Abstract
The lactonase gene of Fusarium oxysporum was expressed in Aspergillus oryzae for optical resolution of dl-pantoyl lactone. When the chromosomal gene encoding the full-length form of the lactonase, which has its own NH2-terminal signal peptide, was introduced in the host cells, the resulting transformant produced an enzyme of 46,600 Da, which corresponded to the wild-type enzyme. In contrast, A. oryzae transformed with the cDNA coding the mature enzyme produced a protein of 41,300 Da. Deglycosylation analysis with an endoglycosidase revealed that the difference in molecular mass arose from the different sugar contents of the recombinant enzymes. The mycelia of the transformant were used as a catalyst for asymmetric hydrolysis of dl-pantoyl lactone. The initial velocity of the asymmetric hydrolysis reaction catalyzed by the transformant was estimated to be 30 times higher than that by F. oxysporum. When the mycelia of the transformant were incubated with a 20% dl-pantoyl lactone solution for 4 h, 49.9% of the racemic mixture was converted to d-pantoic acid (>95% ee).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lactonases, which catalyze the reversible or irreversible hydrolysis of lactone compounds, belong to the esterase family of enzymes. We previously reported that a fungus, Fusarium oxysporum, produces a lactonase that catalyzes the stereoselective hydrolysis of d-pantoyl lactone, which is an important chiral building block for the commercial production of calcium d-pantothenate and its derivatives (Shimizu et al. 1992). This asymmetric reaction has been used for the optical resolution of racemic pantoyl lactone (Kataoka et al. 1995a, b). A novel d-pantothenate synthesis process, which involves the enzymatic resolution of dl-pantoyl lactone with Fusarium lactonase, has been in practical use since 1999. It has been shown that the new process is highly satisfactory not only from an economic aspect but also from an environmental one (Honda et al. 2002). Nowadays, about 30% of the world production of calcium d-pantothenate occurs through this chemo-enzymatic process.
Like this successful example, a number of enzymatic conversion processes have been applied to the commercial synthesis of organic compounds. However, while enzymes show strict substrate specificity and high reactivity under moderate conditions, the enzymatic synthesis of organic compounds is generally inferior to chemical synthesis in terms of the rate of reaction and the concentration of the product. To overcome these disadvantages, recombinant microorganisms, in which an enzyme gene is overexpressed, have been sometimes used. For example, the enzymatic production of chiral alcohols through the asymmetric reduction of prochiral carbonyl compounds with Escherichia coli transformants overexpressing carbonyl reductase genes are recent successful examples of the use of this process (Kataoka et al. 1997, 1999; Kizaki et al. 2001; Yasohara et al. 2001).
We have cloned and sequenced the whole length of the Fusarium lactonase gene, revealing that this gene includes five introns and a function-unknown NH2-terminal signal peptide-coding region (Shimizu et al. 2003; Fig. 1). The signal peptide consists of 20 amino acid residues which were not found in the NH2-terminal amino acid sequencing of the purified mature enzyme. We introduced the cDNA encoding the mature lactonase into E. coli under the control of the lac promoter, but the lactonase productivity of the recombinant E. coli was not sufficient (data not shown). A large fraction of the recombinant enzyme accumulated as inclusion bodies and the low level of production of the recombinant enzyme was considered to be due to the complicated post-translational modification of the eukaryotic protein. So, it was supposed that the enzyme gene might be expressed successfully using a eukaryotic microorganism, such as a yeast or a fungus, as a heterologous host.
In recent years, filamentous fungi have been found to be suitable hosts for the production of heterologous proteins. In particular, Aspergillus oryzae has been industrially used in Japan for sake and soy sauce manufacture for a long time and therefore is considered to be quite safe as a recombinant host. Here, we describe the high-level expression of a Fusarium lactonase gene in A. oryzae, a preliminary structural analysis of the recombinant enzyme, and application of the recombinant microorganism to the optical resolution of racemic pantoyl lactone.
