Abstract
Over the past 15 years the biosynthetic gene clusters for numerous bioactive polyketides have been intensively studied and recently this work has been extended to the antifungal polyene macrolides. These compounds consist of large macrolactone rings that have a characteristic series of conjugated double bonds, as well as an exocyclic carboxyl group and an unusual mycosamine sugar. The biosynthetic gene clusters for nystatin, pimaricin, amphotericin and candicidin have been investigated in detail. These clusters contain the largest modular polyketide synthase genes reported to date. This body of work also provides insights into the enzymes catalysing the unusual post-polyketide modifications, and the genes regulating antibiotic biosynthesis. The sequences also provide clues about the evolutionary origins of polyene biosynthetic genes. Successful genetic manipulation of the producing organisms leading to production of polyene analogues indicates good prospects for generating improved antifungal compounds via genetic engineering.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
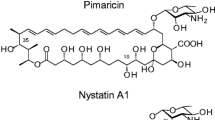
In recent years, the biosynthetic gene clusters for polyene antibiotics FR-008 (Hu et al. 1994), nystatin, pimaricin, amphotericin and candicidin (Fig. 1) have been cloned and methods have been developed for genetic manipulation of the producing organisms. The nystatin, pimaricin and amphotericin gene clusters have been sequenced completely (Fig. 2) (Aparicio et al. 1999, 2000; Brautaset et al. 2000; Caffrey et al. 2001). The DNA sequence encoding ten modules of the candicidin polyketide synthase (PKS) as well as some of the genes involved in transport, regulation and post-polyketide modification have also been disclosed (Fig. 2) (Campelo and Gil 2002). In addition, the complete genome sequence of Streptomyces avermitilis has revealed a PKS gene cluster presumably responsible for the synthesis of a pentaene macrolide (Omura et al. 2001).
Comparison of biosynthetic gene clusters for nystatin, amphotericin, pimaricin and candicidin. The large polyketide synthase (PKS) genes (striped arrows) are not shown on the same scale as the other genes. The functions of the gene products are shown in Table 1. The architecture of the PKS proteins is shown in Fig. 3
The physical characteristics of polyene antibiotics, their mode of action against fungi, and their use in medicine have been extensively reviewed (Bolard 1986; Gil and Martín 1997; Gupte et al. 2002). This mini-review deals with comparative analysis of the four gene clusters involved in polyene biosynthesis, and provides some important insights into the genetics and enzymology behind the synthesis of these antibiotics. It also highlights the perspectives and rationale for genetic manipulation of the polyene biosynthetic genes aimed at production of new, more active, and less toxic polyenes.
PKS proteins
Examination of the polyene macrolide PKS indicates that unimodular, bimodular, trimodular, tetramodular, and hexamodular proteins are all represented (Fig. 3). Only one other hexamodular protein had been detected previously: RAPS2 of rapamycin PKS (Schwecke et al. 1995). Rapamycin was originally isolated as an antifungal antibiotic and was until recently considered a polyene (Omura and Tanaka 1984).
Modular architecture of polyene macrolide synthases. Modules have been classified on the basis of the functional domains that they include. The minimal PKS requires functional ketosynthase (KS), acyl transferase (AT) and acyl carrier protein (ACP) domains. The thioesterase (TE) domain has been considered as an independent entity since it is not always present in other PKSs. The domain composition of the different loading modules is explained in the text. Note that although the third module of AmphC harbours an enoyl reductase (ER) domain it is thought to be partially active. Functionality of such a domain leads to production of amphotericin A instead of amphotericin B. The architecture of the candicidin PKS is incomplete
Nystatin and amphotericin PKS are very similar, as might be expected from the structural similarity between these two polyenes (Fig. 1). The corresponding proteins show about 72% sequence identity. The pimaricin PKS is organized along the same lines except that certain modules are absent, in accordance with the contracted size of the macrolactone ring (Fig. 1). The candicidin PKS is expected to contain 21 extension modules and is the largest aromatic heptaene PKS investigated in detail so far. Ten of these modules have been completely sequenced (Campelo and Gil 2002).
PKS loading modules
The nystatin, amphotericin and pimaricin PKS all have discrete loading modules (Fig. 3). The AmphA and NysA proteins have the domain composition KSS-AT-DH-ACP (ketosynthaseS-acyl transferase-dehydratase-acyl carrier protein). The KSS domains in these loading modules are almost identical to a regular KS domain except that a serine residue replaces the active site cysteine. No other significant differences between the KSS and condensing KS domains have been detected. Protein engineering experiments with rat fatty acid synthase (FAS) have shown that this change converts the KS domain to a weak decarboxylase that retains some condensation activity (Witkowska et al. 1999). Within the AmphA and NysA proteins, the malonate-specific AT domains are thought to acylate the ACP with malonate, which is decarboxylated by the KSS to provide an acetyl starter unit for transfer to KS1 of the first extension module. Decarboxylating loading modules presumably make a more clear-cut decision between acetate and propionate starter units (Bisang et al. 1999). The DH domains in the AmphA and NysA loading modules seem to be redundant. Direct linkage of a DH to an ACP is unusual but has been observed in extension module 4 of the FK520 PKS (Wu et al. 2000). The candicidin-loading module Can P1 (Fig. 3) consists of a CoA ligase (CoAL) domain and an ACP, and is fused to the first extension module (Campelo and Gil 2002). CoAL presumably activates p-aminobenzoic acid for acylation of the loading ACP domain. The pimaricin-loading module PimS0 (Fig. 3) has the domain composition CoAL-ACP- KSS-AT-ACP. It is possible that this loading module can also generate an acetyl starter unit by decarboxylation of a malonyl group. The CoAL and N-terminal ACP may represent the vestiges of domains that activate another starter acid. Alternatively, it is conceivable that the CoAL could activate acetate to acetyl-AMP for eventual transfer to KS1 via the phosphopantetheine thiols of the ACP domains (Aparicio et al. 1999).
