Abstract
An integrated study on cell growth, enzyme activities and carbon flux redistribution was made to investigate how the central metabolism of Escherichia coli changes with the knockout of genes in the oxidative pentose phosphate pathway (PPP). Mutants deficient in glucose-6-phosphate dehydrogenase and 6-phosphogluconate dehydrogenase were constructed by disrupting the zwf and gnd genes and were grown in minimal media with two different carbon sources, such as glucose or pyruvate. It was shown that the knockout of either gnd or zwf gene did not affect the cell growth rate significantly, but the cellular metabolism was changed. While the specific substrate uptake rate and the specific carbon dioxide evolution rate for either mutant grown on glucose were higher than those obtained for the parent strain, these two rates were markedly decreased in mutants grown on pyruvate. The measurement of enzyme activities implied a significant change in metabolism, when alternative pathways such as the Entner–Doudoroff pathway (EDP) and the malic enzyme pathway were activated in the gnd mutant grown on glucose. As compared with the parent strain, the activities of phosphoglucose isomerase were increased in mutants grown on glucose but decreased in mutants grown on pyruvate. The metabolic flux redistribution obtained based on 13C-labeling experiments further indicated that the direction of the flux through the non-oxidative PPP was reversed in response to the gene knockout. Moreover, the knockout of genes caused an increased flux through the tricarboxlic acid cycle in mutants grown on glucose but caused a decrease in the case of using pyruvate. There was also a negative correlation between the fluxes through malic enzyme and isocitrate dehydrogenase in the mutants; and a positive correlation was found between the fluxes through malic enzyme and phosphoenolpyruvate carboxylase.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Escherichia coli is known to be the most commonly used microorganism for the production of industrial chemicals. With the development of metabolic engineering, specific genes involved in the central metabolic pathways (CMP) can be deleted or over-expressed intentionally, so that the metabolic network can channel more carbon fluxes towards the production of the desired chemicals (Berry 1996; Lim et al. 2002). A study of the global cellular response to gene manipulation is necessary for this purpose, since such an investigation can predict the change in metabolism and evaluate the rationality of the specific gene alteration. Several authors have focused on the glycolytic pathways in E. coli and investigated the influence of gene manipulation on growth and metabolism (Emmerling et al. 1999, 2000; Sauer et al. 1999). Metabolic pathway models for pyruvate kinase- or phosphoglucose isomerase-deficient mutants have been successfully developed by combining labeling experiments with metabolite balancing (Canonaco et al. 2001; Emmerling et al. 2002).
In E. coli, the pentose phosphate pathway (PPP) is a major route for intermediary carbohydrate metabolism, besides glycolysis. It plays various roles, including the breakdown of carbon sources, the generation of reducing power (NADPH) and essential metabolites for biosynthesis, and the formation of components of the cell′s lipopolysaccharide layer (Sprenger et al. 1995). Compared with the detailed study on glycolytic pathways, little is known about how substrates are metabolized through the CMP if pathways involved in the PPP are intentionally blocked. Although there is a long history for the selection of E. coli mutants lacking PPP enzyme activities (Fraenkel 1968; Josephson and Fraenkel 1969), most research work has focused on the analysis of their growth phenotypes (Fraenkel 1968; Josephson and Fraenkel 1974). These findings revealed that the genes involved in the oxidative PPP are not essential under certain culture conditions, but they did not indicate how cells actually regulate the entire metabolic network in order to cope with disruption of the PPP. Considering several central roles of the PPP in the metabolism of E. coli, it is expected that a blockage of the PPP may cause flux rerouting and trigger a compensatory mechanism, from which a network model for the cellular response can be developed. This will provide valuable information on further engineering of the PPP for industrial purposes.