Materials and methods
Materials
F. oxysporum (AKU 3702; Graduate School of Agriculture, Kyoto University, Japan) was used as the source of the lactonase gene. E. coli JM109 was used for plasmid construction. A. oryzae (niaD−), which lacks nitrate reductase (niaD) activity for nitrate assimilation, and a plasmid, pNEN142, were used in transformation experiments as the recipient strain and high-level expression vector, respectively. Plasmid pNEN142 contains a niaD gene as a selectable marker, A. oryzae α-glucosidase terminator T-agdA, and an improved A. oryzae enolase promoter, P-enoA142 (Toda et al. 2001), into which 12 copies of the cis-element (region III) interacting with positive regulators for the transcription of A. oryzae genes (Minetoki et al. 1998) were inserted. Restriction enzymes and other DNA-modifying enzymes were products of Takara-Bio Co. (Kyoto, Japan), unless otherwise specified.
Media and culture conditions
F. oxysporum was cultured in medium consisting of 10 g of glycerol, 5 g of Polypepton (Daigo, Osaka, Japan), 5 g of yeast extract (Oriental Yeast, Tokyo, Japan), and 5 g of corn steep liquor (Shono Starch, Suzuka, Japan) per liter of tap water (pH 6.0). The minimal and complete media for A. oryzae were, respectively, Czapek–Dox medium (3% sucrose, 0.2% NaNO3, 0.1% KH2PO4, 0.05% KCl, 0.05% MgSO4·7H2O, 0.001% FeSO4·7H2O, pH 5.5) and dextrin–peptone medium (2% dextrin, 1% Polypepton, 0.5% yeast extract, 0.5% K2HPO4, 0.05% MgSO4·7H2O, pH 5.8). For production of the lactonase, the recombinant microorganisms were grown at 28°C for 1 day with shaking (300 rpm) in dextrin–peptone medium. Aliquots (10 ml) of the cultures were transferred to 2-l shaking flasks containing 500 ml of medium. After cultivation for 4 days at 28°C with shaking (120 rpm), the cells were harvested by filtration and subjected to further analysis.
Construction of expression plasmids for lactonase production
For expression of the Fusarium lactonase gene, we constructed two plasmids. One plasmid, pNEN-PC, which includes the lactonase cDNA lacking the NH2-terminal signal peptide-coding region, was constructed as follows: total RNA was isolated from F. oxysporum cells ground in liquid nitrogen by the arabinogalactan–protein complex method using ISOGEN (Nippon Gene Co., Tokyo, Japan). The 1.2-kb DNA fragment was amplified by RT-PCR with an Access RT-PCR system (Promega, Madison, Wis.), using total RNA as the template. The oligonucleotides used as primers were FusLac0 (5′-TAGTCGACATGGCTAAGCTTCCTTCTACG-3′) containing a SalI site (in italics) and FusLac2 (5′-AATCTAGACTAATCATAGAGCTTGGGAC-3′) containing a XbaI site (in italics). The amplifying reaction was performed under the conditions recommended by the manufacturer. The PCR product was ligated with pT7Blue (Novagen, Madison, Wis.), sequenced to verify correct coding of the lactonase cDNA, and then inserted into the SalI/XbaI sites of pNEN142.
The genomic gene encoding the full-length form of the lactonase was amplified by PCR using total DNA isolated from F. oxysporum by the method of Malardier et al. (1989) as the template and oligonucleotides FusLac1 (5′-AACTCGAGATGCCTTCTTCCATTTCTGTA-3′), containing an XhoI site (in italics), and FusLac2 as the primers. The amplified DNA fragment was inserted into the SalI/XbaI sites of pNEN142 and the resulting plasmid was designated as pNEN-XG.
Transformation experiments
Transformation of E. coli was performed by the method of Hanahan (1983). A. oryzae was transformed as described by Gomi et al. (1987) and the transformants were obtained on Czapek–Dox plates containing 0.8 M NaCl to stabilize the protoplasts. Stable transformants were isolated by repeated transfer of sporulating colonies on selection plates containing Czapek–Dox medium.