PKS extension modules
Methylmalonate-specific AT domains (Haydock et al. 1995) are present in extension modules 1, 2 and 11 of amphotericin and nystatin PKS, in module 7 of pimaricin PKS, and in modules 2, 3 and 4 of candicidin PKS (Fig. 3). This agrees with the pattern expected from examination of the polyene structures (Fig. 1), and from feeding studies carried out on the amphotericin producer (McNamara et al. 1998). These chemical-labelling studies provided the first direct evidence that the exocyclic carboxyl group results from oxidation of a methyl branch derived from a methylmalonyl extender unit. A methylmalonate-specific AT domain is therefore anticipated for module 13 of the candicidin PKS. All other modules appear to contain malonate-specific AT domains. In principle, there is a possibility that the hydroxyl groups at C10 in nystatin and C8 in amphotericin could result from incorporation of hydroxymalonate extender units in cycles 14 and 15 of the respective pathways. Malonate-specific and methoxymalonate-specific AT domains are not easily distinguishable from sequence data alone (Wu et al. 2000). On the other hand, genes for hydroxymalonyl CoA production are absent from both clusters. It therefore seems more likely that amphotericin C8 and nystatin C10 hydroxyl groups are introduced by cytochrome P450s as post-PKS modifications. This would be analogous to the biosynthesis of pimaricin, as the epoxide group at C4–C5 was shown to be introduced by a dedicated cytochrome P450 epoxidase in a post-PKS modification reaction (Mendes et al. 2001).
The AmphB and NysB proteins are bimodular and catalyse the first two cycles of chain extension. The trimodular CanP2 protein incorporates three propionates in cycles 2, 3, and 4. In pimaricin biosynthesis, the acetate starter unit is transferred directly to the tetramodular PimS1 protein that assembles most of the tetraene region of the polyketide chain.
With amphotericin and nystatin PKS, extension modules 3–8 are organized into hexamodular proteins, AmphC and NysC. These modules assemble most of the polyene unit and all contain functional DH domains (Bevitt et al. 1993). In both proteins, module 5 also contains an enoyl reductase (ER) domain. The DH5-ER5 interdomain region in amphotericin PKS is shorter than its counterpart in nystatin PKS. It has been suggested that the shortened linker restricts movement of the ER5 domain so that it normally fails to operate and the main product is amphotericin B. If ER5 were to act intermittently, the PKS could also produce the precursor of amphotericin A, in which the C28–C29 double bond is reduced (Caffrey et al. 2001). This proposition could be tested by relocating the reduction loop to a new context and determining whether a mixture of products is formed. It is not yet possible to exclude other explanations for the production of amphotericins A and B by Streptomyces nodosus.
The AmphC protein is shorter than NysC by about 186 amino acids. In AmphC, modules 5, 6, 7 and 8, the interdomain regions that follow the DH domains, are slightly shorter than their counterparts in NysC. The domain organisation of AmphC and NysC closely matches that of hexamodular protein RAPS2 of rapamycin PKS (Schwecke et al. 1995), although RAPS2 has methylmalonate-specific AT domains in its second, third and sixth modules and non-functional reduction domains in its first and second modules. The CanP3 protein catalyses six of the seven cycles that assemble the heptaene region. The domain organisation is identical to that of AmphC or NysC except that no ER domain is present (Fig. 3). It is still uncertain whether CanP3 catalyses cycles 5–10 or cycles 6–11. With nystatin, amphotericin and pimaricin biosynthesis, the last double bond in the polyene unit is introduced by the first module of the next PKS protein.
The hexamodular AmphI, NysI and PimS2 proteins (Fig. 3) each assemble another structural feature that is characteristic of polyene polyketides. Ultimately this region contains the exocyclic carboxyl group and the hemiketal ring. All three hexamodular proteins contain a non-functional KR domain in module 13 (AmphI and NysI) or module 9 (PimS2) but are otherwise free of inactive domains.