Recent advances in isotopic tracer experiment enable a more comprehensive analysis of the metabolic profile by way of labeled cellular amino acids which are detectable by mass spectrometry (MS; Wittmann and Heinzle 1999) or nuclear magnetic resonance (NMR) spectroscopy (Marx et al. 1996; Winden et al. 2001). Currently, this tracer technique in combination with extracellular flux measurements is considered to be the most powerful method for obtaining intracellular fluxes (Schmidt et al. 1999a; Dauner et al. 2001). In the case of gene-disruption mutants, however, additional enzyme activity analysis is usually required as the first step for flux calculations (Park et al. 1997; Canonaco et al. 2001), since it can bring insight into which enzymes are active and which ones are not active and whether there is an unknown reaction taking place in the specific mutants under the investigated culture condition. By combining the inspection of enzyme activities with biochemical knowledge, the network structure used for the flux analysis can be identified.
In the present study, the physiology and central carbon metabolism of E. coli mutants were investigated through the study of specific PPP-disruption mutants. The global cellular response was obtained by combining physiological, enzymatic, and metabolic studies. For this, we considered the deletion of glucose-6-phosphate dehydrogenase (G6PDH) and 6-phosphogluconate dehydrogenase (6PGDH) by disrupting the zwf and gnd genes. To understand the condition-dependent utilization of the CMP for the growth and metabolism of the mutants, we also considered the effect of different carbon sources on the metabolic phenotypes of the mutants.
Materials and methods
Strains and plasmids
E. coli BW25113 (lacI q rrnB T14 ΔlacZ WJ16 hsdR514 ΔaraBAD AH33 ΔrhaBAD LD78 ) was used as the parent strain for the construction of deletion mutants. The mutants, JWK 2011 (with the gnd gene deletion) and JWK 1841 (with the zwf gene deletion), were obtained by a one-step inactivation protocol using PCR primers (Datsenko and Wanner 2000). The gene-knockout mutants were verified by PCR and enzyme activity analysis.
Media and culture condition
For the construction of the mutants, E. coli cells were cultivated in Luria–Bertani medium. In the case of using antibiotics, ampicillin (Wako, Osaka, Japan) at 50 μg ml−1 or kanamycin (Wako) at 30 μg ml−1 was used. Batch and chemostat cultivations were performed with minimal medium prepared as described by Sauer et al. (1999) with the following concentrations of carbon sources: 5 g l−1 for batch cultivation and 4 g l−1 for chemostat culture. Batch and chemostat cultures were conducted at 37°C in a 2-l reactor (BMJ-02 PI; ABLE Co., Japan) with pH controlled at 7.0. Airflow was maintained at 0.5 l min−1 and the dissolved oxygen concentration was kept above 30% air saturation. The dilution rate (D) for chemostat culture was 0.2 h−1.
Labeling experiments were initiated after the chemostat culture reached a steady state, which was inferred from stable O2 and CO2 concentrations in the off-gas and a stable optical density at 600 nm (OD600) in the effluent medium for at least twice the residence time. The feed medium containing 4 g of unlabeled carbon source per liter was then replaced by an identical medium containing a mixture of either (per liter): (a) 0.4 g of [U-13C] glucose, 0.4 g of [1-13C] glucose, and 3.2 g of natural glucose, or (b) 0.5 g of [2-13C] sodium pyruvate and 3.5 g of natural sodium pyruvate. Biomass samples for C-13 tracer analysis were taken after one residence time in the case of glucose and two residence times in the case of pyruvate. The labeling measurements were corrected for the remaining original (nonlabeled) biomass that was present at the end of the labeling experiment (Dauner et al. 2001).
Analytical procedures
Cellular dry weight (CDW) from batch and continuous cultures was monitored by OD600 and calculated from previously determined OD-to-CDW correlations. Glucose concentration was determined using commercial kits (Wako, Japan). Acetate, pyruvate, and lactate concentrations in the culture broth were measured by HPLC (Waters Co., USA). Oxygen and carbon dioxide concentrations in the off-gas were measured by gas analyzer (DEX-2562; ABLE Co., Japan). Protein concentration was measured by the method of Lowry et al. (1951). Physiological parameters were calculated as described by Sauer et al. (1999).