Enzyme assay
The reaction mixture, comprising 1 M Tris-HCl buffer (pH 7.4), 1 mM CaCl2, 307 mM (4%, w/v) d-pantoyl lactone, and an appropriate amount of the enzyme solution, in a final volume of 250 μl, was placed in a microtube. It was then incubated at 30°C for 15 min. The reaction was terminated by the addition of 250 μl of methanol containing 2 mM EDTA and then the mixture was analyzed for pantoyl lactone and pantoic acid by HPLC as described by Shimizu et al. (1992). Whole cell activity was determined in 1 ml of the reaction mixture in a test tube. The composition of the mixture was the same as described above, except that 10 mg of wet cells were used as the catalyst instead of the enzyme solution. The reaction was performed at 30°C for 15 min with reciprocal shaking (150 rpm). After removal of the cells by centrifugation, the supernatant was analyzed by HPLC. One unit was defined as the amount of enzyme catalyzing the hydrolysis of 1 μmol/min of d-pantoyl lactone under the standard assay conditions.
SDS-PAGE, NH2-terminal amino acid sequencing, and protein determination
SDS-PAGE was performed in a 10% polyacrylamide slab gel using the Tris-glycine buffer system described by King and Laemmli (1971). The molecular mass of the subunit was calculated from its mobility relative to that of standard molecular markers. Sequencing analysis was carried out with a model 491HT pulsed liquid protein sequencer equipped with an on-line phenylthiohydantoin analyzer (Applied Biosystems, Foster, Calif.). Proteins were determined by the method of Bradford (1966) with bovine serum albumin as the standard.
Purification of the enzyme
All procedures were carried out at 4°C, unless specified otherwise. Potassium phosphate (pH 7.0) and centrifugation at 14,000 g for 10 min were usually used throughout the enzyme purification procedure. Wet cells of the pNEN-PC transformant (2.5 g) were ground in liquid nitrogen, suspended in 10 ml of 20 mM buffer containing 1 M KCl (pH 7.0), and then gently shaken at 4°C for 10 days to extract the soluble proteins. Residual cells were removed by filtration, followed by centrifugation, and the supernatant was retained as the crude extract. For preparation of a crude extract of the pNEN-XG transformant, wet cells (2.5 g) were suspended in 10 ml of 20 mM buffer containing 1 M KCl (pH 7.0) without being homogenized. Extraction of soluble proteins from the homogenate and removal of the residue were carried out in the same manner as described above. The crude extracts were dialyzed against 2 l of 20 mM buffer for 8 h.
Each extract was put on a MonoQ HR 5/5 anion-exchange column (Amersham Biosciences, Buckinghamshire, UK) equilibrated with 20 mM buffer, separately. After the column had been washed with the buffer, the enzyme was eluted with a linear gradient of NaCl (0–0.6 M, 25 ml). Each eluted enzyme solution was concentrated by ultrafiltration using a Centricon-10 (Millipore, Bedford, Mass.).
Concentrated enzyme solution (250 μl) was put on a Superdex 200 HR 10/30 gel filtration column and the enzyme was eluted with 20 mM buffer supplemented with 0.15 M NaCl. The fractions containing enzyme activity were used for characterization of the enzyme. The recombinant lactonases obtained from pNEN-PC and pNEN-XG transformants were designated as PC and XG, respectively. The wild-type lactonase (WT) was purified from F. oxysporum as reported by Shimizu et al. (1992).
Deglycosylation assay
The reaction mixture comprised a purified lactonase (WT, PC, or XG, 10 μg), an endoglycosidase (EndoHf, 0.5 μg; New England BioLabs, Beverly, Mass.), and 50 mM sodium phosphate buffer (pH 5.5) in a final volume of 25 μl and was incubated at 37°C for 40 h. After the reaction, 2.5 μl of the mixture was subjected to SDS-PAGE. Both WT and XG gave two bands on SDS-PAGE, so these bands were chromatographically separated as follows. The solvent in the reaction mixture was changed to potassium phosphate (pH 7.0) using a Fast Desalting PC 3.2/10 packed column (Amersham Biosciences). The mixture was then put on a MonoQ PC 1.6/5 column (Amersham Biosciences) and the proteins were eluted with a linear gradient of NaCl (0–0.5 M). The fractions containing the protein corresponding to the lower band observed on SDS-PAGE were combined and then subjected to mass spectral analysis as the sugar chain-free lactonase.
