Abstract
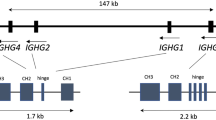
Eleven genomic porcine Cγ gene sequences are described that represent six putative subclasses that appear to have originated by gene duplication and exon shuffle. The genes previously described as encoding porcine IgG1 and IgG3 were shown to be the IgG1a and IgG1b allelic variants of the IGHG1 gene, IgG2a and IgG2b are allelic variants of the IGHG2 gene, while “new” IgG3 is monomorphic, has an extended hinge, is structurally unique, and appears to encode the most evolutionarily conserved porcine IgG. IgG5b differs most from its putative allele, and its CH1 domain shares sequence homology with the CH1 of IgG3. Four animals were identified that lacked either IgG4 or IgG6. Alternative splice variants were also recovered, some lacking the CH1 domain and potentially encoding heavy chain only antibodies. Potentially, swine can transcribe >20 different Cγ chains. A comparison of mammalian Cγ gene sequences revealed that IgG diversified into subclasses after speciation. Thus, the effector functions for the IgG subclasses of each species should not be extrapolated from “same name subclasses” in other species. Sequence analysis identified motifs likely to interact with Fcγ receptors, FcRn, protein A, protein G, and C1q. These revealed IgG3 to be most likely to activate complement and bind FcγRs. All except IgG5a and IgG6a should bind to FcγRs, while all except IgG6a and the putative IgG5 subclass proteins should bind well to porcine FcRn, protein A, and protein G.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
IgG can be considered the “flagship” antibody of mammals, since it is unique to this vertebrate class. Its origin is obscure although amphibian IgF is a candidate, since it is phylogenetically the first Ig with a hinge exon like IgG (Zhao et al. 2006). Mammalian IgG is characterized by a diversity of subclasses ranging from 1 to 7 (Butler 1997, 2006) and a range of functions mediated through Fc receptors (Ravetch and Kinet 1991). Subisotype diversification is not restricted to IgG in mammals; a similar process may explain the 13 subclasses of IgA in rabbits (Burnett et al. 1989) and the two subclasses of IgA in humans (Mestecky et al. 2004). Different subclasses possess different biological characteristics. Human IgG1 (IGHG1) and IgG3 (IGHG3) decorate phagocytic cells because of their high affinity for FcγR1 (Ravetch and Kinet 1991). Therefore, IgG1 and IgG3 are especially important in: (a) the removal of small IgG–Ag complexes and (b) positively or negatively stimulating B cell development and antibody production. Human IgG3 is a long-hinged antibody and a potent activator of complement (Lefranc and Lefranc 2001). IgG1 comprises two thirds of total human serum IgG, whereas IgG3 and IgG4 together comprise only about 10%. As in humans, there are differences in relative IgG subclass concentrations in the serum of mice and in the affinity of various mouse IgG subclasses for FcγRs (Ravetch and Kinet 1991). In cattle, IgG1 is the predominant IgG in all exocrine fluids and, because it is preferentially transported across the acinar epithelial cells of the mammary glands, comprises 90% of the IgG in colostrum but only 50% of serum IgG (Butler 1969, 1974). Physiological differences are also seen among IgA subclasses; human IgA1 (IGHA1) is relatively more prevalent in blood than secretions (Mestecky et al. 2004), but IgA1 is more susceptible to bacterial IgA proteases than IgA2 (Plaut et al. 1975; Kilian and Russell 2004; IGHA2). The 13 rabbit IgA subclasses also differ in their distribution among tissue (Spieker-Polet et al. 1993) and porcine IgG3 comprises >60% of IgG transcripts in the ileal Peyers patches (IPP) and mesenteric lymph nodes of fetal piglets (Butler and Wertz 2006).
