Abstract
Microbial community DNA was extracted from activated sludge samples taken from a chemical bioflocculation process and a chemical coagulation process in Shanghai, China. 16S rDNA of ammonia-oxidizing bacteria (AOB)was amplified by nested polymerase chain reaction and fingerprinted by denaturing gradient gel electrophoresis for microbial structure analysis. The Shannon diversity index of each sample was determined. The results indicated that the microbial structure of AOB in chemical bioflocculation process was comparable at two operational conditions. The ammonia-oxidizing bacterial communities were similar in three channels of the chemical bioflocculation process and in three serial tanks in the chemical coagulation process at the same condition. The diversity of microbial structures in the chemical bioflocculation process was higher than in the chemical coagulation process, in which the microbial structure was similar to that in the influent. Although the microbial study provides insights to the nitrification removal, higher microbial diversity of AOB does not necessarily mean higher ammonia oxidization. Molecular analysis should be combined with chemical assays to optimize operational conditions.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
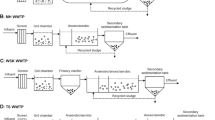
Chemical bioflocculation is an enhanced primary wastewater treatment process in which particulates are precipitated through chemical coagulation and biological flocculation. The chemical bioflocculation process consists of three plug-flow channels with declining aeration rate. Air was supplied to the channels through microhole aeration pipes, and the aeration rate can be adjusted separately in each channel. The total hydraulic retention time in all three channels was about 30 min. The process produces less sludge and is cost-effective (Wu et al. 2003; Zhang et al. 2001).
Although primary treatment processes were traditionally designed to remove particulates in wastewater, the deteriorating environments have required researchers to explore the potential of these inexpensive processes for biological oxygen demand/chemical oxygen demand (BOD/COD) and nutrients removal. Among the nutrients, nitrogen (N) is a major element to cause eutrophication. Nitrification and denitrification are the main mechanisms of N removal in wastewater. However, before nitrification takes place, ammonia-N has to be oxidized by AOB. Research indicated that the distribution of AOB is related to nitrification efficiency of two treatment processes, and the quantity of AOB is proportional to the ammonia oxidization rate (You et al. 2003). AOB are in the category of chemolithotroph. The difficulty in culturing chemolithotrophic bacteria has limited our knowledge of these bacteria. The development of molecular biotechnology in recent years, however, provides a new tool to study AOB in environmental samples. Indeed, many researchers have taken advantage of various molecular techniques available to study AOB in various wastewater treatment processes. For instance, (Sofia et al. 2004) investigated the nitrogen-removing bacterial community in an anoxic/oxic membrane bioreactor (A/O MBR) using fluorescence in situ hybridization (FISH) technique. The results indicated that Nitrosospira spp. and Nitrospira spp. were the most dominant of the ammonia- and nitrite-oxidizing group, and their preferred dissolved oxygen (DO) was from 2 to 5 mg/l (Sofia et al. 2004). In another study, terminal restriction fragment length polymorphism (T-RFLP) followed by cloning and sequencing were applied to study the ammonia-oxidizing bacterial population in an aerated-anoxic Orbal process, and the study suggested that Nitrosospira-like organisms were one of the major contributors to ammonia oxidization (Park et al. 2002). The study further revealed that the AOB groups varied with ambient temperature. Detailed review in this aspect could be found in the work of (Limpiyakorn et al. 2004), in which the distribution of AOB in 12 different sewage-activated sludge systems was evaluated.
Chemolithotroph usually requires longer hydraulic retention time to grow in wastewater treatment processes due to their competitive disadvantage with other microorganisms for carbon source. The hydraulic retention time in the chemical bioflocculation process was designed to be as short as 30 min in this case. The growth and the distribution of AOB in such a short period of time were unclear. Besides, it was unclear how the AOB bacterial groups would change when operational conditions are changed. Therefore, the purpose of this work was to determine the phylogenic positions of AOB in the chemical bioflocculation process at different operational conditions. For comparison, the phylogenic analysis of AOB in a traditional chemical coagulation process was also performed. The chemical coagulation process included three serially connected tanks. In each tank, the wastewater was completely mixed by mechanical force.
Materials and methods
Activated sludge samples
Activated sludge samples were obtained from the pilot-scale chemical bioflocculation and coagulation reactors located in Anting wastewater treatment plant, Shanghai. The detailed configuration of the chemical bioflocculation reactor was described elsewhere (Xia et al. 2005). Mixed liquor suspended solids (MLSS) were collected with plastic sampling bottles at two typical operational conditions in each process, and the detailed sampling positions are shown in Fig. 1. The first sampling occurred on 20 May 2004 and the second on 5 June 2004. Detailed information of DO, BOD, and other parameters in each condition are listed in Table 1. The sampling points were located at the end of each channel. The samples were allowed to precipitate for a few hours in a cooler to remove large particles. After the samples were transported to the lab at Tongji University, the supernatant was centrifuged at 12,000×g for 1 min, and the pellet was stored at −20°C for future use.