The preparation of crude cell-free extracts and analysis of key enzyme activities involved in the CMP were based on standard or modified enzymological methods (Colowick 1963). EDP activities were determined from combined edd (EC 4.2.1.12) and eda (EC 4.1.2.14) reactions (Canonaco et al. 2001).
The preparation of biomass hydrolysates and recording of GC-MS (PerkinElmer) and 2-D NMR (Bruker) spectra were made as described by Szyperski (1995) and Zhao and Shimizu (2003). The Turbomass Gold program (PerkinElmer) was used for peak assignment and MS data processing. The skewing effect of natural isotopes was corrected based on the algorithm proposed by Paul Lee (1991). In the case of 2-D NMR spectra processing, the assignment of carbon signals was performed according to the protocol described by Schmidt et al. (1999b) and the WINNMR (Bruker) program was used to quantify the relative contributions of singlet, doublet, and doublet of doublet signals to the overall multiplet patterns. Since a high concentration of amino acids is a prerequisite to NMR acquisition, due to its low sensitivity, 2-D NMR was used here only in the case when glucose was used as a carbon source. It was incorporated into the algorithm together with the GC-MS analysis. For the case when pyruvate was used as a carbon source, only the sensitive GC-MS was employed in the tracer detection.
Mathematical modeling for flux calculation
The CMP network considered here was based on Neidhardt et al. (1990) and the Internet-accessible EcoCyc (http://biocyc.org:1555/server.html). In addition, enzyme activity analysis was made to identify the network structure in terms of the activities of the pathways that were not necessarily required by the parent strain. Since lactate was below the level of detection during culture, methylglyoxal bypass was not considered in the network. The flux calculation was accomplished by a full isotopomer model (Schmidt et al. 1999a) together with the previous model proposed by Zhao and Shimizu (2003). The isotopomer model was used to simulate the carbon transformation through the metabolic reaction. The best-fit intracellular fluxes were then estimated by minimization of the deviation between experimental data and the simulated values, using the iterative scheme in the minimization procedure. A set of intracellular fluxes (including net fluxes and exchange fluxes) that gives the minimum deviation can be taken as the best estimate for the intracellular flux distribution. Matlab language (Math Co., USA) was used to perform all the calculations.
A statistical analysis based on a simulation of the measurement data (Schmidt et al. 1999b) was made to extract the confidence regions for the individual flux estimates. The strategy was to generate 100 simulated measurement data sets (including GC-MS, 2-D NMR, extracellular flux data) by the addition of normally distributed measurement errors to the simulated data set, which corresponds to the best-fit flux distribution. The probability distributions of the random measurement errors were chosen according to the assumed probability distribution of the measurement errors in the actual experiment. The standard deviation for the mass distribution measurement was 0.006, which was computed from multiple GC-MS analysis. A standard deviation of 2.0% was assumed for 2-D NMR (Dauner et al. 2001) and the standard deviation values in the measurement of extracellular fluxes were assumed to be 5%. The same optimization procedure as was used for the estimation of the best-fit flux distribution was applied to estimate the flux distribution from the 100 simulated measurement data sets. Then, from the probability distribution of these flux distributions, confidence limits were obtained from the estimated parameters.
Results
Growth characteristics of the mutants
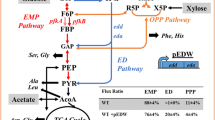
Batch cultures were used to determine the specific growth rates. The results are presented in Table 1. To identify the carbon balance during cellular growth, several yields were calculated for continuous cultures, as shown in Table 2. It is indicated that the nature of the carbon source has a significant effect on growth behavior, whereas genetic manipulation gave either similar or slightly reduced growth rates, compared with the parent strain. In spite of the similar growth characteristics, the metabolic patterns were significantly changed in response to genetic manipulation. As can be seen in Table 2, the specific substrate uptake rate and specific carbon dioxide evolution rate obtained for either mutant grown on glucose were somewhat higher than those obtained for the parent strain. This is contrary to what was observed for mutants grown on pyruvate, where these two rates were markedly decreased. Our results also revealed differences in acetate excretion between the parent strain and the mutants. Gene deletion led to increased acetate production; and this effect was more significant when glucose was used as sole carbon source.