The studies described were undertaken because of the emerging role of swine in understanding the: (a) role of commensal flora in driving development of adaptive immunity (Butler et al. 2002, 2005), (b) immune response to influenza which infects both humans and pigs (Vincent et al. 2006), (c) action of immunoregulatory viruses on the development of immune homeostasis (Lemke et al. 2004; Butler et al. 2007, 2008a), and (d) causes of cystic fibrosis in a CFTR knockout piglet model (Rogers et al. 2008). Swine are also important in xenotransplantation (Auchincloss and Sachs 1998) and in generating humanized antibodies (Waltz 2006; Butler et al. 2008a; Mendicino et al 2008). Unlike the major ruminant and guinea pig IgG subclass proteins, those from swine cannot be purified biochemically (Butler 1969, 1983), and there are no naturally occurring or induced plasmacytomas. However, Cγ diversity in swine was suggested from ion exchange and immunoelectrophoretic (Metzger and Fougereau 1967; Kaltreider and Johnson 1972) differential monoclonal antibody specificity (Bianchi et al. 1990) allotypic studies (Rapacz and Hasler-Rapacz 1982) and from cDNA sequences and genomic blots (Kacskovics et al. 1994) although the extent of diversity was unknown. Here we report the sequences of 11 porcine genomic Cγ sequences that appear to comprise six subclasses. A comparison with IgG sequences from various mammals agrees with the early prediction that subclass evolution occurred after major mammalian speciation (Kehoe and Capra 1974). Because of this, subclass function for “same name subclasses” cannot be extrapolated among mammals. Therefore, we analyzed the porcine Cγ gene sequences for binding motifs for C1q, FcγR, FcRn, as well as protein A (SpA) and G (SpG). Further characterization of the porcine IgG complex and confirmation of our predictions will require gene mapping, designed mating studies, in vitro expression of purified subclass proteins, and preparation of subclass-specific mAbs.
Materials and methods
Source of tissue
Animal tissues were by-products of studies on gut colonization in isolator piglet (Butler et al. 2000, 2002, 2005) studies on the effect of neonatal viral infection (Lemke et al. 2004; Butler et al. 2007, 2008a) and studies on IgA allotype distribution (Navarro et al. 2000a). Solid tissues were immediately frozen in liquid nitrogen, and blood samples were collected in EDTA and processed for the recovery of PBLs as described previously (Lemke et al. 2004; Kacskovics et al. 1994; Butler et al. 2000; Navarro et al. 2000b). Additional DNA samples from NIH minipigs were kindly provided by Dr. Joan Lunney, ARS, Beltsville, MD, USA from a variety of swine of different breeds and species by Dr. Max Rothschild, Department of Animal Science, Iowa State University, Ames, IA, USA and from purebred Meishan pigs (Dr. Larry Shook, University of Illinois; Champaign-Urbana, IL, USA).
Preparation of DNA, RNA, and cDNA
DNA, total RNA, and first strain cDNA were prepared as previously described (McAleer et al. 2005; Butler et al. 2001; Sun and Butler 1996) except that a primer for the 3′ UTR shared by all known Cγ genes and an antisense Cα3 primer (Navarro et al. 2000b), as well as random hexamers, were added (Table 1).
Identification of Cγ diversity among transcripts
Cγ-containing gene products were recovered using a pan-specific CH1–CH3 primer set common to all known swine IgG genes (Table 1; Fig. 1). The product was evaluated by electrophoresis in 1.5% agarose gel, the appropriate-sized Cγ-containing polynucleotide band removed and used as the target for a second round polymerase chain reaction (PCR) that utilized a heminested primer set containing an antisense primer that recognized a conserved sequence in the CH2 domain (Fig. 1; Table 1). The product of the second round PCR was cloned into PCR TOPO 2.1, grown in TOP TEN cells overnight at 37°C and plated on Luria–Bertani agar containing 50 μg/ml kanamycin (LB–Kan). The KAN-resistant clones were transferred to wells of round bottom 96-well microtiter plates (Costar 3799, Corning, NY, USA) and grown overnight at 37°C in LB–Kan. Plasmid DNA, harvested by alkaline lysis and centrifugation, was transferred and cross-linked to nylon membranes (Schleicher & Schull, Keene, NH) as previously described (McAleer et al. 2005; Butler et al. 2001; Sun and Butler 1996; Sun et al. 1998). Membranes were hybridized with Cγ probes specific for the originally described Cγ genes (Kacskovics et al. 1994) and a pan-specific Cγ probe (Butler and Wertz 2006; Table 1). cDNA clones that hybridized with the pan-specific probe but with none of the original subclass-specific probes, were grown out and sequenced. This process lead to the discovery of six additional Cγ sequences which necessitated that additional Cγ probes be prepared (Table 1).