The main difference between nystatin and amphotericin PKS is seen in the trimodular NysJ and AmphJ proteins that catalyse cycles 15–17 (see Figs. 1, 3). In the NysJ protein, module 15 contains a full complement of reduction domains [DH-ER-ketoreductase (KR)] whereas module 16 contains only KR. In the AmphJ protein, module 16 contains a complete reduction loop, whereas module 15 contains a KR plus a DH domain, which appears to be non-functional although it has an apparently intact active site motif. Both PKS contain a DH domain in module 17 that is inactive because the active site His is replaced by Arg in AmphJ and Tyr in NysJ. There is no trimodular protein in pimaricin PKS, and the penultimate cycle of chain extension (cycle 11) is carried out by a unimodular protein, PimS3, containing a functional DH domain. This domain is responsible for the formation of the pimaricinolide C4–C5 double bond (Fig. 1) that will constitute the substrate for PimD epoxidase (see below).
Unimodular proteins, NysK, AmphK, and PimS4 catalyse the last cycles in the biosyntheses of the amphotericin, nystatin, and pimaricin polyketides. These proteins all have a thioesterase (TE) domain. It is not clear how each long precursor polyketide chain folds back on itself to allow correct cyclisation. Formation of the hemiketalic ring could assist this process. Deletion of a module from NysC converted nystatin PKS to a hexaene synthase (Brautaset et al. 2002). This suggests that the TE can lactonise shortened polyketide chains if appropriate hydroxyl groups are accessible.
Post-polyketide modification and secretion
In general, polyenes undergo relatively few post-polyketide modifications. The majority contain only a single sugar, mycosamine, which has not been found anywhere else in nature. The conversion of a methyl branch to a carboxyl group is also unique to polyenes, although an ethyl branch is oxidised to an aldehyde in tylosin and related compounds (Omura 1985). Amphotericin and nystatin are thought to undergo hydroxylation in the polyol region whereas pimaricin has an epoxide. The sequences have provided some clues on how these modifications are brought about.
Mycosamine biosynthesis and polyene glycosylation
It was initially thought that mycosamine is synthesised from a dTDP-glucose, as is the case with most neutral and amino sugars that are added to polyketide structures (Martin 1985; Stockmann and Piepersberg 1992). However, each of the four polyene clusters has been found to contain a gene for a GDP-mannose dehydratase (AmphDIII, CanM, NysDIII, PimJ) (Table 1). This suggests that the mycosamine sugar (3-amino-3,6-dideoxy-d-mannose) is synthesised from GDP-mannose, which can be channelled from primary metabolism (Fig. 4). A straightforward pathway to mycosamine starting from GDP-mannose would involve the GDP-mannose dehydratase to GDP-4-keto-6-deoxy-d-mannose, a 3, 4 isomerisation to give GDP-3-keto-6-deoxy-d-mannose, followed by a transamination to form GDP-mycosamine (Fig. 4). All four clusters encode putative transaminases (AmphDII, CanA, NysDII, PimC) that are homologous to various PLP-dependent perosamine synthases that convert GDP-4-keto-6-deoxy-d-mannose to GDP-perosamine (4-amino,4,6-dideoxy-d-mannose) in lipopolysaccharide biosynthesis in Vibrio cholerae O1 (Stroeher et al. 1995) and Escherichia coli O157 (Bilge et al. 1996).
There is no gene common to the nystatin, pimaricin, amphotericin and candicidin clusters that might encode a GDP-4-keto-6-deoxy-d-mannose 3, 4 isomerase. Almost nothing is known about enzymes that catalyse 3, 4 isomerisation of NDP-4-keto-sugars. The biosynthesis of desosamine involves 3, 4 isomerisation of dTDP-4-keto-6-deoxy-glucose. The eryCII gene is required for the desosaminylation of mycarosyl-erythronolide B in Saccharopolyspora erythraea and has been designated as the isomerase gene, although the evidence is largely circumstantial (Salah-Bey et al. 1998). The EryCII protein shows some homology with cytochrome P450 monooxygenases, although the cysteine residue that normally co-ordinates the haem iron is absent. The TylORF1 and DnrQ proteins are homologous to EryCII and consequently are thought to catalyse the same 3,4 isomerisation steps in the biosyntheses of dTDP-mycaminose and dTDP-daunosamine. It is not clear why dTDP-4-keto-6-deoxy-glucose 3, 4 isomerases should resemble P450s. Interestingly, non-enzymatic ketoisomerisation of dTDP-4-keto-6-deoxy-glucose to dTDP-3-keto-6-deoxy-glucose has been observed in vitro (Naundorf and Klaffke 1996). The isomerisation reaction can apparently be catalysed by a basic Dowex 2-X8 anion exchange resin and goes to completion after 10 h incubation at 4°C. This suggests that in mycosamine biosynthesis, it is possible that GDP-4-keto-6-deoxy-d-mannose could isomerise to GDP-3-keto-6-deoxy-d-mannose in the absence of a conventional enzyme. This would be consistent with the absence of isomerase genes in all the polyene clusters so far analysed.
All of the clusters contain genes (amphDI, canG, nysDI, pimK) (Table 1) encoding proteins with homology to UDP-glucuronate transferases. Further studies should reveal whether these transferases are absolutely specific for GDP-mycosamine, or whether other GDP-sugars could be used in the absence of GDP-mycosamine. Attempts to produce polyene analogues by engineered biosynthesis will presumably succeed in making structural alterations near the glycosylation site. Some of these experiments will test the aglycone specificity of the glycosyl transferases, and also establish whether the post-polyketide modifications occur in a preferred order.