DNA extraction and purification
DNA extraction was slightly modified from the protocols published in literature (Muriel et al. 1999; Purohit et al. 2003; Sam Brook 1996; Tsai and Olson 1991). The community DNA was extracted by adding approximately 100 mg activated sludge into a 1.5-ml sterilized microcentrifuge tube. Extraction buffer (0.8 ml) and proteinase K (10 μl, 10 mg/ml) were added to the microcentrifuge tube, and the tube was incubated on a shaker (200 rpm) at 37°C for 30 min. Twenty percent sodium dodecyl sulfate (SDS) (100 μl) was added to the tube, and the tube was incubated at 37°C for 2 h. The tube was shaken every 15–20 min during the period of incubation. Following the incubation, the tube was centrifuged at 8,000×g for 10 min, and the supernatant was transferred to a new 1.5-ml tube. The same volume of chloroform and isoamyl alcohol (ratio 24:1) was added to the supernatant, and the mixture was centrifuged for 10 min at 8,000×g. The supernatant was transferred to another 1.5-ml tube, and 0.6 vol isopropanol was added. The tube was left in the room temperature for 1 h and centrifuged again for 10 min at 12,000×g. The supernatant was dumped, and 1 ml 70% ethanol was added to the pellet. The tube was centrifuged for 10 min at 12,000×g, and the liquid was removed. After the pellet was dry, 300 μl TE buffer was added to the tube to resuspend DNA. The community DNA was examined with 0.8% agarose electrophoresis. The marker, ë-HindIII digest, was purchased from Bao Biotechnology Company (Dalian).
Polymerase chain reaction (PCR) amplification
PCR primers and thermo-cycling programs
All PCR reactions were carried out in 50 μl volume. PCR mastermix solution was prepared as follows: 0.2 M of each primer, 200 μM dNTP, 1 U/50 μl Taq DNA polymerase, 1× PCR buffer, 2 mM MgCl2, and (μl) distilled water. The primers (Boya Biotechnology Engineering Company, Shanghai) and thermo-cycling conditions are listed in Table 2. PCR products were examined with 1% agarose gel electrophoresis. The DNA marker DL2000 was obtained from Bao (Dalian).
Condensation of PCR products
The first PCR was carried out with primers CTO189fAB, CTO189fC (mixture of two forward primers) and CTO653r. The first-round PCR product was amplified in six tubes simultaneous with the second amplification that was done with the primers P338f and P518r. The products were then combined together and sealed with Parafilm. The combined product was frozen at −70°C overnight and dried with a freeze dryer for 4 h. The dried powder was dissolved in 15 μl deionized (DI) water and stored at −20°C.
Denaturing gradient gel electrophoresis (DGGE)
PCR products after second amplification were separated with DGGE through Decode System (Bio-Rad Laboratories) on an 8% acrylamide 1-mm gel and 0.5× TAE buffer [20 mM Tris, 10 mM acetic acid, 0.5 mM ethylenediaminetetraacetic acid (pH 8.0)]. The gradient of the gel was from 45 to 60% (100% denaturant stock solution was made of 7 M urea and 40% deionized formamide). Gels were run at 60°C for 16 h at 40 V. Gels were photographed after staining with 1× TAE buffer containing 0.5 mg/l ethidium bromide.
Sequencing of DGGE fractions
The gel fraction with targeted DNA was cut off and transferred to a microcentrifuge tube. Thirty microliters DI water was added, and the product was stored overnight at 4°C. The product was used as a PCR template and amplified. The PCR products were sequenced in Boya (Shanghai). The sequences were examined in blastn for similarity comparison. The most similar sequences were chosen to draw phylogenic tree with ClustalX and Phylip. DGGE products were analyzed with Smartview, and biodiversity of the samples was calculated with the Shannon biodiversity index formula, as described in the following equation (Cox 1979):
,where S is the Shannon biodiversity index, Pi is the ratio of one specific group of bacteria to the total microorganisms in the samples, and i is the total number of microbial species in the samples.
Results and discussion
Total community DNA extraction and nested PCR
The total community DNA was successfully extracted with the modified protocol as indicated by gel analysis. In the new method, only 100–200 mg of sample was needed. The small amount of sample shortened the pretreatment time and avoided complicated extraction procedures and errors in extractions. The size of the raw DNA was about 23 kb after the analysis with Smartview. After dilution, the DNA samples were successfully amplified with PCR, showing that the modified extraction method worked well in extracting and purifying DNA from activated sludge. The DNA size after the first amplification was about 460 bp, and the DNA size after the second amplification was about 220 bp. Both the PCR products are the targeted DNA fractions, and no unspecific bands were observed. After condensation, the PCR products of the second amplification were successfully fingerprinted with DGGE as demonstrated in Fig. 2. Multiple bands could be seen in all the samples, indicating that the AOB were abundant in the samples.