The complete genomic sequence of 11 porcine Cγ genes using the sequence of porcine IgG1a as a reference and numbering from the first codon of CH1. Intron nucleotides are in lower case and the deduced amino acid sequences for the exons are in lower case. The upper (99–105), middle or core hinge (106–114), and lower hinges (115–121) are separated by vertical lines. Dots indicate that the reference nucleotide sequence (IgG1a) is shared, whereas solid bold horizontal lines indicate the absence of the corresponding nucleotide. Cysteine codons involved in intrachain loops are underlined and in boldface. Cysteines believed to be involved in H–H covalent bonds are circled. Cysteine 14 in CH1 is believed to be involved in L–H bonds. Potential N-linked glycosylation motifs are boxed. Motifs considered to be important in interactions with C1q, SpA, SpG, FcRn, and FcγR are enclosed in vertical rectangles and their importance indicated at the base of the rectangles. Annealing sites for the PCR primers used to recover cDNA and DNA sequences are indicated in horizontal rectangles
Characterization of genomic Cγ genes
The same primer set used for first round PCR on cDNA was used to generate near full-length genomic Cγ genes, ca. 1 kb. The genomic PCR product was then cloned and sequenced to allow coding and intron sequences to be obtained (Fig. 1). Sequence analysis identified 11 genomic Cγ gene sequences, the exons of which corresponded to those recovered from cDNA. The extreme 5′ portion of the CH1 domain (78 nt) was obtained from cDNA sequences recovered using a VH gene FR 1 primer and an antisense CH2 primer (Table 1). The extreme 3′ region of CH3 was recovered from DNA using the common CH1 sense and an anti-sense 3′ UTR primer set (Table 1).
Identification of allotypic variants
Evidence suggesting the existence of allotypic variants was obtained in expression studies in which certain animals were found to lack clones hybridizing with the gene originally described as IgG1 by Kacskovics et al. (1994). This initiated a series of genomic dot-blots using gene- and pan-specific probes to test for the allelic nature of the various Cγ genes recovered. Dot blots were further verified by restriction fragment length analysis (RFLP) followed by Southern blots using appropriate probes. This form of analysis was then extended to other putative allelic variants.
Identification and frequency of alternatively spliced IgG transcripts
Sequence analysis of 184 transcripts revealed various alternatively spliced transcripts. While splice variants were seen for all Cγ genes, they were especially prevalent in IgG5b. To estimate the frequency of occurrence in IgG5b, we sequenced 25 randomly selected IgG5b clones recovered from four different pigs.
Allotyping of porcine IgA
The IgAb allotype of porcine IgA differs from IgAa by a 12-nt deletion in the hinge region due to mutation of the splice acceptor site (Brown et al. 1995). This allows for the development of a typing system based on IgA transcripts as previously described by Navarro et al. (2000b). In the case of genomic DNA, the presence or absence of a Dde I site also allows the allotypes to be distinguished by RFLP. In the case of animals for which only DNA was available, this method was used (Navarro et al. 2000a).
Immunoglobulin nomenclature
The original and familiar system used for immunoglobulins (e.g., IgM) immunoglobulin subclass (e.g., IgG1) or allotypic variants (e.g., IgG1a) is well established in the literature including the one for swine. In addition to this nomenclature, we also designated the Cγ genes according to the IMGT system (Lefranc and Lefranc 2001; Table 2).
Sequence analysis
Genomic and cDNA clones were grown up overnight, and restriction digested to confirm the insert size. All insert-containing clones with appropriate-sized inserts were sequenced in the DNA Core Facility of the University of Iowa using the four-color ABI PRISM DNA Analyzer (Applied Biosystems). Amino acid sequences were deduced and analyzed in the Omiga program (Accelyrs) and added to Fig. 1.