Modifications catalysed by cytochrome P450 monooxygenases
The biosynthesis of macrolides often requires the action of specific oxidases in order to construct the final bioactive compound. Such oxidases belong to a family of enzymes that is one of the most widely distributed in nature: cytochromes P450. These are b-type cytochromes that carry out oxygenation reactions on an enormous array of substrates (Munro and Lindsay 1996), among them the polyketide precursors of polyene antibiotics.
As already mentioned, several chemical groups on the nystatin, amphotericin, candicidin and pimaricin aglycones could result from cytochrome P450 reactions. In particular, the hydroxyl groups at C10 in nystatin and C8 in amphotericin, the epoxide group at C4–C5 in pimaricin, and the characteristic exocyclic carboxyl group present in the hemiketal ring of the four polyenes (Fig. 1). As expected, several genes of the clusters studied appear to encode P450-like monooxygenases (Table 1, Fig. 2) that might introduce such functionalities. P450 monooxygenase genes in the polyene clusters all encode proteins with the oxygen-binding site and the haem-binding pocket, with the invariant cysteine characteristic of this protein family.
Comparative analysis of the seven P450 proteins showed a high degree of identity among all of them, but also allowed the distinction of two phylogenetic groups, one formed by AmphN, NysN, PimG and CanC, and the other by AmphL, NysL and PimD. There are two potential target sites—the exocyclic methyl branch and the polyol region—for the cytochrome P450 reactions on the amphotericin, pimaricin and nystatin precursors, while there is only one (exocyclic methyl branch) present on the candicidin precursor. Therefore, it is conceivable that CanC, and also its counterparts AmphN, NysN and PimG, could participate in the oxidation of the methyl branch to the carboxyl group, whereas AmphL, NysL and PimD would introduce oxidations in the polyol region. This hypothesis was strengthened after disruption of pimD, which led to the synthesis of 4,5 deepoxypimaricin (Mendes et al. 2001).
The action of P-450 monooxygenases requires electron transfer from NADH, mediated by NADH:ferredoxin oxidoreductase and/or a ferredoxin (O'Keefe and Harder 1991). Several prokaryotic P450 monooxygenase-encoding genes have been found in an operon with such electron transport components, as is the case for pimG, amphN, nysN and canC. The genes pimF, amphM, nysM and canF lie immediately downstream of the afore-mentioned cytochrome P450-encoding genes (Fig. 2, Table 1), and encode small acidic proteins with convincing similarity to ferredoxins containing (3Fe-4S) clusters. Interestingly, the ferredoxin gene is always linked to a P450 of the AmphN group, which would ensure their coordinated expression. Either these ferredoxins also partner the second cytochrome P450 (AmphL group) or some general-purpose ferredoxin may be recruited from elsewhere in cellular metabolism and serve as the electron carrier for these monooxygenases.
Export genes
The analysis of the four clusters revealed the presence of genes possibly involved in the export of the final antibiotics. These encode proteins with a high degree of identity to members of the ATP-binding cassette (ABC) transporter superfamily, particularly to several from Streptomyces species involved in the efflux of antibiotics. Two different ABC transporters have been found in Streptomyces nodosus (AmphG and AmphH), Streptomyces noursei (NysG and NysH), and Streptomyces natalensis (PimA and PimB), but only one ABC transporter protein (CanRA) was found in the candicidin cluster (Table 1,Fig. 2). All these proteins possess Walker A and Walker B motifs (Walker et al. 1982) and the duplicate proteins show a 25–28% identity over their full length. It is still unclear why two different transporter proteins are present in the pimaricin, nystatin and amphotericin clusters. Since some bacterial ABC-transporters are thought to function as homodimers (Bolhuis et al. 1997), it could be that the amphotericin, nystatin and pimaricin transport proteins associate to form heterodimers that export the antibiotics from the producing cell.
The organisation of the putative candicidin export genes is different from the genes involved in the secretion of pimaricin, amphotericin or nystatin. PimA, PimB, NysG, NysH, AmphG and AmphH are all around 600 amino acid residues in length, larger than the 335-residue CanRA protein. CanRA is homologous to the C-terminal regions of these proteins. In the candicidin cluster, the canRA gene is close to canRB, which encodes a protein homologous to transmembrane proteins typically associated with members of a group of ABC transporters that includes one of the two oleandomycin exporters (Rodríguez et al. 1993). CanRA and CanRB might therefore cooperate in ATP-dependent efflux of candicidin, constituting a Type I ABC transporter (similar to OleC plus OleC5 or DrrA plus DrrB), whereas nystatin, amphotericin or pimaricin systems might be Type II ABC transporters (constituted by proteins with both transmembrane and nucleotide-binding domains) similar to the 170-kDa P-glycoprotein that confers multidrug resistance on eukaryotic tumour cells (Mendez and Salas 1998). It is tempting to speculate that these differences between export of candicidin and the rest of the polyenes might be due to the presence of an aromatic moiety on the candicidin molecule.