Mass spectral analysis
Matrix-assisted laser desorption time-of-flight mass spectrometry (MALDI-TOF MS) analysis was performed to determine the precise molecular masses of the recombinant and wild-type lactonases. For XG and WT, both the native and deglycosylated forms were analyzed. Each protein (approx. 100 pmol) was desalted and concentrated with a ZipTip C4 (Millipore). Then, a sample was mixed with a saturated solution of sinapic acid matrix in 50% acetonitrile containing 0.1% trifluoroacetic acid. The spectrum was measured with an AXIMA curved field reflectron MALDI-TOF spectrometer (Shimadzu Co., Kyoto, Japan) operated in the linear mode. Bovine serum albumin prepared in the same matrix was used to calibrate the mass spectrometer.
Preparative-scale reaction for asymmetric hydrolysis of racemic pantoyl lactone
For the reaction involving wet cells as the catalyst, 5 g (wet weight) of cells were incubated with 95 ml of a 20% (w/v) dl-pantoyl lactone aqueous solution in a thermostatically controlled reaction vessel (200 ml, 30°C), with gentle stirring. The pH of the mixture was automatically controlled at pH 6.8–7.5 with 2 M NaOH. Samples, which were withdrawn periodically, were analyzed by HPLC to determine the amounts of pantoyl lactone and products. The ratio of d-pantoic acid to l-pantoic acid was also determined by HPLC under the same conditions as those reported by Kataoka et al. (1995a)
Nucleotide sequence accession number
The sequence of the lactonase gene described here was deposited in the DDBJ/EMBL/GeneBank databases under accession number AB164638.
Results
Expression of the lactonase gene in A. oryzae
The lactonase genomic gene of F. oxysporum contains five introns and a function-unknown NH2-terminal signal peptide, which consists of 20 amino acid residues and is cleaved through processing (Fig. 1). In order to determine whether or not A. oryzae can recognize these splice junctions and this signal sequence, we constructed two expression plasmids. The pNEN-PC plasmid includes the cDNA of the lactonase, which lacks the NH2-terminal signal peptide-coding region and introns, and the pNEN-XG one includes the chromosomal DNA encoding the full-length form of the lactonase, including the signal peptide. These plasmids were introduced individually into A. oryzae and then the lactonase activity of each transformant was examined (Table 1). The recombinant A. oryzae transformed with each plasmid exhibited lactonase activity. The pNEN-XG transformants showed higher lactonase activity (mean = 1.70 units/mg dry cells). This indicates that the host strain could recognize the splice junctions in the nucleotide sequence of the Fusarium lactonase gene. The activity of the pNEN-PC transformants was 0.347 units/mg dry cells on average. The highest lactonase activity was 1.96 units/mg dry cells, which was obtained from one of the pNEN-XG transformants. No lactonase activity was detected in the negative control experiment, in which the transformant harboring only pNEN142 was assayed. The transformant that showed the highest lactonase activity among the transformants with each plasmid was subjected to further investigation as a representative of that group.
Comparison of the recombinant enzymes
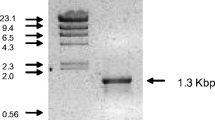
When the crude extract of the pNEN-XG transformant was subjected to SDS-PAGE, a smeared thick band corresponding to 54.2 kDa was observed. In contrast, the crude extract of the pNEN-PC transformant gave an extra band corresponding to 51.0 kDa on SDS-PAGE. These extra bands were not detected for the crude extract of the pNEN142 transformant (data not shown). Their molecular masses corresponded to those of purified lactonases XG and PC, respectively (Fig. 2, lanes 4, 6). In addition, the NH2-terminal amino acid sequences of both PC and XG was AKLPS, which is identical to that of WT. Thus, it was confirmed that these proteins were certainly lactonases and that the NH2-terminal signal peptide of XG was entirely cleaved.
Deglycosylation of the wild-type and recombinant lactonases. Lanes 1, 2 WT (0.8 μg), lanes 4, 5 XG (0.8 μg), lanes 6, 7 PC (0.8 μg) electrophoresed after incubation with (lanes 2, 5, 7) or without (lanes 1, 4, 6) EndoHf. Lanes 3, 8 were loaded with the following molecular mass standards: phosphorylase b (94 kDa), bovine serum albumin (67 kDa), ovalbumin (43 kDa), and carbonic anhydrase (29 kDa)
Purified recombinant lactonases PC and XG showed similar specific activities under the standard assay conditions (881 units/mg protein for PC, 940 units/mg protein for XG). However, a significant difference was observed in their stability. When XG was incubated at various temperatures for 30 min at pH 7.0, it was found to be stable below 45°C and 86% and 75% of the initial activity was retained at 50°C and 55°C, respectively. In contrast, the residual activity of PC at 50°C and 55°C was, respectively, 13% and 5.9% of the initial level (Fig. 3).