Cladogram and dendogram construction
Alignments were calculated with CLUSTAL X (version 1.8) using the GONNET series protein matrices. The gap opening penalties for the pairwise and the multiple alignments were set to 10.00. The gap extension penalties were set to 0.1 and 0.2 for the pairwise and multiple alignments, respectively. From this alignment, rooted phylogenetic trees were constructed by using the neighbor-joining (NJ) method of MEGA 4.0 and Poisson distance correction (gaps ignored). Bootstrapping of the NJ tree was performed with 1,000 replicates (Tamura et al. 2007). The sequence data for species Cγ genes are available under the following accession numbers: Bovine Cγ1 = X16701; bovine Cγ2 = X16702; bovine Cγ3 = U63638; canine Cy1 = AF354264; canine Cγ2 = AF354265; canine Cγ3 = AF354266; canine Cγ4 = AF 354267; Equine Cγ1; 2; 3; 4; 5; 6 and 7 = AJ1302055; AJ302056; AJ312379; AJ302057; AJ312380; AJ312381; AJ302058, respectively. Human Cγ1; 2; 3 and 4 = J00228; J00230; X03604; K01316; respectively. Mouse Cγ1; 2a; 3; 2c and 2b = J00453; V00825; X00915; J00479; V00763; respectively. Sheep Cγ1 = X69797; Sheep Cψ2 = X70983; Rabbit Cγ = L29172; Camel 1a = AJ421266; Camelid 1b = unpublished; Opossum = AF035195; Possum = AF157619.
Binding motifs for C1q, FcγRs, FcRn, SpA, and SpG
The analysis of potential interaction sites with C1q was based on the interaction of human IgG1 and the B subunit of the human C1q (Gaboriaud et al. 2003) and the sequence of the B subunit of porcine C1q (GenBank accession code: AY349420) and the mouse orthologue (NP_033907). Human FcγRs (GenBank accession codes: P12314, AAD00641, and AAH33678, respectively) were compared to the sequences of porcine FcγRI, FcγRIIB, and FcγRIII (GenBank accession codes: AAZ20792; NP_001028185 and Q28942, respectively). Putative FcγR interaction motifs in porcine Cγ sequences were identified according to those present in human IgG (Radaev et al. 2001). IgG-FcRn interactions were analyzed using the model described for rat IgG2a and the rat FcRn crystal structure (Martin et al. 2001). The comparison of porcine FcRn to its orthologues has been previously published (Kacskovics et al. 2006). Motifs identified in the porcine Cγ sequences predicted to bind SpA and SpG were evaluated according to previously published models (Deisenhofer 1981; Sauer-Eriksson et al. 1995).
Results
Genomic sequences of porcine Cγ genes group into six pairs
The complete genomic sequences of the porcine Cγ genes recovered from DNA (Fig. 1) segregate into six groups (Fig. 2) of which allelic variants were identified for five (see below). Data for Fig. 2 are based on 88 cloned genomic sequences (Table 2).