Enzymatic analysis of mutants
Key enzymes that are located at the key branch points and those involved in NADPH formation were measured. As can be seen in Table 3, a remarkable increase in phosphoglucose isomerase (PGI) activity was observed for the glucose-grown mutant, as compared with the parent strain. The change in enzyme activities in the tricarboxylic acid (TCA) cycle is complicated, as the activity is due not only, in part, to the level of the Embden–Meyerhof pathway (EMP), but also to the level of other branches. Thus, although a significant increase in the specific activity of PGI was observed for the glucose-grown gnd mutant, the activity of TCA-related enzymes (e.g., isocitrate dehydrogenase; ICDH) was only a little higher than that of the parent strain. However, in the case of the zwf mutant, a remarkable increase in the activity of ICDH was observed.
The specific activity of PGI decreased 3.7-fold in the parent strain grown on pyruvate, as compared with the case grown on glucose. Moreover, the PGI activity was significantly lower in both mutants than in the parent strain. This phenomenon is opposite to what was observed in glucose metabolism, where the EMP pathway was more active for the mutants as a consequence of gene deletion.
It is worth noting that the enzymatic study detected two pathways that are usually not required by the parent strain grown on glucose. Namely, a high level of EDP enzymes was detected for the gnd mutant (398±29 units of activity), while the activity was below the level of detection in the parent strain. Likewise, an active pathway through malic enzyme (MAE) was observed for both mutants. To identify the route by which glucose is converted to 6-PG before entering the EDP, we measured two glucose dehydrogenase pathway (GCDP) enzymes, glucose dehydrogenase and gluconate kinase, in both the supernatant fraction of the cell extracts and in the pellet fraction. It was found that these two enzyme activities were both below the level of detection, illustrating that the tested strains do not possess the GCDP pathway.
Metabolic flux analysis of mutants
An analysis of 95% confidence limits was made for the estimated net and exchange fluxes to evaluate the quality of the estimates. Here, only those that are located at key branch points of the CMP are shown as the absolute net fluxes with lower and upper bounds (Tables 4). Others are shown in Figs. 1 and 2 as the best-fit flux distribution relative to the specific uptake rates of substrates. As indicated by the deviation between the experimental and predicted data (see Electronic Supplementary Material), there was no set of measurements with a particularly large deviation between the predicted and the measured signals, proving that mathematical modeling is reliable for characterizing these metabolic fluxes in vivo.
Metabolic flux distributions in chemostat culture of glucose-grown Escherichia coli parent strain (upper values), gnd (middle values), and zwf (lower values) mutants at D=0.2 h−1. Fluxes are given relative to the specific glucose consumption rate and are expressed as the net fluxes. The exchange coefficients are shown in brackets for the reactions that were considered reversible. Negative values indicate the reversed pathway direction. See text for codes used
Metabolic flux distribution in chemostat culture of pyruvate-grown E. coli parent strain (upper values), gnd (middle values), and zwf (lower values) mutants at D=0.2 h−1. Fluxes are given relative to the specific pyruvate consumption rate and are expressed as the net fluxes. For other descriptions, see Fig. 1
Based on the flux distribution profile, certain important trends can be seen. The first observation that can be made is the utilization of PPP in response to genetic and environmental changes. For mutants grown on either glucose or pyruvate, the direction of the flux through non-oxidative PPP was reversed. While the flux through G6PDH was decreased in the gnd mutant grown on glucose, this pathway was totally blocked in mutants grown on pyruvate, although the constitutive enzyme, G6PDH, remained expressed.
Two other key branches of the CMP network, the EMP and TCA cycle, also exhibited variations with the genetic and environmental alterations. Since the flux through oxidative PPP was significantly decreased or totally blocked in the mutants grown on glucose, the CMP network has to give a higher flux through EMP or EDP to the TCA cycle. The opposite relations are seen in mutants grown on pyruvate, where the deletion of genes caused a reduction in fluxes through the TCA cycle.