Effect of temperature on the stability of the recombinant lactonases. Recombinant lactonases PC and XG (1 μg) in 10 mM potassium phosphate buffer (pH 7.0) were incubated for 30 min at various temperatures. The residual activity was measured under the standard assay conditions. Relative activity is expressed as a percentage of that of the enzyme incubated at 0°C. Open squares, dotted lines PC, filled squares, solid lines XG. Results are means of triplicate assays ± standard deviation
Deglycosylation assay
When lactonases XG and WT were treated with an endoglycosidase, EndoHf, which cleaves the chitobiose core from N-linked glycoproteins, the molecular masses of both proteins were obviously decreased and each gave two bands (50.0 kDa, 48.0 kDa) on SDS-PAGE (Fig. 2). This indicates that XG possesses N-linked sugar chains like WT. The upper bands are suggested to represent proteins whose sugar chains could not be removed entirely. The proteins corresponding to the upper and lower bands could be chromatographically separated. In contrast, the molecular mass of PC was not affected by incubation with EndoHf, which suggests that lactonase PC possesses no sugar chain.
MALDI-TOF MS analysis
MALDI-TOF MS analysis allowed us to determine precisely the subunit molecular masses of the enzymes. The molecular mass of PC measured by MALDI-TOF MS, 41,320 Da, is consistent with the predicted mass (41,343 Da). Deglycosylated XG and WT gave peaks at m/z 42,149 and 42,128, respectively. When a N-linked sugar chain of a glycoprotein is digested by EndoHf, one GlcNAc molecule is left on the polypeptide chain. In the deduced amino acid sequence of the lactonase, there are four asparagine residues, which presumably bind oligosaccharide (Fig. 1). The observed values show good agreement with the molecular mass of the lactonase possessing four GlcNAc molecules and thus support this assumption.
While the oligosaccharide-free lactonases gave sharp and high intensity peaks, the native forms of WT and XG gave multiple and broad peaks. This indicates the variety of oligosaccharides linked to the lactonase molecule. The main peak was observed at m/z 45,444 for WT and at m/z 46,617 for XG.
Asymmetric hydrolysis of racemic pantoyl lactone by the recombinant A. oryzae cells
F. oxysporum and pNEN-XG transformant cells were incubated with a 20% dl-pantoyl lactone solution, the pH being controlled in the range pH 6.8–7.5. The pNEN-XG transformant exhibited much higher hydrolysis activity than F. oxysporum (Fig. 4). The initial velocity for the lactonase reaction catalyzed by the pNEN-XG transformant was determined to be 1.87 units/mg dry cells (approx. 30 times higher than that for F. oxysporum). After reaction for 4 h with the transformant, about 50% of the initial amount of dl-pantoyl lactone was hydrolyzed, i.e., the d-form in the racemic substrate was completely converted to d-pantoic acid. Actually, the optical purity of the reaction product was as high as that produced using F. oxysporum as catalyst (>95% ee).
Asymmetric hydrolysis of dl-pantoyl lactone with the lactonase. Reactions were carried out as described in the Materials and methods, using mycelia of the A. oryzae pNEN-XG transformant (filled squares) or F. oxysporum (filled circles) as the catalyst. Results are means of triplicate assays ± standard deviation
Discussion
For production of the recombinant lactonase in A. oryzae, we constructed two plasmids: pNEN-PC including the cDNA encoding the lactonase without the NH2-terminal signal peptide and pNEN-XG with the genomic DNA encoding the lactonase including the signal peptide. Although both the pNEN-PC and pNEN-XG transformants exhibited lactonase activity, structural analysis revealed that the molecular masses of the recombinant lactonases were quite different. The pNEN-XG transformant produced a recombinant lactonase that had almost the same molecular mass as that of WT, while PC was obviously smaller than both of them. A deglycosylation assay with EndoHf revealed that the difference in molecular mass between the recombinant lactonases arose from the different sugar contents of the enzymes. Although the location of the lactonase in Fusarium cells has not been revealed, the enzyme should mature via post-translational modification in the endoplasmic reticulum (ER), in which proteins are glycosylated. In the NH2-terminal signal peptide of the lactonase, successive hydrophobic amino acid residues were found (-Val-Leu-Ala-Gly-Val-Leu-Val-), which is the typical sequence of the signal peptide of an ER protein. These facts suggests that the signal peptide is essential for the immature lactonase to be transferred to the ER after translation. The recombinant lactonase produced by the pNEN-PC transformant might be translated and localized in the cytoplasm, without being transferred to the ER or glycosylated, because of the absence of the signal peptide.
