1997); and both VC and cis-1,2-dichloroethylene (cis-DCE) are US EPA priority pollutants (Bradley and Chapelle 1998). Chloroform is also a suspected carcinogen (McClay et al. 1996). We have shown that toluene-o-xylene monooxygenase (ToMO) from Pseudomonas stutzeri OX1 degrades chlorinated aliphatics, including chloroform, trichloroethylene (TCE), 1,1-dichloroethylene (1,1-DCE), cis-DCE, trans-DCE, and VC, individually and as mixtures (Chauhan et al. 1998; Ryoo et al. 2000; Shim and Wood 2000), and that ToMO is the only known enzyme which degrades PCE aerobically (Ryoo et al. 2000). Aerobic bioremediation of chlorinated aliphatics avoids the formation of VC, which frequently is the product of anaerobic degradation of these pollutants as one chlorine at a time is removed, with the last most difficult (He et al. 2003; McCarty 1997). Under aerobic conditions, the chlorinated ethenes may be completely dechlorinated, as we have shown with ToMO for all the chlorinated aliphatics (Shim et al. 2001; Shim and Wood 2000), after the formation of a probable chlorinated epoxide that is formed by the monooxygenase (van Hylckama Vlieg et al. 1996). It has also been shown that PCE and TCE induce their own degradation in P. stutzeri OX1 (Ryoo et al. 2000, 2001) and that this strain is chemotactic toward PCE and other less-chlorinated compounds (Vardar et al. 2005a). Hence, P. stutzeri OX1 expressing ToMO is very attractive for bioremediating chlorinated aliphatics.
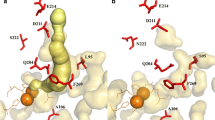
P. stutzeri OX1, isolated from the activated sludge of a wastewater treatment plant in Italy (Baggi et al. 1987), grows on o-xylene, toluene, cresols, 2,3-dimethylphenol (2,3-DMP), and 3,4-DMP via initial oxidation with ToMO (Bertoni et al. 1996). ToMO hydroxylates toluene in the ortho, meta, and para positions, hydroxylates o-xylene in both the meta and para positions, and, along with chlorinated aliphatics, oxidizes many substrates including m-xylene, p-xylene, benzene, ethyl-benzene, styrene, and naphthalene (Bertoni et al. 1996). ToMO is also capable of three successive hydroxylations of benzene, forming 1,2,3-trihydroxybenzene (THB; Vardar and Wood 2004) along with other toluene monooxygenases, including toluene ortho-monooxygenase (TOM) of Burkholderia cepacia G4, toluene-para-monooxygenase of Ralstonia pickettii PKO1, and toluene-4-monooxygenase (T4MO) of P. mendocina KR1 (Tao et al. 2004b). The six proteins coding for ToMO are TouABE (three-component hydroxylase with two catalytic oxygen-bridged dinuclear centers, A2B2E2), TouC (ferredoxin), TouD (mediating protein), and TouF (NADH-ferredoxin oxidoreductase; Bertoni et al. 1996; Cafaro et al. 2002). The alpha subunit of ToMO (TouA) consists of the carboxylate-bridged diiron center where the substrate hydroxylation occurs (Sazinsky et al. 2004). The crystal structure of the ToMO hydroxylase shows that the alpha (TouA) and beta (TouE) subunits have almost identical structure with the soluble methane monooxygenase (sMMO) alpha (MmoX) and beta (MmoY) subunits of Methylococcus capsulatus, especially near the active site, whereas the gamma subunits (TouB for ToMO, MmoZ for MMO) differ from each other in terms of location and folding (TouB consists mostly of beta sheets, whereas MmoZ consists mostly of helices; Elango et al. 1997; Rosenzweig et al. 1997; Sazinsky et al. 2004).