In correspondence with the observation in the enzymatic study, the metabolic flux redistribution indicated an activation of EDP in the gnd mutant grown on glucose and an increase in the flux through MAE in both glucose- and pyruvate-grown mutants. Further inspection of NADPH-generating reactions revealed a negative correlation between the MAE and ICDH pathways in the mutants, i.e., a higher flux of the MAE pathway was always accompanied by a lower flux though the ICDH pathway. Moreover, the conversion of carbon skeletons from the TCA cycle through MAE enabled the cells to respond to TCA carbon depletion by regulating the carbon flux through phosphoenolpyruvate carboxylase (PPC). This can be shown in the positive correlation between the MAE and PPC pathways.
Discussion
The present study indicates the importance of using integrated information from intracellular metabolic flux redistributions, enzyme activities, and other physiological phenotypes to investigate the global cellular response to genetic manipulations. Growth characteristics indicate that neither zwf nor gnd is essential under the tested culture condition, which is in accordance with the study by Fraenkel et al. (1968). However, further inspection of the metabolic parameters revealed a unique alteration in the utilization of the metabolic pathways for optimal growth of the mutants under different culture conditions. This was further supported by the enzymatic study, in which the enzyme activities related to PPP, EMP, and the TCA cycle were found to be strongly dependent on the genetic background and environmental condition.
The most important function of the enzymatic study is to identify the network structure in terms of the activities of the pathways. This identification is indispensable for further flux analysis of mutants, since it can indicate the aberrant regulation of pathway enzymes and has the potential to reveal pathways that are not necessarily required by the parent strain. In the present work, the enzymatic study identified that two pathways, the EDP and MAE pathways, were active in the gnd mutant grown on glucose. As we know, the carbon can be fed into the EDP through two routes in E. coli grown on glucose. One route (the so-called GCDP) consists of the oxidation of d-glucose to d-gluconate by glucose dehydrogenase and the phosphorylation of d-gluconate to 6-phosphogluconate by gluconate kinase. The other route is such that glucose is utilized via glucose-6-phosphate dehydrogenase to 6-phosphogluconate (the G6PDH pathway). The mechanism for activating GCDP in E. coli remains quite intriguing. Our enzymatic study is consistent with the opinion (Matsushita et al. 1997) that E. coli may not be able to produce gluconate from glucose via GCDP, since it cannot synthesize pyrroloquinoline quinone (PQQ), a cofactor of glucose dehydrogenase. Although it may be possible for E. coli to scavenge the surroundings for PQQ, our enzyme analysis excluded this possibility when cells were grown in minimal medium.
It should be noted that the metabolic analysis has to be made carefully when it is based only on the enzyme activities. Since not all in vivo effector concentrations are clearly known, the extent of the pathway change at branch points cannot be quantified by utilizing only the enzyme activity analysis. Moreover, there are constitutive enzymes or isoenzymes involved in the central metabolism, which may complicate the interpretation of the results. This can be seen in Table 3, as an example. It can be seen that disruption of the specific pathway through G6PDH does not affect the expression of 6GPDH, although this pathway is actually inactive because the substrate for 6GPDH is absent.
Normally, any blocks in the CMP network may cause an accumulation of metabolic intermediates in close proximity to the enzyme blocked. If other pathways cannot reduce this accumulation, it may cause growth suppression. Actually, a negative effect of the gnd deletion has been observed for Saccharomyces cerevisiae grown on glucose, since intermediates such as 6-phosphogluconate are reported to be toxic in high concentrations (Holger et al. 1996). It is clear that EDP, which is not possessed by S. cerevisiae, serves as the route to relieve the toxic level of 6-phosphogluconate in the E. coli mutant grown on glucose. Because of the activation of this potential bypass reaction, the flux via G6PDH is reduced but not blocked. The activity of the oxidative PPP is therefore partly maintained.