DGGE pattern of AOB. Samples 1, 2, and 3, chemical bioflocculation process (operation condition 1); samples 4, 5, and 6, chemical coagulation process (operation condition 1); samples 7, 8, and 9, chemical bioflocculation process (operation condition 2); samples 10, 11, and 12, chemical coagulation process(operation condition 2); sample 13, influent at operation condition 2
The microbial structure of AOB changed partly at different operational conditions. In the study of Park et al. (2002), ammonia oxidizing bacterial populations in an aerated–anoxic Orbal process switched when operational parameters were changed. Two major parameters they studied were temperature and DO. In this work, the temperatures and the chemical dosages of polyaluminum-ferric chloride (PAFC) were the same at the two sampling conditions. Hence, the difference of microbial communities at different treatment processes might result from the difference of the DO and the sludge returning ratio that is the main difference between chemical bioflocculation process and chemical coagulation process, which resulted in different MLSS and their microbial characteristics in the reactors. At the same operational condition, the microbial structures were similar in the three channels of the chemical bioflocculation reactors (samples 1, 2, 3 and samples 7, 8, 9) and three tanks of coagulation processes (samples 4, 5, 6 and samples 10, 11, 12; Table 3 and Fig. 2). The fingerprinting of the three samples had similar bands and densities, demonstrating that microorganisms were distributed evenly in the channels. The microbial structures were different in chemical bioflocculation reactors and chemical coagulation reactors. Both reactors had their specific bands in the fingerprinting figures (Table 3). The result was in accordance with earlier studies that the AOB community was contingent upon the treatment processes.
The Shannon diversity index was calculated with the Shannon formula and is shown in Fig. 3. The diversity index in the chemical bioflocculation process at the second operational condition (sample 7, 8, 9) was higher than at the first one (sample 1, 2, 3), As more aiding flocculants were supplied at condition 2, the biodiversity index only increased slightly, indicating that the addition of aiding flocculent polyacrylamide (PAM) at the range of our experiment on the biodiversity of AOB in the chemical bioflocculation reactors was negligible. The biodiversity index in the three tanks of chemical coagulation reactors did not differ significantly from each other. However, the biodiversity index decreased from 1.05 in the influent to 1.01, 0.91, and 0.98 at the second condition. The biodiversity index in the first condition was also lower than that of the influent. The decrease was contributed to the addition of coagulants to the reactors, which inhibited some species of microorganisms. The diversity of AOB in the chemical bioflocculation reactors was higher than in the chemical coagulation reactors as demonstrated by the Shannon index (Fig. 3). The analysis of influent at the second operational conditions showed that the microbial structures in the influent and the chemical coagulation reactors were practically the same (Fig. 2). The high degree of similarity was ascribed to the fact that the chemical coagulation reactors were not aerated and no sludge was returned to the reactors. Therefore, all the microorganisms in the chemical coagulation reactors were from the influent.
Shannon diversity index of AOB in all samples. Samples 1, 2, and 3, chemical bioflocculation process (condition 1); samples 4, 5, and 6, chemical coagulation process (condition 1); samples 7, 8, and 9, chemical bioflocculation process (condition 2); samples 10, 11, and 12, chemical coagulation process (condition 2); sample 13, influent at operation condition 2
Of all the influencing factors, oxygen is probably the single most important factor which affects the AOB. Although traditional wisdom has it that nitrification will not happen efficiently unless DO is maintained above 1.5 mg/l (Wanner 1997), more recent studies have demonstrated that nitrification could take place at anoxic conditions (Park et al. 2002). The fact that AOB were detected in the third tank of the chemical coagulation process where the DO was as low as 0.3 mg/l underpinned the latest findings. Because the diversity index in the third tank was higher than in the second tank, the detection of AOB could not be simply accounted for by the carryovers from the second thank. Other than DO, the main difference of the two operational conditions was the sludge returning rate. The higher returning rate was thought to be a contributing factor to the lower removal rate of ammonia-N, although it did not switch the microbial community significantly. The caveat was that the higher returning rate brought back more non-AOB which competed with AOB for substrates and contained AOB activities. Because DNA did not reveal the activities of bacteria, other analyses such as rRNA analysis and lipid analysis would be desirable. Nevertheless, the study was insightful for the operations and maintenance of the treatment processes to maximize the N removal efficiency.