DNA shuffling is a common and powerful method for protein mutagenesis (Stemmer 1994) in which hot spots of an enzyme may be discovered. All other possible amino acid substitutions can then be checked via saturation mutagenesis (Sakamoto et al. 2001) at the hot spots identified via DNA shuffling. Using DNA shuffling, the alpha subunit (TomA3) mutation V106A in TOM of B. cepacia G4 was identified as a gate residue (corresponding to I100 of the alpha subunit TouA of the hydroxylase in ToMO), which resulted in a 2.0-fold enhancement for TCE degradation, a 6.0-fold enhancement in naphthalene oxidation, and enhanced degradation of phenanthrene, fluorene, and anthracene due to greater access to the diiron site (Canada et al. 2002). Through saturation mutagenesis, variant TomA3 A106E was identified with 2.0-fold enhancement in 1-naphthol synthesis from naphthalene (Rui et al. 2004). Another variant TomA3 A106F degraded chloroform 2.8-fold faster than the wild-type TOM (Rui et al. 2004). The alpha subunit positions TmoA Q141, T201, and F205 in the related T4MO of P. mendocina KR1 were also shown to be important for regiospecificity (Pikus et al. 2000, 1997). We recently discovered that position TouA E214 in ToMO influences the rate of reaction by controlling substrate entrance/product efflux as another gate residue (Vardar and Wood 2005); the rate of oxidation of p-nitrophenol to 4-nitrocatechol was inversely related to the size of the residue at position E214. Hence, we reasoned that ToMO residues I100, Q141, T201, F205, and E214 may be important for controlling the oxidation of chlorinated aliphatics.
In this work, our goals were to enhance the degradation of chlorinated aliphatics and to alter the oxidation regiospecificity of o-xylene and naphthalene, using both DNA shuffling of touA and saturation mutagenesis of TouA at positions I100, Q141, T201, F205, and E214 of ToMO. It was discovered that TouA positions I100 and E214, the two gate residues in the hydroxylase subunit, play a role in enhancing the degradation of TCE and cis-DCE and that TouA positions A107 and T201 play a role in altering the regiospecificity of o-xylene and naphthalene oxidation. This is the first random mutagenesis of this enzyme for chlorinated substrates.
Materials and methods
Bacterial strains, growth conditions, and SDS-PAGE
Escherichia coli strain TG1 [supEhsdΔ5 thi Δ(lac-proAB) F′(traD36 proAB+lacIqlacZΔM15); Sambrook et al. 1989] was used to host pBS(Kan)ToMO (Vardar and Wood 2004) and its variants which expressed the touABCDEF genes from a constitutive lac promoter. Cells were initially streaked from −80°C glycerol stocks on Luria–Bertani (LB) agar plates (Sambrook et al. 1989) containing 100 μg ml−1 kanamycin and incubated at 37°C. After growth on LB agar plates, cells were cultured from a fresh single colony in LB medium (Sambrook et al. 1989) supplemented with 100 μg ml−1 kanamycin at 37°C with shaking at 250 rpm (New Brunswick Scientific Co., Edison, N.J.). Specific growth-rate measurements were conducted with 25 ml exponentially grown cells (in 250-ml shake flasks) with an optical density at 600 nm (OD600) between 0.1 and 0.7 and calculated from the slope of ln OD600 versus time. The relative expression of TouA, TouE, and TouF from E. coli TG1/pBS(Kan)ToMO was evaluated using SDS-PAGE (Sambrook et al. 1989) and a 12% Tris-HCl gel both with and without 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG; Fisher Scientific Co., Fairlawn, N.J.).
Chemicals
Naphthalene, chloroform, TCE, p-cresol, toluene, and o-xylene were purchased from Fisher Scientific Co. (Pittsburgh, Pa.), cis-DCE and trans-DCE were purchased from TCI America (Portland, Ore.), VC were purchased from Supelco (Bellefonte, Pa.), and 1-naphthol, 2-naphthol, PCE, 1,1-DCE, o-cresol, m-cresol, 2,3-DMP, and 3,4-DMP were purchased from Sigma Chemical Co. (Milwaukee, Wis.). All materials used were of the highest purity available and were used without further purification.
ToMO mutagenesis and dechlorination screen
Saturation mutagenesis at positions I100, Q141, T201, F205, and E214 of the alpha subunit (touA) of ToMO and DNA shuffling of 90% of touA of ToMO were performed as described by Vardar and Wood (2004, 2005). The inorganic chloride generated from the dechlorination of the chlorinated aliphatics by whole cells was measured spectrophotometrically in 96-well plates (Canada et al. 2002). Cells in 96-well plates were contacted with shaking at 37°C in an airtight chamber, 23×20×23 cm, with TCE and chloroform vapors (0.5 ml) for 24 h.