Metabolic flux analysis suggested significant differences in carbon flux distribution over the TCA cycle between glucose- and pyruvate-grown mutants. The deletion of genes caused an enhanced activity of the TCA cycle in the former but a decrease in the latter. While it is understandable that the glucose metabolism requires a higher flux through the TCA cycle if the other branch is blocked, the response of the TCA cycle in pyruvate metabolism is surprising. Using the obtained flux redistribution and growth parameters presented in Table 2, it is reasonable to postulate that, although the PPP flux is considerably lower during gluconeogenesis, expression of the zwf gene may coordinate with other genes to control the substrate uptake. The mutant directed a lower carbon flux through the TCA cycle and, therefore, produced CO2at relatively low rates, as compared with those of the parent strain. This cellular response is necessary because the network has to conserve more carbon for biosynthesis through the reduction in CO2 production, so that the mutant can grow at the same rate as the parent strain when the carbon source uptake rate decreases.
In E. coli, the oxidative PPP plays a major role in the generation of NADPH. ICDH is also found to play an important role in producing NADPH (Choi et al. 2003). Although the shortage of NADPH due to disruption of the oxidative PPP can be partially compensated by an increased flux through ICDH, it appears that the flux via ICDH alone could not enable the apparent shortage of NADPH to be met under certain circumstances. For this, malate was deviated out of the TCA cycle through MAE to function as the route in which an adequate supply of NADPH is generated to meet the biosynthesis requirements. This postulation can explain the negative correlation between the fluxes through MAE and ICDH, as occurred in the mutants. When glucose was used as a sole carbon source, a higher flux through ICDH was observed for the zwf mutant. This could generate more NADPH than in gnd mutant, thus rendering the pathway via MAE less active. In the case of pyruvate used as a carbon source, however, zwf gene deletion led to a lower flux through ICDH, as compared with the case of gnd gene deletion. Therefore, the flux via MAE significantly increased to complement the shortage of NADPH. Activation of the MAE pathway changed the flux distribution around the oxaloacetate (OAA) node. The drain of carbon skeletons from the TCA cycle through MAE enabled the cells to replenish the OAA pool by regulating the carbon flux through PPC. To increase the synthesis of OAA from phosphoenol pyruvate, PPC was therefore up-regulated in accordance with the activity of the MAE pathway.
In addition to the generation of NADPH, the other important function of the oxidative PPP is to drive the carbon flow to the non-oxidative branch, where the strain recruits intermediates such as erythrose-4-phosphate (E4P) and ribose-5-phosphate (R5P) for the biosynthesis of nucleic acids, amino acids, and vitamins and the generation of components of the cell′s lipopolysaccharide layer. In the mutants grown on glucose, all directions of the fluxes through non-oxidative PPP were reversed, indicating that the mutant tried to compensate for the lack of E4P and R5P through the glycolytic metabolites triose-3-phosphate (T3P) and fructose-6-phosphate (F6P). In the mutants grown on pyruvate, an increased flux through tkt (F6P+T3P→X5P+E4P) was found, suggesting a similar function for the non-oxidative PPP as in the glucose-grown mutants. In this way, the non-oxidative branch can function as an important metabolic route without the participation of the oxidative PPP. This can explain why the mutant can grow well in the minimal medium used in this study.
One of the goals of a gene-knockout study is the engineering of metabolic pathways for enhanced production of industrial chemicals. Genetically altering metabolic pathways, however, often causes undesirable changes, such as reduced growth, decreased glycolytic flux, and the creation of futile cycles, which may limit its utility (Holger et al. 1996). Our results indicate that E. coli grown on glucose with genetic disruption in the oxidative PPP can serve as a good candidate for industrial production, since the mutations have little effect on cell growth and the increased activities of EMP and TCA may benefit the production of useful chemicals which are synthesized from precursors involved in these two branches. The genetic alteration of zwf is preferred, since mutation in the other gene, gnd, activates EDP from which only one molecule of ATP is produced. In view of energy metabolism, EDP is less efficient than EMP with a net gain of two molecules of ATP for each molecule of glucose.