This also indicates that the host strain, A. oryzae, could recognize the signal peptide of a heterologous protein. Various eukaryotic heterologous genes, not only from fungi but also from plants and animals, possessing their own signal coding regions have been successfully expressed as mature proteins in aspergilli (Archer et al. 1990; Juge et al. 1998). This indicates the wide applicability of aspergilli as heterologous hosts for eukaryotic protein production.
The lactonase activity was not detected in culture broth of either F. oxysporum or the pNEN-XG transformant of A. oryzae; and thus the mature lactonase was considered to be an intracellular protein. However, the enzyme easily leaked out from the cells by soaking the mycelia for several days in buffer. It was assumed that the enzyme localized in the juxta-membrane region of the cell and leaked out because of autolysis of the cells. Further investigations on the localization of the enzyme in the cell are required.
MALDI-TOF MS analysis gave the precise molecular mass of the enzyme. Interestingly, the mass determined by SDS-PAGE and the m/z values observed on MALDI-TOF MS are quite different from each other. For example, while the mass of PC was estimated to be 47,300 Da, MS analysis gave a peak at m/z 41,320, which is consistent with the predicted mass (41,344 Da). Some glycoproteins and acidic proteins do not bind to SDS to the same extent as a “normal” protein. Consequently, their mobility on SDS-PAGE is decreased and their molecular masses may easily be overestimated.
The physiological roles of oligosaccharides of glycoproteins are quite divergent and have not been completely revealed yet, but two of their most important roles are assumed to be maintenance of the structure of proteins and stabilization of the enzyme activities. Actually, the stability of oligosaccharide-free lactonase PC was significantly lower than that of XG.
The transformant that exhibited the highest lactonase activity (1.96 units/mg dry cells, under standard assay conditions) among all the transformants was obtained from one of the pNEN-XG transformants. When wet cells of this transformant were incubated with 20% dl-pantoyl lactone, about 50% of the initial amount of dl-pantoyl lactone, i.e., all of the d-form in the racemic substrate, was converted to d-pantoic acid in 4 h. In comparison with the current process involving F. oxysporum cells as the catalyst, the reaction time was dramatically shortened. Thus, it was shown that the pNEN-XG transformant is quite promising as a catalyst for the optical resolution of racemic pantoyl lactone. Further studies on application of the recombinant enzyme to practical operations, i.e., optimization of the reaction and culture conditions, etc., are in progress.