Extent and rates of dechlorination
Experiments were conducted with exponentially-grown cells with an OD600 of 1.0. The cells were washed three times at 6,000 g for 5 min at 25°C (JA-17 rotor in a J2 series centrifuge; Beckman, Palo Alto, Calif.) and resuspended with 1 vol. Tris-HNO3 buffer (50 mM, pH 7.0), to an OD600 of 5.0. Cell suspensions (2.5 ml) were sealed with a Teflon-coated septum and aluminum seal in 15-ml glass vials and the chlorinated aliphatic compounds (chloroform, PCE, TCE, cis-DCE, trans-DCE, 1,1-DCE, VC) were added from dimethyl formamide (DMF) stock solutions at 200 μM. After contacting at 37°C, 250 rpm in a KS250 benchtop shaker (IKA Laboratories, Cincinnati, Ohio), the supernatant chloride ion concentrations generated from mineralizing the chlorinated aliphatic compounds were measured after specific times of incubation (22 h for PCE, 17 h for chloroform, 2 h for trans-DCE and VC, 90 min for cis-DCE, 60 min for 1,1-DCE, 30 min for TCE ). PCE, TCE, and the three dichloroethylenes were added at an initial concentration of 200 μM and chloroform at 80 μM, assuming all the volatile organics were in liquid phase (DMF as diluent); the actual initial liquid concentrations, based on Henry’s law (Bradley and Chapelle 1998; Chauhan et al. 1998; Dolfing et al. 1993), were 34 μM for PCE, 67 μM for TCE, 108 μM for cis-DCE, 69 μM for trans-DCE, 32 μM for 1,1-DCE, 34 μM for VC, 50 μM for chloroform. After contact, the enzyme activity was quenched by heating the vials in boiling water for 2 min. The chloride ion concentrations in 500 μl supernatant were measured spectrophotometrically as described by Canada et al. (2002), using the negative control of DMF addition (no chlorinated aliphatics) for each strain.
The initial degradation rates at 200 μM for wild-type ToMO and selected mutants were determined by monitoring substrate depletion via gas chromatography (GC), using a cell density around OD600=10. The protein content was 0.22 mg protein ml−1 for each OD600 unit for recombinant E. coli TG1, as determined using the Sigma protein assay kit (Sigma Diagnostics, St. Louis, Mo.). The whole-cell catalytic parameters (apparent V max, K m) were determined for the degradation of cis-DCE (at 13.5, 27, 54, 108, 216, 540 μM actual liquid concentration using Henry’s law; Bradley and Chapelle 1998) and TCE (at 8.3, 16.7, 33.4, 66.7, 133.4, 333.4 μM actual liquid concentration using Henry’s law; Chauhan et al. 1998) for wild-type ToMO and variants I100Q or E214G/D312N/M399V, from the initial rate at each concentration. For PCE and cis-DCE degradation, 0.5 mM IPTG was used during growth at OD600=2.5–3.0 for 2 h and 1% of succinate was added during contact with PCE.
Regiospecificity of toluene, o-xylene, and naphthalene oxidation
To determine the toluene and o-xylene regiospecificities, the products of the monooxygenase reactions were analyzed via GC, using ethyl acetate extraction and a hexadecane internal standard (Vardar et al. 2005b; Vardar and Wood 2004). To determine the naphthalene regiospecificities, the products of the monooxygenase reactions were analyzed via high performance liquid chromatography (HPLC) and similar conditions were used as for nitrobenzene (Vardar et al. 2005b) with slight changes: the gradient for naphthalene oxidation was 65:35 (H2O with 0.1% formic acid/acetonitrile) for 0–5 min, then gradients of 35:65 at 12 min and 65:35 at 20 min. The retention times for naphthalene, 1-naphthol, and 2-naphthol were 17.2, 13.4, and 12.7 min, respectively.
DNA sequencing
A dideoxy chain-termination technique (Sanger et al. 1977) with the ABI Prism BigDye terminator cycle sequencing ready reaction kit (PerkinElmer, Wellesley, Mass.) and the PE Biosystems ABI 373 DNA sequencer (PerkinElmer) were used to determine the TouA variant nucleotide sequences, using sequencing primers ToMO 1, 2, and 3 (Vardar and Wood 2004). The sequencing data were analyzed using the Vector NTI program (InforMax, Frederick, Md.).