The problem in utilizing the zwf mutant in industrial production is that the gene deletion causes an enhanced production of acetate. Acetate accumulation should be avoided in industrial-scale production, since a high acetate concentration in the culture medium has been found to decrease the yield of recombinant protein. As we know, this type of metabolism overflow usually occurs when high fluxes from the pyruvate pool exceed the capacity for respiratory metabolism and the balance is therefore excreted as acetate. This can be manifested in the zwf mutant where the flux entering the pyruvate pool is extremely high due to the high activity of EMP. Further blockage of the acetate-producing pathway is expected to solve this problem. Since the glucose uptake pathways, EMP and oxidative PPP, are connected to a common substrate, G6P, a blockage of oxidative PPP will inevitably channel more carbon fluxes towards EMP. Therefore, it is reasonable to expect that fluxes through EMP and the TCA cycle will still be high when a second mutation in the acetate-producing pathways is introduced in the zwf mutant. The approach regarding simultaneous mutation in the primary acetate pathway genes (ackA, the gene encoding acetate kinase, or pta, the gene encoding phosphotransacetylase) is being considered and will be employed to further modify the metabolic network of the zwf mutant for industrial purposes.
Finally, it should be noted that the reason for acetate excretion in the gnd mutant was somewhat different from the zwf mutant when glucose was used as a sole carbon source. The flux profile of the former demonstrated that the increase in the flux through the early step of EMP was actually counteracted later by enhanced fluxes entering into non-oxidative PPP through T3P and F6P. The increase in the flux through pyruvate dehydrogenase was mainly due to the supply from the EDP and MAE pathways. Thereafter, the increased flux was partly channeled towards acetate excretion as the result of metabolism overflow.
References
Berry A (1996) Improving production of aromatic compounds in Escherichia coli by metabolic engineering. Trends Biotechnol 14:250–256
Canonaco F, Hess TA, Heri S, Wang T, Szyperski T, Sauer U (2001) Metabolic flux response to phosphoglucose isomerase knock-out in Escherichia coli and impact of overexpression of the soluble transhydrogenase UdhA. FEMS Microbiol Lett 204:247–252
Choi IY, Sup KI, Kim HJ, Park JW (2003) Thermosensitive phenotype of Escherichia coli mutant lacking NADP(+)-dependent isocitrate dehydrogenase. Redox Rep 8:51–56
Colowick SP (1963) Preparation and assay of enzymes. Methods Enzymol 6:1–640
Datsenko K A, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K12 using PCR products. Proc Natl Acad Sci USA 97:6640–6645
Dauner M, Bailey JE, Sauer U (2001) Metabolic flux analysis with a comprehensive isotopomer model in Bacillus subtilis. Biotechnol Bioeng 76:144–156
Emmerling M, Bailey JE, Sauer U (1999) Glucose catabolism of Escherichia coli strains with increased activity and altered regulation of key glycolytic enzymes. Metab Eng 1:117–127
Emmerling M, Bailey JE, Sauer U (2000) Altered regulation of pyruvate kinase or cooverexpression of phosphofructokinase increases glycolytic fluxes in resting Escherichia coli. Biotechnol Bioeng 67:623–627
Emmerling M, Dauner M, Ponti A, Fiaux J, Hochuli M, Szyperski T, Wuthrich K, Bailey JE, Sauer U (2002) Metabolic flux responses to pyruvate kinase knockout in Escherichia coli. J Bacteriol 184:152–164
Fraenkel DG (1968) Selection of mutants lacking glucose-6-phosphate dehydrogenase or gluconate-6-phosphate dehydrogenase. J Bacteriol 95:1267–1271
Holger J, Bernhard K, Peter K, Karl-Dieter E (1996) Mutants that show increased sensitivity to hydrogen peroxide reveal an important role for the pentose phosphate pathway in protection of yeast against oxidative stress. Mol Gen Genet 252:456–464
Josephson BL, Fraenkel DG (1969) Transketolase mutants of Escherichia coli. J Bacteriol 100:1289–1275
Josephson BL, Fraenkel DG (1974) Sugar metabolism in transketolase mutants of Escherichia coli. J Bacteriol 118:1082–1089
Lim SJ, Jung YM, Shin HD, Lee YH (2002) Application of the NADPH-related genes zwf and gnd for the Oddball biosynthesis of PHB in an E. coli transformant harbouring a cloned phbCAB operon. J Biosci Bioeng 93:543–549
Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with the Folin phenol reagent. J Biol Chem 193:265–275
Marx A, Graaf AA, Wiechert W, Eggeling L, Sahm H (1996) Determination of the fluxes in the central metabolism of Corynebacterium glutamicum by nuclear magnetic resonance spectroscopy combined with metabolite balancing. Biotechnol Bioeng 49:111–129
Matsushita K, Arents JC, Bader R, Yamada M, Adachi O, Postma PW (1997) Escherichia coli is unable to produce pyrroloquinoline quinine (PQQ). Microbiology 143:3149–3156
Neidhardt FC, Ingraham JL, Schaechter M (1990) Physiology of the bacterial cell: a molecular approach. Sinauer Associates, Sunderland, Mass.