An extra gene, pimH, coding for a putative integral membrane protein (Aparicio et al. 2000) is located at the left end of the pimaricin cluster and its role in the export of pimaricin remains unclear.
Regulatory genes
A majority of the antibiotic biosynthesis gene clusters in streptomycetes contain pathway-specific regulatory genes that directly control the expression of the structural genes within the cluster. The polyene gene clusters are no exception. The regulatory genes were found in the nystatin (Brautaset et al. 2000), candicidin (Campelo and Gil 2002), amphotericin (P. Caffrey, unpublished), and pimaricin (J.F. Aparicio, unpublished) gene clusters (Fig. 3, Table 1). Although detailed functional analysis of these regulatory genes has not yet been carried out, some interesting features of the proteins encoded by these genes can be discussed. To date, the most comprehensive information is available only for the regulatory genes found in the vicinity of the nystatin biosynthesis structural genes in S. noursei. nysRI, nysRII and nysRIII seem to constitute an operon, and encode large proteins belonging to the LAL (large ATP-binding regulator of the LuxR) family of transcriptional regulators (De Schrijver and De Mot 1999). A nysRI gene disruption experiment generated a mutant that was unable to produce nystatin (Brautaset et al. 2000). Although this mutation most probably had a polar effect on the nysRII and nysRIII genes, it still proves the importance of this operon for nystatin biosynthesis. Similar organisation of the regulatory genes closely resembling nysRI-RIII has been shown for the candicidin (orf1, orf2 and orf3) and amphotericin (amphRI, RII, RIII) gene clusters (Campelo and Gil 2002; P. Caffrey, unpublished). Interestingly, partial sequence data indicate that regulatory genes homologous to amphRI-RIII are also present at the other end of the amphotericin gene cluster (P. Caffrey, unpublished).
Polyene antibiotic gene clusters contain very large genes encoding PKSs, some of which, based on cluster organisation, seem to be co-transcribed. Such transcripts could be extremely long, encompassing more than 47 kb on mRNA synthesized from the nysA, nysB and nysC genes in S. noursei, or the corresponding amph genes in S. nodosus. An operon of exceptional size (more than 42 kb) has also been predicted to include pimS2, pimS3 and pimS4 genes in S. natalensis. How the stability of these mRNAs is maintained is unclear, but it seems likely that transcriptional regulators might be involved.
The putative products of nysRI-III and can-orf1–3 contain a LuxR-type helix-turn-helix motif at their C-termini, while only NysRI, NysRIII and can-ORF3 possess distinct N-terminally located Walker A and B motifs implicated in ATP/GTP binding (the sequence of can orf1 is incomplete). The latter suggests that whatever activities NysRI, NyRIII and can-ORF3 possess are probably dependent on ATP/GTP hydrolysis. The importance of an NTP-binding motif for the functionality of LAL regulatory proteins has recently been proved for the pikromycin gene cluster regulator PikD (Wilson et al. 2001). Besides these features, both NysRI and NysRIII seem to contain tetratricopeptide repeats implicated in protein-protein interactions (Blatch and Lassle 1999). It seems plausible that at least these two proteins are engaged in such interaction, and form a regulatory protein complex required for activation of the nystatin structural genes. This would explain why individual expression of nysRI, nysRII and nysRIII in S. noursei has no significant effect on nystatin biosynthesis (O.N. Sekurova, personal communication).
The nys-orf4 gene, located 404 bp downstream of nysRIII in the nystatin gene cluster (Fig. 2) encodes a different type of regulator not reported so far for the candicidin, pimaricin and amphotericin clusters. The nys-ORF4 protein contains a N-terminally located PAS-like domain, which has been ascribed a function of a signalling domain in many sensor kinases and in some transcriptional regulators. The PAS domain seems to be responsible for sensing light, oxygen, redox potential, small ligands, and overall energy level in the cells (Taylor and Zhulin 1999). Interestingly, the DNA binding domain of nys-ORF4 is of the same type as in NysRI, NysRII and NysRIII. Recently, nys-orf4 was inactivated in S. noursei, leading to severe impairment of nystatin biosynthesis, while inactivation of two other regulatory genes linked to the nystatin cluster had no significant effect (O.N. Sekurova, personal communication).
Putative regulatory proteins of the LAL family encoded within the polyene antibiotic gene clusters closely resemble transcriptional regulators found in the vicinity of the cholesterol oxidase/cytochrome P450 operon in Streptomyces sp. SA-COO (Molnár and Murooka 1993). Interestingly, a gene encoding putative cholesterol oxidase (pimE) has been found within the pimaricin gene cluster (Fig. 2, Table 1), although the function of this gene in pimaricin biosynthesis remains unknown (Aparicio et al. 2000). Also, a cholesterol oxidase-encoding gene along with the LAL-family transcriptional regulator gene was identified within the polyene antibiotic gene cluster in S. avermitilis (Omura et al. 2001). It seems plausible that the regulatory genes that control cholesterol oxidase expression might have been acquired in the process of evolution by the polyene antibiotic gene clusters. The rationale behind such acquisition would be to provide a prompt response to the presence of fungi (whose cell walls contain ergosterol) in the environment via expression of the polyene antibiotic biosynthesis genes.