Results
Screening
DNA shuffling of 90% of touA and saturation mutagenesis of five codons in touA (I100, Q141, T201, F205, E214) were performed to create mutations in ToMO to enhance the degradation rate of the chlorinated aliphatics TCE, cis-DCE, trans-DCE, 1,1-DCE, VC, and chloroform and to alter the regiospecific oxidation of o-xylene and naphthalene. A library of 8,000 mutants was generated for saturation mutagenesis. The number of colonies from saturation mutagenesis that must be screened to ensure a 99% probability that all possible substitutions at a single codon are sampled was 293 (Rui et al. 2004); hence, around 500 colonies from positions TouA I100, Q141, T201, and F205 (2,000 colonies in total) were screened for chloride generation from TCE and chloroform, and 500 colonies from position TouA E214 were screened for chloride generation from cis-DCE, using whole cells in a 96-well plate format. For DNA shuffling of touA, an additional 4,000 colonies were screened.
Mutants with enhanced chloride generation from the 96-well plate format were identified via DNA sequencing. Interestingly, amino acid changes were found at TouA positions I100 and E214 (via saturation mutagenesis) and at E214/D312/M399 and K160 (via DNA shuffling). No mutants with enhanced activities for chlorinated aliphatic degradation were found from saturation mutagenesis at TouA positions Q141, T201, or F205, whereas mutants with altered regiospecific activities for o-xylene and naphthalene were found at TouA positions A107/E214 and T201. Note, variant A107T/E214A arose as an unexpected spontaneous mutant with two amino acid changes during saturation mutagenesis at position E214; both it and variant M180T/E284G were identified previously from a nitrobenzene agar plate assay (Vardar et al. 2005b) and were characterized here for the oxidation of chlorinated aliphatics, o-xylene, and naphthalene.
Characterization of the best mutants
The best mutants identified initially by screening for chloride release from TCE, cis-DCE, and chloroform in 96-well plates were further examined in 15-ml vials for chloride release from dechlorination of TCE, cis-DCE, trans-DCE, 1,1-DCE, VC, and chloroform. The amount of chloride produced with wild-type ToMO and TouA variants I100Q, M180T/E284G, E214G/D312N/M399V, and K160N are shown in Table 1. Wild-type ToMO produced 73 μM chloride from TCE, whereas TouA variant I100Q produced 108 μM (1.5-fold increase). From cis-DCE, TouA variants I100Q, K160N, M180T/E284G, E214G/D312N/M399V, and E214G produced 20–40% more chloride (Table 1). The specific growth rates of the best mutants were approximately the same (0.50±0.02 h−1 in LB +kanamycin medium) except for mutant I100Q, which had a 2.0-fold increase compared with wild-type ToMO (1.0 h−1).
Based on the enhanced degradation of TCE and cis-DCE by several of the ToMO variants, the initial degradation rates at 200 μM (concentration assuming all the organic volatile was in the liquid phase) for wild-type ToMO and TouA variants E214G/D312N/M399V, I100Q, and K160N were determined by monitoring TCE and cis-DCE depletion via GC (Table 2; Fig. 1a). Variants I100Q and K160N degraded TCE faster (207% and 130%, respectively) than the wild-type ToMO. Variants I100Q and E214G/D312N/M399V degraded cis-DCE faster (141% and 196%, respectively) than the wild-type ToMO.
Representative time-course experiments (a) and Lineweaver–Burk plot (b) for TCE (67 μM actual liquid phase concentration) degradation by TG1 cells expressing wild-type ToMO (○) and TouA I100Q (•), with regression lines shown. Error bars represent the mean standard deviations from two independent experiments. From the slope (a) and biomass of each experiment, the initial degradation rates in Table 2 were calculated. Total TCE concentration shown (a) includes nanomoles of TCE in both the gas and liquid phases. The kinetic constants reported were calculated from plot b
To investigate more fully the enhanced rate of cis-DCE degradation by E214G/D312N/M399V relative to wild-type ToMO, the apparent V max values were measured for whole cells and found to be 8.4 nmol min−1 mg−1 protein−1 and 3.3 nmol min−1 mg−1 protein−1, respectively, whereas the apparent K m was 59.8 μM for E214G/D312N/M399V and 14.7 μM for wild-type ToMO. Hence, the apparent V max was enhanced 2.5-fold for E214G/D312N/M399V for cis-DCE degradation. To investigate the enhanced rate of TCE degradation by I100Q relative to wild-type ToMO, the apparent V max values were measured for whole cells and found to be 1.77 nmol min−1 mg−1 protein−1 and 0.63 nmol min−1 mg−1 protein−1, respectively, whereas the apparent K m was 72.7 μM for I100Q and 59.5 μM for wild-type ToMO (Fig. 1b). Hence, the apparent V max was enhanced 2.8-fold for ToMO variant I100Q for TCE degradation.