Sequencing and comparison of DGGE fragments
To further investigate the ammonia-oxidizing bacterial community, several typical bands (bands 1–13) were sequenced, and the results were aligned with previously published sequences in the NCBI database (Table 4). Some sequences were analyzed with ClustalX and PhyloDraw software, and the phylogenic tree is shown in Fig. 4.
The 13 bands chosen for sequencing showed more than 98% similarity to those identified bacteria, indicating that the primers used in the first round of PCR were specific. All 13 sequenced species were from Betaproteobacteria. Bands 1, 2, 3, 5, and 8 were assigned to the family of Nitrosomonadaceae nitrosomonas in the order of Nitrosomonadales, and bands 4 and 6 were assigned to the family of Nitrosomonadaceae nitrosospira in the same order. Bands 9 and 11 were from the family of Rhodocyclaceae dechloromonas, and band 10 fell in the family of Rhodocyclaceae. Bands 7, 12, and 13 stood for bacteria from Betaproteobacteria, but could not be assigned to any known order. The sequences indicated higher percentage of Nitrosomonas than Nitrosopira. Nitrosopira is widespread in the environment and is primarily associated with low DO concentration environments such as rice paddies and sediments. The operational conditions in this work, where DO was above 1 mg/l except for the third tank at the second condition, favored the growth of Nitrosomonas.
In conclusion, the results demonstrated that AOB were abundant in both chemical bioflocculation and coagulation processes. The AOB community structures were similar in the subunits of each process at the same condition. The community structures and diversity changed when operational conditions were changed, but the difference was insignificant. The microbial structure in the chemical bioflocculation was less susceptible to the addition of aiding flocculants and the sludge returning rate; however, those factors indeed had some impacts on the removal of ammonia-N. It is likely that microbial structures varied at varying addition amounts of adding flocculants and sludge returning rates, but the difference was smaller than the detection resolution of DGGE. The molecular analysis should combine with chemical analysis to provide more comprehensive knowledge on the operation of wastewater treatment processes.
References
Cox GW (1979) General ecology experimental manual. Science, Beijing, pp 120–124
Limpiyakorn T, Shinohara Y, Kurisu F, Yagi O (2004) Distribution of ammonia-oxidizing bacteria in sewage activated sludge: analysis based on 16S rDNA sequence. Water Sci Technol 50:9–14
Muriel B, Wafa A, Vincent U, Thierry H (1999) DNA extraction from activated sludges. Curr Microbiol 38:315–319
Park H, Regan J, Noguera D (2002) Molecular analysis of ammonia-oxidizing bacterial populations in aerated–anoxic orbal processes. Water Sci Technol 46:273–280
Purohit H, Kapley A, Moharikar A, Narde G (2003) A novel approach for extraction of PCR-compatible DNA from activated sludge samples collected from different biological effluent treatment plants. J Microbiol Methods 52:315–323
Sam Brook J (1996) Molecular clone experimental manual. Science, Beijing, pp 919–929
Sofia A, Liu W, Ong S, Ng W (2004) In-situ characterization of microbial community in an A/O submerged membrane bioreactor with nitrogen removal. Water Sci Technol 50:41–48
Tsai Y, Olson B (1991) Rapid method for direct extraction of DNA from soil and sediments. Appl Environ Microbiol 57:1070–1074
Wanner J (1997) Microbial population dynamics in biological wastewater treatment plants. In: Cloete TE, Muyima NYO (eds) Microbial community analysis: the key to the design of biological wastewater treatment systems. IAQW, London, pp 35–59
Wu Z, Xia S, Yang D, Gao T (2003) Comparative research of chemical and biological flocculation process to municipal wastewater. Chongqing Environmental Sciences 25:12–14
Xia S, Wang F, Fu Y, Yang D, Ma X (2005) Biodiversity analysis of microbial community in the chem-bioflocculation treatment process. Biotechnol Bioeng 89(6):656–659
You S, Chuang S, Ouyang C (2003) Nitrification efficiency and nitrifying bacteria abundance in combined AS-RBC and A2O systems. Water Res 37:2281–2290
Zhang Z, Wei Q, Zhang G (2001) Technology and development trend of the enhanced primary treatment of municipal wastewater. Chongqing Environmental Sciences 23:46–49
Øvreas L, Forney L, Daae F, Torsvik V (1997) Distribution of bacterioplankton in meromictic lake Saelevannet, as determined by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl Environ Microbiol 63:3367–3373
Acknowledgements
This article is supported by the National High Technology Research and Development Program of China (863 Program) No. 2002AA601320 and the Shuguang Program of Shanghai.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Xia, S., Shi, Y., Fu, Y. et al. DGGE analysis of 16S rDNA of ammonia-oxidizing bacteria in chemical–biological flocculation and chemical coagulation systems. Appl Microbiol Biotechnol 69, 99–105 (2005). https://doi.org/10.1007/s00253-005-0035-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-005-0035-5