Evolutionary relationships
Detailed bioinformatic analysis of the sequences of the polyene biosynthetic genes is outside the scope of this mini-review. However, preliminary comparisons reveal that the amph, can, nys, and pim genes show a considerable degree of sequence similarity, especially the amph and nys PKS genes, which share 75% sequence identity over their entire length. This high degree of sequence homology is even more striking considering that some of these genes are the largest ever found in bacteria. The amphC, amphI, canP3, nysC, nysI and pimS2 genes encode hexamodular PKS proteins and range between 28.5 and 33.3 kb in length. Global alignments of these exceptionally large protein sequences divide them into two groups, one comprising AmphC, NysC and CanP3, and the other PimS2, AmphI and NysI (S.B. Zotchev, unpublished data)
Gene duplication appears to have been important in the evolution of the AmphC, NysC and CanP3 group. Each of these proteins assembles a polyketide precursor containing conjugated double bonds and has at least five modules of the type KS-AT(ac)-DH-KR-ACP (Fig. 3). The amphC, canP3 and nysC genes contain short internal direct repeats of 257, 164 and 742 bp, which most probably represent a relatively recent duplication event.
The AmphI, NysI and PimS2 proteins are also closely related and presumably have a common ancestor. Each of these proteins contains a series of modules that assembles a region specifying additional features that are also characteristic of glycosylated polyene macrolides. These are the glycosylation site, adjacent to one of the conjugated double bonds, and a methyl branch that is later oxidised to give the exocyclic carboxyl group that is unique to polyene macrolides. This region also forms the hemiketal ring in the cyclised macrolactone. These structural features appear to be important for antibiotic activity. It follows that the same series of six modules should appear in many different polyene PKSs.
The late genes also show a high degree of sequence similarity, and the order of genes is virtually identical in the nystatin and amphotericin clusters (Fig. 2). It is difficult to escape the conclusion that polyene clusters have common evolutionary origins. Presumably, synthesis of polyenes enables streptomycetes to compete with fungi in soil environments. Selective pressure could have driven the evolution of large polyene-producing systems from smaller PKSs like the 6- and 7-module synthases that assemble 14- and 16- membered macrolides that inhibit prokaryotic ribosomes.
Prospects for engineered biosynthesis of new polyene antibiotics
During the last decade, a large number of new polyketides have been produced by manipulation of PKS genes (Rodriguez and McDaniel 2001). The PKSs involved in polyene antibiotic biosynthesis, because of their unprecedented size and number of modules, represent the most challenging system in terms of creating chemical diversity via genetic engineering. The post-PKS modification enzymes responsible for the biosynthesis and attachment of mycosamine, and formation of additional hydroxyl, epoxide, and carboxyl groups on the polyene macrolactone ring must be considered as well, since these moieties seem to be important for activity and toxicity of the polyenes (Gary-Bobo 1989; Cybulska et al. 1995).
Genetic manipulation of any organism requires the establishment of a reliable gene transfer system. The polyene-producing Streptomyces turned out to be difficult organisms in this respect. So far, only interspecific conjugation and phage-mediated gene transfer allow the efficient introduction of recombinant DNA into these strains (Aparicio et al. 1999; Zotchev et al. 2000; Caffrey et al. 2001; Campelo and Gil 2002). It is possible, however, that a gene transfer system based on an autonomously replicating plasmid, pSNA1, found in the pimaricin producer S. natalensis (Mendes et al. 2000) could be established.
The first example of manipulation of a polyene biosynthetic gene is represented by the inactivation of the pimD gene (encoding a P450 monooxygenase) in the pimaricin cluster of S. natalensis (Mendes et al. 2001). The pimD mutant constructed via gene disruption was shown to accumulate 4,5-deepoxypimaricin at high yield, thus establishing a role for PimD as 4,5-deepoxypimaricin epoxidase. The biological activity studies on the 4,5-deepoxypimaricin clearly demonstrated the importance of the epoxy group for the pimaricin antifungal activity.
Manipulation of the nystatin PKS in S. noursei resulting in the production of a novel polyene antibiotic has recently been reported (Brautaset et al. 2002). In this case an attempt was made to convert the nystatin tetraene molecule to a heptaene via deletion of the ER domain in NysC PKS protein. Although the desired mutant was obtained, it produced very little of the expected heptaene derivative. However, another mutant selected in the same screening was shown to produce significant amounts of hexaene nystatin derivatives. Genetic analysis of this mutant revealed an in-frame deletion in the nysC gene resulting in elimination of one complete module from the protein. Purification and analysis of the completely post-PKS modified hexaene derivative suggest that it has a contracted macrolactone ring and is substantially less active that nystatin.
Although the manipulations described above yielded less active polyene antibiotics, they demonstrate the potential for engineering of polyene biosynthetic genes. With the genetic tools at hand it seems now possible to rationally engineer the polyene biosynthetic pathways in order to produce more active and less toxic polyene antibiotics. Changes in the polyene structural features that might be of interest in this respect would include the conjugated double bonds, hydroxy, epoxy, and carboxy groups, as well as the mycosamine moiety. Besides the possibility for creating polyenes with improved pharmacological properties, these attempts will provide important information regarding the structure-activity relationships of this group of antibiotics.