The best variants (I100Q, E214G/D312N/M399V) and wild-type ToMO were further examined in 15-ml vials for chloride release from PCE degradation. PCE degradation (200 μM) was confirmed for wild-type ToMO as seen previously (Ryoo et al. 2000; Shim et al. 2001) but no enhanced activity was found with TouA variants I100Q and E214G/D312N/M399V. Wild-type ToMO produced 15 μM chloride from PCE, whereas TouA variants I100Q and E214G/D312N/M399V produced 12 μM and 10 μM, respectively, when DMF was used as a negative control.
Oxidation regiospecificity for the natural substrate toluene by wild-type ToMO and mutants I100Q, A107T/E214A, T201G, T201S, M180T/E284G, and E214G/D312N/M399V (Table 3) were reported by Vardar et al. (2005b) and Vardar and Wood (2004). A107T/E214A mutation converted ToMO into a para-enzyme and made 2% o-cresol, 5% m-cresol, and 93% p-cresol from toluene, whereas wild-type ToMO made 32% o-cresol, 21% m-cresol, and 47% p-cresol (Vardar et al. 2005b). The I100Q and T201G mutations caused a shift in product distribution and made 22% and 53% o-cresol, 44% and 12% m-cresol, and 34% and 35% p-cresol, respectively. Mutants M180T/E284G, T201S, and E214G/D312N/M399V gave no substantial shift in product distribution (Vardar et al. 2005b; Vardar and Wood 2004). Oxidation regiospecificities for the other natural substrate o-xylene by wild-type ToMO and mutant I100Q were also reported by Vardar et al. (2005b). Wild-type ToMO oxidized o-xylene and made 82% 3,4-DMP and 18% 2,3-DMP, whereas variant I100Q gave a slight shift and made 76% 3,4-DMP and 24% 2,3-DMP.
Here, the toluene hydroxylation regiospecificity of mutant K160N and the o-xylene and naphthalene regiospecificities of mutants A107T/E214A, K160N, M180T/E284G, T201G, T201S, and E214G/D312N/M399V were determined (Table 3). Variant A107T/E214A produced 97% 3,4-DMP and 3% 2,3-DMP from o-xylene; and, hence, there was a 6-fold change in regiospecificity for 2,3-DMP formation (3% vs 18%), whereas variant T201S produced 2-fold more 2,3-DMP from o-xylene than the wild-type ToMO produced (35% vs 18%). However, variant T201G was more regiospecific than variant A107T/E214A for o-xylene oxidation and produced 100% 3,4-DMP (Table 3).
Also, the naphthalene hydroxylation regiospecificities of wild-type ToMO and TouA mutants I100Q, A107T/E214A, K160N, T201G, T201S, M180T/E284G, and E214G/D312N/M399V were determined (Table 3). Variant A107T/E214A produced 3-fold more 2-naphthol from naphthalene than wild-type ToMO produced (36% vs 12%), whereas variant T201S produced 3-fold less 2-naphthol (4% vs 12%). Variant T201G was more regiospecific than variant T201S for naphthalene oxidation and produced >99% 1-naphthol. Mutants I100Q, K160N, M180T/E284G, and E214G/D312N/M399V produced the same products (Table 3).
Enzyme expression level
The expression levels of TouA variants I100Q, E214G/D312N/M399V, E214G, and M180T/E284G were reported previously, since these variants were identified using screens for toluene, p-nitrophenol, and nitrobenzene, respectively (Vardar et al. 2005b; Vardar and Wood 2005). TouA variant I100Q is an expression-down mutant, as evidenced by SDS-PAGE, with a single nucleotide change in one codon leading to less-elevated protein expression (1.5- to 2-fold decrease). The expression levels of variants M180T/E284G, E214G/D312N/M399, and E214G remained approximately the same as that of wild-type ToMO. Hence, the increase in the activity of mutants E214G/D312N/M399V, E214G, and M180T/E284G derives from the amino acid substitutions rather than protein expression level changes, and, for TouA mutant I100Q, the activity might be 2-fold more than that reported. Similar changes in expression levels due to mutation have been seen with naphthalene dioxygenase and para-nitrobenzyl esterase (Moore and Arnold 1996; Sakamoto et al. 2001). The variation in protein expression level could be due to either a modification of the primary amino acid sequence (leading to an increase in protein lability) or a single nucleotide change (leading to increased lability of the transcript), while the ribosome-binding site and promoter remain unaltered.