References
Archer DB, Jeenes DJ, Mackenzie DA, Brightwell G, Lambert N, Lowe G, Radford SE, Dobson CM (1990) Hen egg white lysozyme expressed in, and secreted from, Aspergillus niger is correctly processed and folded. Biotechnology 8:741–745
Bradford MM (1966) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal Biochem 72:248–254
Gomi K, Iimura Y, Hara S (1987) Integrative transformation of Aspergillus oryzae with a plasmid containing the Aspergillus nidulans argB gene. Agric Biol Chem 51:2549–2555
Hanahan D (1983) Studies on transformation of Escherichia coli with plasmids. J Biol Chem 166:557–580
Honda K, Kataoka M, Shimizu S (2002) Functional analyses and application of microbial lactonohydrolases. Biotechnol Bioprocess Eng 7:130–137
Juge N, Svensson B, Williamson G (1998) Secretion, purification, and characterization of barley alpha-amylase produced by heterologous gene expression in Aspergillus niger. Appl Microbiol Biotechnol 49:385–392
Kataoka M, Shimizu K, Sakamoto K, Yamada H, Shimizu S (1995a) Optical resolution of racemic pantolactone with a novel fungal enzyme, lactonohydrolase. Appl Microbiol Biotechnol 43:974–977
Kataoka M, Shimizu K, Sakamoto K, Yamada H, Shimizu S (1995b) Lactonohydrolase-catalyzed optical resolution of pantoyl lactone: selection of a potent enzyme producer and optimization of culture and reaction conditions for practical resolution. Appl Microbiol Biotechnol 44:333–338
Kataoka M, Rohani LPS, Yamamoto K, Wada M, Kawabata H, Kita K, Yanase H, Shimizu S (1997) Enzymatic production of ethyl (R)-4-chloro-3-hydroxybutanoate: asymmetric reduction of ethyl 4-chloro-3-oxobutanoate by Escherichia coli transformant expressing the aldehyde reductase gene from yeast. Appl Microbiol Biotechnol 48:699–703
Kataoka M, Yamamoto K, Kawabata H, Wada M, Kita K, Yanase H, Shimizu S (1999) Stereoselective reduction of ethyl 4-chloro-3-oxobutanoate by Escherichia coli transformant cells coexpressing the aldehyde reductase and glucose dehydrogenase genes. Appl Microbiol Biotechnol 51:486–490
King J, Laemmli UK (1971) Polypeptides of the tail fibers of bacteriophage T4. J Mol Biol 62:465–477
Kizaki N, Yasohara Y, Hasegawa J, Wada M, Kataoka M, Shimizu S (2001) Synthesis of optical pure ethyl (S)-4-chloro-3-hydroxybutanoate by Escherichia coli transformant cells coexpressing the carbonyl reductase and glucose dehydrogenase genes. Appl Microbiol Biotechnol 55:590–595
Malardier L, Daboussi MJ, Julien J, Roussel F, Scazzocchio C, Brygoo Y (1989) Cloning of the nitrate reductase gene (niaD) of Aspergillus nidulans and its use for transformation of Fusarium oxysporum. Gene 78:147–156
Minetoki T, Kumagai C, Gomi K, Kitamoto K, Takahashi K (1998) Improvement of promoter activity by the introduction of multiple copies of the conserved region III sequence, involved in the efficient expression of Aspergillus oryzae amylase-encoding genes. Appl Microbiol Biotechnol 50:459–467
Shimizu S, Kataoka M, Shimizu K, Hirakata M, Sakamoto K, Yamada H (1992) Purification and characterization of a novel lactonohydrolase, catalyzing the hydrolysis of aldonate lactones and aromatic lactones, from Fusarium oxysporum. Eur J Biochem 209:383–390
Shimizu S, Kataoka M, Sakamoto K (2003) Preparation of lactonase and its application. Jpn patent 2003-371750
Toda T, Sano M, Honda M, Rimoldi OJ, Yang Y, Yamamoto M, Takase K, Hirozumi K, Kitamoto K, Minetoki T, Gomi K, Machida M (2001) Deletion analysis of the enolase gene (enoA) promoter from the filamentous fungus Aspergillus oryzae. Curr Genet 40:260–267
Yasohara Y, Kizaki N, Hasegawa J, Wada M, Kataoka M, Shimizu S (2001) Stereoselective reduction of alkyl 3-oxobutanoate by carbonyl reductase from Candida magnoliae. Tetrahedron Asymmetry 12:1713–1718
Acknowledgements
This work was supported in part by Grants-in-Aid for Scientific Research, numbers 1729 (to K.H.) and 14360054 (to M.K.), from the Japan Society for the Promotion of Science. This work was also supported in part by the “COE for Microbial-Process Development Pioneering Future Production System”, J-3 (to S.S.), of the COE Program of the Ministry of Education, Culture, Sports, Science and Technology, Japan.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Honda, K., Tsuboi, H., Minetoki, T. et al. Expression of the Fusarium oxysporum lactonase gene in Aspergillus oryzae: molecular properties of the recombinant enzyme and its application. Appl Microbiol Biotechnol 66, 520–526 (2005). https://doi.org/10.1007/s00253-004-1758-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-004-1758-4