References
Aparicio JF, Colina AJ, Ceballos E, Martín JF (1999) The biosynthetic gene cluster for the 26-membered ring polyene macrolide pimaricin. J Biol Chem 274:10133–10139
Aparicio JF, Fouces R, Mendes MV, Olivera N, Martín JF (2000) A complex multienzyme system encoded by five polyketide synthase genes is involved in the biosynthesis of the 26-membered polyene macrolide pimaricin in Streptomyces natalensis. Chem Biol 7:895–905
Bevitt DJ, Staunton J, Leadlay PF (1993) Mutagenesis of the dehydratase active site in the erythromycin-producing polyketide synthase. Biochem Soc Trans 21:30S
Bilge SS, Vary JC, Dowell SF, Tarr PI (1996) Role of the Escherichia coli O157:H7 O side chain in adherence and analysis of an rfb locus. Infect Immunol 64:4795–4801
Bisang C, Long PF, Cortes J, Westcott J, Crosby J, Matharu AL, Cox RJ, Simpson TJ, Staunton J, Leadlay PF (1999) A chain initiation factor common to both aromatic and modular polyketide synthases. Nature 401:502–505
Blatch GL, Lassle M (1999) The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays 21:932–939
Bolard J (1986) How do the polyene macrolide antibiotics affect the cellular membrane properties? Biochim Biophys Acta 864:257–304
Bolhuis H, van Veen HW, Poolman B, Driessen AJM, Konings WN (1997) Mechanisms of multidrug transporters. FEMS Microbiol Rev 21:55–84
Brautaset T, Sekurova ON, Sletta H, Ellingsen TE, Strom AR, Valla S, Zotchev SB (2000) Biosynthesis of the polyene antifungal antibiotic nystatin in Streptomyces noursei ATCC 11455: analysis of the gene cluster and deduction of the biosynthetic pathway. Chem Biol 7:395–403
Brautaset T, Bruheim P, Sletta H, Hagen L, Ellingsen TE, Strom AR, Valla S, Zotchev SB (2002). Hexaene derivatives of nystatin produced as a result of an induced rearrangement within the nysC polyketide synthase gene in Streptomyces noursei ATCC 11455. Chem Biol 9:367–373
Caffrey P, Lynch S, Flood E, Finnan S, Oliynyk M (2001) Amphotericin biosynthesis in Streptomyces nodosus: deductions from analysis of polyketide synthase and late genes. Chem Biol 8:713–723
Campelo AB, Gil JA (2002) The candicidin gene cluster from Streptomyces griseus IMRU 3570. Microbiology 148:51–59
Cybulska B, Bolard J, Seksek O, Czerwinski A, Borowski E (1995). Identification of the structural elements of amphotericin B and other polyene macrolide antibiotics of the hepteane group influencing the ionic selectivity of the permeability pathways formed in the red cell membrane. Biochim Biophys Acta 1240:167–178
De Schrijver A, De Mot R (1999). A subfamily of MalT-related ATP-dependent regulators in the LuxR family. Microbiology 145:1287–1288
Gary-Bobo CM (1989) Polyene-sterol interaction and selective toxicity. Biochimie 71:37–47
Gil JA, Martín JF (1997) Polyene antibiotics. In: Strohl WR (ed) Biotechnology of antibiotics, 2nd edn. Dekker, New York, pp 551–575
Gupte M, Kulkarni P, Ganguli BN (2002) Antifungal antibiotics. Appl Microbiol Biotechnol 58:46–57
Haydock SF, Aparicio JF, Molnar I, Schwecke T, Khaw LE, Konig A, Marsden AF, Galloway IS, Staunton J, Leadlay PF (1995) Divergent sequence motifs correlated with the substrate specificity of (methyl)malonyl-CoA:acyl carrier protein transacylase domains in modular polyketide synthases. FEBS Lett 374:246–248
Hu H, Bao K, Zhou X, Hopwood DA, Kieser T, Deng Z (1994) Repeated polyketide synthase modules involved in the biosynthesis of a heptaene macrolide by Streptomyces sp. FR-008. Mol Microbiol 14:163–172
Martin JF (1985) Biosynthesis, regulation, and genetics of polyene macrolide antibiotics. In: Omura S (ed) Macrolide antibiotics. Chemistry, biology and practice. Academic Press, New York
McNamara CM, Box S, Crawforth JM, Hickman BS, Norwood TJ, Rawlings BJ (1998) Biosynthesis of Amphotericin B. J Chem Soc Perkin Trans 1:83–87
Mendez C, Salas JA (1998) ABC transporters in antibiotic-producing actinomycetes. FEMS Microbiol Lett 158:1–8
Mendes MV, Aparicio JF, Martin JF (2000) Complete nucleotide sequence and characterization of pSNA1 from pimaricin-producing Streptomyces natalensis that replicates by a rolling circle mechanism. Plasmid 43:159–165
Mendes MV, Recio E, Fouces R, Luiten R, Martin JF, Aparicio JF (2001) Engineered biosynthesis of novel polyenes: a pimaricin derivative produced by targeted gene disruption in Streptomyces natalensis. Chem Biol 8:635–644