Discussion
It is shown clearly in this paper that saturation mutagenesis of ToMO at the key site I100 in the alpha subunit TouA and DNA shuffling of TouA result in enhanced degradation of TCE and cis-DCE (e.g., TouA I100Q, TouA E214G/D312N/M399V). Previously, we showed the importance of TouA positions E214/D312/M399 for increasing the oxidation of nitro aromatics (Vardar et al. 2005b). TouA DNA shuffling variant E214G/D312N/M399V oxidized nitrobenzene 6-fold faster than wild-type ToMO and produced m- and p-nitrophenol as well as 4-nitrocatechol, whereas wild-type ToMO produced only m- and p-nitrophenol. Also, the oxidation rate of nitrophenols was increased 4- to 20-fold by the mutant E214G/D312N/M399V, compared with the wild-type ToMO. We then showed that the rates of oxidation by variant E214G for o-nitrophenol, p-nitrophenol, m-nitrophenol, and nitrobenzene were similar to variant E214G/D312N/M399V. Therefore, positions D312 and M399 did not appear to play an important role in catalysis; and TouA E214G was responsible for the enhanced rate of oxidation of the nitro aromatics (Vardar and Wood 2005).
The same TouA variant I100Q that was identified here for increased TCE degradation, based on the chloride screen, was isolated independently by us for significantly altered hydroxylation regiospecificities for o-cresol, m-cresol, phenol, catechol, and m-nitrophenol, allowing for the novel formation of methylhydroquinone, nitrohydroquinone, hydroquinone, and 1,2,4-THB (Vardar et al. 2005b; Vardar and Wood 2004). For example, TouA mutant I100Q produced catechol and hydroquinone (major peak) from benzene and methylcatechols and methylhydroquinone (major peak) from toluene, whereas wild-type ToMO produced only catechol from benzene and methylcatechols from toluene (Vardar and Wood 2004). Similarly, wild-type ToMO produced only 4-nitrocatechol from m-nitrophenol, but TouA I100Q produced both 4-nitrocatechol (37%) and nitrohydroquinone (63%; Vardar et al. 2005b). In addition, ToMO TouA variant I100Q oxidized nitrobenzene and nitrophenols 2-fold faster than the wild-type ToMO. Here, we show that the I100Q mutation of ToMO increases the degradation of TCE and cis-DCE, and this result is similar to the V106A mutation that was found previously in the gate residue of the alpha subunit of TOM of B. cepacia G4 (Canada et al. 2002; V106 corresponds to position TouA I100 in ToMO). Therefore, the enhanced chlorinated aliphatic degradation rates with TouA variants I100Q and E214G corroborate the importance of these gate residues for monooxygenase activity.
Previously, we showed the effect of the A107T/E214A, T201G, and T201S mutations on nitro aromatics and toluene hydroxylation regiospecificities (Vardar et al. 2005b). By showing TouA position E214 does not influence regiospecificity (Vardar and Wood 2005), we determined that it is the A107T mutation that is relevant for the A107T/E214A double mutant. TouA mutant A107T/E214A acted like a para-enzyme and formed 21% m-nitrophenol and 79% p-nitrophenol from nitrobenzene, and it formed p-cresol as the major product (93%) from toluene. In contrast, variant T201G had a slight shift in product distribution from toluene. Here, we show that variant A107T/E214A also acted like a para-enzyme for o-xylene oxidation, since it formed 97% 3,4-DMP and 3% 2,3-DMP from o-xylene, whereas wild-type ToMO formed 82% 3,4-DMP and 18% 2,3-DMP from o-xylene. Similarly, variant A107T/E214A favored hydroxylation of naphthalene in the C2 position (like wild-type T4MO; Tao et al. 2004a) and produced 3-fold more 2-naphthol than wild-type ToMO (36% vs 12%). In contrast, the T201S mutation enhanced 2,3-DMP formation from o-xylene 2-fold and altered the oxidation regiospecificity of naphthalene 3-fold, and the T201G mutation caused ToMO to be even more specific, by forming 100% 3,4-DMP from o-xylene and >99% 1-naphthol from naphthalene (Table 3).