Park SM, Sinskey AJ, Stephanopoulos G (1997) Metabolic and physiological studies of Corynebacterium glutamicum mutants. Biotechnol Bioeng 55:864–878
Paul Lee WN (1991) Mass isotopomer analysis: theoretical and practical consideration. Biol Mass Spectrom 20:451–458
Sauer U, Lasko DR, Fiaux J, Hochuli M, Glaser R, Szyperski KW, Bailey JE (1999) Metabolic flux ratio analysis of genetic and environmental modulations of Escherichia coli central carbon metabolism. J Bacteriol 181:6679–6688
Schmidt K, Nielsen J, Villadsen J (1997) Modeling isotopomer distributions in metabolic networks using isotopomer mapping matrices. Biotechnol Bioeng 55:831–840
Schmidt K, Nielsen J, Villadsen J (1999a) Quantitative analysis of metabolic fluxes in Escherichia coli using two-dimensional NMR spectroscopy and complete isotopomer models. J Biotechnol 71:175–190
Schmidt K, Norregaard LC, Pedersen B, Meissner A, Nielsen JQ (1999b (✉) Quantification of intracellular metabolic fluxes from fractional enrichment and 13C–13C coupling constraints on the isotopomer distribution in labeled biomass components. Metab Eng 1:166–179
Sprenger GA (1995) Genetics of pentose-phosphate pathway enzymes of Escherichia coli K12. Arch Microbiol 164:324–330
Szyperski T (1995) Biosynthetically directed fractional 13C-labeling of proteinogenic amino acids—an efficient analytical tool to investigate intermediary metabolism. Eur J Biochem 232:433–448
Winden V, Schipper D, Verheijen P, Heijnen J (2001) Innovations in generation and analysis of 2D [(13)C,(1)H] COSY NMR spectra for metabolic flux analysis purposes. Metab Eng 3:322–343
Wittmann C, Heinzle E (1999) Mass spectrometry for metabolic flux analysis. Biotechnol Bioeng 62:739–750
Zhao J, Shimizu K (2003) Metabolic flux analysis of Escherichia coli K12 grown on 13C-labeled acetate and glucose using GC-MS and powerful flux calculation method. J Biotechnol 101:101–117
Acknowledgement
This research was supported in part by a grant from New Energy and Industrial Technology Development Organization of the Ministry of Economy, Trade and Industry of Japan (Development of a Technological Infrastructure for Industrial Bioprocess Project).
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Rights and permissions
About this article
Cite this article
Zhao, J., Baba, T., Mori, H. et al. Global metabolic response of Escherichia coli to gnd or zwf gene-knockout, based on 13C-labeling experiments and the measurement of enzyme activities. Appl Microbiol Biotechnol 64, 91–98 (2004). https://doi.org/10.1007/s00253-003-1458-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-003-1458-5