Molnár I, Murooka Y (1993) Nucleotide sequence analysis of a region upstream of the cholesterol oxidase-cytochrome P450 operon of Streptomyces sp. SA-COO revealing repeating units coding for putative transmembrane and DNA-binding proteins. J Ferment Bioeng 76:257–264
Munro AW, Lindsay JG (1996) Bacterial cytochromes P-450. Mol Microbiol 20:1115–1125
Naundorf A, Klaffke W (1996) Substrate specificity of native dTDP-d-glucose-4,6-dehydratase: chemo-enzymatic syntheses of artificial and naturally occurring deoxy sugars. Carbohydr Res 285:141–150
O'Keefe DP, Harder PA (1991) Occurrence and biological function of cytochrome P450 monooxygenases in actinomycetes. Mol Microbiol 5:2099–2105
Omura S (ed) (1985) Macrolide antibiotics. Chemistry, biology and practice. Academic Press, New York
Omura S, Tanaka H (1984) Polyene production, structure and activity. In: Omura S (ed) Macrolide antibiotics. Chemistry, biology and practice. Academic Press, New York
Omura S, Ikeda H, Ishikawa J, Hanamoto A, Takahashi C, Shinose M, Takahashi Y, Horikawa H, Nakazawa H, Osonoe T, Kikuchi H, Shiba T, Sakaki Y, Hattori M (2001) Genome sequence of an industrial microorganism Streptomyces avermitilis: deducing the ability of producing secondary metabolites. Proc Natl Acad Sci USA 98:12215–12220
Rodríguez AM, Olano C, Vilches C, Méndez C, Salas JA (1993) Streptomyces antibioticus contains at least three oleandomycin-resistant determinants, one of which shows similarity with proteins of the ABC-transporter superfamily. Mol Microbiol 8:571–582
Rodriguez E, McDaniel R (2001) Combinatorial biosynthesis of antimicrobials and other natural products. Curr Opin Microbiol 4:526–534
Salah-Bey K, Doumith M, Michel JM, Haydock S, Cortes J, Leadlay PF, Raynal MC (1998) Targeted gene inactivation for the elucidation of deoxysugar biosynthesis in the erythromycin producer Saccharopolyspora erythraea. Mol Gen Genet 257: 542–553
Schwecke T, Aparicio JF, Molnar I, Konig A, Khaw LE, Haydock SF, Oliynyk M, Caffrey P, Cortes J, Lester JB, Boehm GA, Staunton J, Leadlay PF (1995) The biosynthetic gene cluster for the polyketide immunosuppressant rapamycin. Proc Natl Acad Sci USA 92:7839–7843
Stockmann M, Piepersberg W (1992) Gene probes for the detection of 6-deoxyhexose metabolism in secondary metabolite-producing streptomycetes. FEMS Microbiol Lett 90:185–190
Stroeher UH, Karageorgos LE, Brown MH, Morona R, Manning PA (1995) A putative pathway for perosamine biosynthesis is the first function encoded within the rfb region in Vibrio cholerae O1. Gene 166:33–42
Taylor BL, Zhulin IB (1999) PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev 63:479–506
Walker JE, Saraste M, Runswick MJ, Gay NJ (1982) Distantly related sequences in the α- and β-subunits ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J 1:945–951
Wilson DJ, Xue Y, Reynolds KA, Sherman DH (2001) Characterization and analysis of the PikD regulatory factor in the pikromycin biosynthetic pathway of Streptomyces venezuelae. J Bacteriol 183:3468–3475
Witkowska A, Joshi AK, Lindqvist Y, Smith S (1999) Conversion of a β-ketoacyl synthase to a malonyl decarboxylase by replacement of the active site cysteine with glutamine. Biochemistry 38:11643–11650
Wu K, Chung L, Revill WP, Katz L, Reeves CD (2000) The FK520 gene cluster of Streptomyces hygroscopicus var ascomyceticus (ATCC 14891) contains genes for biosynthesis of unusual polyketide extender units. Gene 251:81–90
Zotchev S, Haugan K, Sekurova O, Sletta H, Ellingsen TE, Valla S (2000) Identification of a gene cluster for antibacterial polyketide-derived antibiotic biosynthesis in the nystatin producer Streptomyces noursei ATCC 11455. Microbiology 146:611–619
Acknowledgements
This work was supported by grants from the Comisión Interministerial de Ciencia y Tecnología, Plan Nacional de Biotecnología (Spain) and the European Commission (BIO93-0831, BIO96-0583, 1FD97-1419-C02-01 and BIO2001-0040) to J.A.G. and J.F.A., a grant from the Research Council of Norway to S.B.Z., and grants from the Wellcome Trust and Enterprise Ireland to P.C.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Aparicio, J.F., Caffrey, P., Gil, J.A. et al. Polyene antibiotic biosynthesis gene clusters. Appl Microbiol Biotechnol 61, 179–188 (2003). https://doi.org/10.1007/s00253-002-1183-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-002-1183-5