These results corroborate that positions A107 and T201, which are respectively ∼6 Å and ∼7 Å away from the diiron center, are important amino acids for converting the unspecific ToMO to a more regiospecific enzyme. In contrast, gate residues I100 and E214 of ToMO are most important for controlling the rate of oxidation of chlorinated aliphatics and nitroaromatics (residues which are ∼7 Å and 23 Å away, respectively, from the diiron center). I100 can also influence the regiospecificity of the products (e.g., benzene, methyl aromatics, nitroaromatics), but E214 cannot, due to its distance from the active site. These results should also apply to the intractable sMMO.
References
Baggi G, Barbieri P, Galli E, Tollari S (1987) Isolation of a Pseudomonas stutzeri strain that degrades o-xylene. Appl Environ Microbiol 53:2129–2132
Bertoni G, Bolognese F, Galli E, Barbieri P (1996) Cloning of the genes for and characterization of the early stages of toluene and o-xylene catabolism in Pseudomonas stutzeri OX1. Appl Environ Microbiol 62:3704–3711
Bradley PM, Chapelle FH (1998) Effect of contaminant concentration on aerobic microbial mineralization of DCE and VC in stream-bed sediments. Environ Sci Technol 32:553–557
Cafaro V, Scognamiglio R, Viggiani A, Izzo V, Passaro I, Notomista E, Piaz FD, Amoresano A, Casbarra A, Pucci P, Donato AD (2002) Expression and purification of the recombinant subunits of toluene/o-xylene monooxygenase and reconstitution of the active complex. Eur J Biochem 269:5689–5699
Canada KA, Iwashita S, Shim H, Wood TK (2002) Directed evolution of toluene ortho-monooxygenase for enhanced 1-naphthol synthesis and chlorinated ethene degradation. J Bacteriol 184:344–349
Carter SR, Jewell WJ (1993) Biotransformation of tetrachloroethylene by anaerobic attached-films at low temperatures. Water Res 27:607–615
Chauhan S, Barbieri P, Wood TK (1998) Oxidation of trichloroethylene, 1,1-dichloroethylene, and chloroform by toluene/o-xylene monooxygenase from Pseudomonas stutzeri OX1. Appl Environ Microbiol 64:3023–3024
Dolfing J, Van den Wijngaard AJ, Janssen DB (1993) Microbiological aspects of the removal of chlorinated hydrocarbons from air. Biodegradation 4:261–282
Elango N, Radhakrishnan R, Froland WA, Wallar BJ, Earhart CA, Lipscomb JD, Ohlendorf DH (1997) Crystal structure of the hydroxylase component of methane monooxygenase from Methylosinus trichosporium OB3b. Protein Sci 6:556–568
He J, Ritalahti KM, Yang K-L, Koenigsberg SS, Löffler FE (2003) Detoxification of vinyl chloride to ethene coupled to growth of an anaerobic bacterium. Nature 424:62–65
Hylckama Vlieg JET van, Koning W de, Janssen DB (1996) Transformation kinetics of chlorinated ethenes by Methylosinus trichosporium OB3b and detection of unstable epoxides by on-line gas chromatography. Appl Environ Microbiol 62:3304–3312
McCarty PL (1997) Breathing with chlorinated solvents. Science 276:1521–1522
McClay K, Fox BG, Steffan RJ (1996) Chloroform mineralization by toluene-oxidizing bacteria. Appl Environ Microbiol 62:2716–2722
Moore JC, Arnold FH (1996) Directed evolution of a para-nitrobenzyl esterase for aqueous–organic solvents. Nat Biotechnol 14:458–467
Pikus JD, Studts JM, McClay K, Steffan RJ, Fox BG (1997) Changes in the regiospecificity of aromatic hydroxylation produced by active site engineering in the diiron enzyme toluene 4-monooxygenase. Biochemistry 36:9283–9289
Pikus JD, Mitchell KH, Studts JM, McClay K, Steffan RJ, Fox BG (2000) Threonine 201 in the diiron enzyme toluene 4-monooxygenase is not required for catalysis. Biochemistry 39:791–799
Rosenzweig AC, Brandstetter H, Whittington DA, Nordlund P, Lippard SJ, Frederick CA (1997) Crystal structures of the methane monooxygenase hydroxylase from Methylococcus capsulatus (Bath): implications for substrate gating and component interactions. Proteins Struct Funct Genet 29:141–152
Rui L, Kwon YM, Fishman A, Reardon KF, Wood TK (2004) Saturation mutagenesis of toluene ortho-monooxygenase from Burkholderia cepacia G4 for enhanced 1-naphthol synthesis and chloroform degradation. Appl Environ Microbiol 70:3246–3252
Ryoo D, Shim H, Canada K, Barbieri P, Wood TK (2000) Aerobic degradation of tetrachloroethylene by toluene-o-xylene monooxygenase of Pseudomonas stutzeri OX1. Nat Biotechnol 18:775–778
Ryoo D, Shim H, Arenghi FLG, Barbieri P, Wood TK (2001) Tetrachloroethylene, trichloroethylene, and chlorinated phenols induce toluene-o-monooxygenase activity in Pseudomonas stutzeri OX1. Appl Microbiol Biotechnol 56:545–549
Sakamoto T, Joern JM, Arisawa A, Arnold FH (2001) Laboratory evolution of toluene dioxygenase to accept 4-picoline as a substrate. Appl Environ Microbiol 67:3882–3887
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning, a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
Sanger F, Nicklen S, Coulson AR (1977) DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA 74:5463–5467
Sazinsky MH, Bard J, Donato AD, Lippard SJ (2004) Crystal structure of the toluene/o-xylene monooxygenase hydroxylase from Pseudomonas stutzeri OX1: insight into the substrate specificity, substrate channeling and active site tuning of multicomponent monooxygenases. J Biol Chem 279:30600–30610
Shim H, Wood TK (2000) Aerobic degradation of mixtures of chlorinated aliphatics by cloned toluene-o-xylene monooxygenase and toluene o-monooxygenase in resting cells. Biotechnol Bioeng 70:693–698
Shim H, Ryoo D, Barbieri P, Wood TK (2001) Aerobic degradation of mixtures of tetrachloroethylene, trichloroethylene, dichloroethylenes, and vinyl chloride by toluene-o-xylene monooxygenase of Pseudomonas stutzeri OX1. Appl Microbiol Biotechnol 56:265–269
Stemmer WPC (1994) DNA shuffling by random fragmentation and reassembly: in vitro recombination for molecular evolution. Proc Natl Acad Sci USA 91:10747–10751
Tao Y, Bentley WE, Wood TK (2004a) Regiospecific oxidation of naphthalene and fluorene by toluene monooxygenases and engineered toluene 4-monooxygenase of Pseudomonas mendocina KR1. Biotechnol Bioeng (in press)
Tao Y, Fishman A, Bentley WE, Wood TK (2004b) Oxidation of benzene to phenol, catechol, and 1,2,3-trihydroxybenzene by toluene 4-monooxygenase of Pseudomonas mendocina KR1 and toluene 3-monooxygenase of Ralstonia pickettii PKO1. Appl Environ Microbiol 70:3814–3820
Vardar G, Wood TK (2004) Protein engineering of toluene-o-xylene monooxygenase from Pseudomonas stutzeri OX1 for synthesizing 4-methylresorcinol, methylhydroquinone, and pyrogallol. Appl Environ Microbiol 70:3253–3262
Vardar G, Wood TK (2005) Alpha subunit positions methionine 180 and glutamate 214 of Pseudomonas stutzeri OX1 toluene-o-xylene monooxygenase influence catalysis. J Bacteriol 187:1511–1514
Vardar G, Barbieri P, Wood TK (2005a) Chemotaxis of Pseudomonas stutzeri OX1 and Burkholderia cepacia G4 toward chlorinated ethenes. Appl Microbiol Biotechnol 66:696–701
Vardar G, Ryu K, Wood TK (2005b) Protein engineering of toluene-o-xylene monooxygenase from Pseudomonas stutzeri OX1 for oxidizing nitrobenzene to 3-nitrocatechol, 4-nitrocatechol, and nitrohydroquinone. J Biotechnol 115:145–156
Acknowledgement
This study was supported by the U.S. Army Research (DAAD19-00-1-0568).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Vardar, G., Wood, T.K. Protein engineering of toluene-o-xylene monooxygenase from Pseudomonas stutzeri OX1 for enhanced chlorinated ethene degradation and o-xylene oxidation. Appl Microbiol Biotechnol 68, 510–517 (2005). https://doi.org/10.1007/s00253-005-1923-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-005-1923-4