The allelic nature and occurrence of porcine Cγ genes
The examination of >9,000 cDNA clones from outbred animals revealed that IgG1 (now called IgG1b) and “old” IgG3 (Kacskovics et al. 1994; now called IgG1a) were the most prominently expressed Cγ genes in all newborn piglets and were expressed in statistically equal amounts as early as 40 days of gestation (Table 2; Butler and Wertz 2006). Since fetal piglet no. 5 and adult pig 8266 failed to express the IgG1 described by Kacskovics et al. (1994; now designated IgG1b), we examined their DNA. Neither animal possessed a Cγ gene that hybridized with a probe specific for the IgG1 of Kacskovics et al. (1994; now designated IgG1b) while hybridizing with the probe for the gene designated as IgG3 by Kacskovics et al. (1994; now designated IgG1a; Table 2; Fig. 3b,c). Furthermore, DNA from piglet no. 5 digested with Pci I did not yield the products of 560 and 291 nt predicted from the sequence of IgG1b (Fig. 3a). The digested DNA of piglet no. 5 did not hybridize with the IgG1b probe (Fig. 3b). This suggested that piglet no. 5 and swine 8266 were either deficient in the gene encoding IgG1 or that the original IgG1 and IgG3 were allelic variants or that piglet nos. 5 and 8266 were homozygous for IgG1a. Previous immunogenetic studies on IgA had shown that all of >40 NIH minipigs tested were homozygous for IgAa (Navarro et al. 2000a). Since the IGHC genes in other species are closely linked, we hypothesized that IGHG and IGHA genes of these minipigs were part of a common haplotype. When minipigs were examined for their transcripts or the possession of hybridizing Cγ genes, animals that were homozygous IgAa/a lacked the Cγ gene originally designated IgG1 (now designated IgG1b) and expressed only the subclass described by Kacskovics et al. (1994) as IgG3 (now called IgG1a; Fig. 3C). Because IgAa/a and “old IgG3” appear to be part of the same haplotype, we designated the originally described IgG1 as IgG1b (IGHG1*02) and the originally described IgG3 as IgG1a (IGHG1*01; Fig. 1; Table 2). A similar pattern of equal expression (320/275) was seen with the genes initially designated IgG2a and IgG2b (Kacskovics et al. 1994; Table 2). We have therefore renamed them as the IgG2a and IgG2b alleles of the IGHG2 gene.
Southern blot of restriction digested and intact Cγ DNA. A Cγ DNA of three outbreed piglets (nos. 3, 4, and 5) digested with Pci I (C) or undigested (U) and stained with ethidium bromide after separation in a 1.5% agarose gel. The predicted size of the IgG1b polynucleotides after Pci I digestion are depicted by light arrows while the heavy arrows indicate the size of the undigested Cγ DNA. S nt length standards. B Southern blot analysis of agarose gel in (A) after hybridization with an IgG1b-specific probe. Arrows indicate the same polynucleotides as in (A). Piglet no. 5 did not express IgG1b and yields no product after Pci I digestion. C Southern dot-blot of Cγ-containing DNA. Top hybridization with the probe for IgG1a, middle hybridization with the probe for IgG1b, bottom code for animal identification (given below the diagram)
In the same manner as described for the putative IgG1 alleles, animals which failed to transcribe certain Cγ genes were selected for analysis of their DNA. The PCR products were first tested for their ability to hybridize with subclass-specific pan-specific probes. The products were then cloned and the individual clones identified by hybridization using the putative subclass probes described in Table 1. Table 3 shows recovery data for a selected panel of animals that lacked clones expressing particular putative alleles. Results are based on 29–36 clones for each animal. Data reveal animals homozygous and heterozygous for the putative alleles of IgG1, IgG2, and IgG5. In the examples shown, no animal homozygous for either allele of IgG6 was identified. Surprisingly, neither variant of IgG2 was recovered from animal nos. 5 nor 755, and neither variant of IgG6 was recovered from animal nos. 476 nor 481. Two of these “deficient “animals were Meishans, one an inbred NIH minipig and the other a mixed-breed farm pig.
Nomenclature revision for porcine Cγ genes
Since the original IgG3 has now been renamed IgG1a, we have proposed that the recently described long-hinged Cγ gene (Fig. 1; Table 2) be redesignated as IgG3. IgG4, IgG5, and IgG6 and their putative alleles, complete the list of IgG subclass genes in swine. We have proposed designations for these Cγ genes that are consistent with both the familiar and the IMGT nomenclature systems (Lefranc and Lefranc 2001; Table 2).
Analysis of porcine IgG sequences
Figure 1 shows the genomic nucleotide sequence for the various porcine Cγ genes and the deduced amino acid sequence of the domain exons, while Fig. 2 compares the complete genomic sequences, showing that the putative allelic variants pair together. The deduced amino acid sequences of the individual domains are compared in Fig. 4 and their relationships discussed below.
CH1 domain
All CH1 domains share a potential 56 amino acid intradomain disulfide loop (Cys 27 to Cys 83). Position 14 is believed to provide the cysteine for the L–H disulfide bond as in human IgG2, IgG3 and IgG4. The CH1 domain is the most conserved among the different IgGs (Fig. 4A; note genetic distance scale). IgG5b and IgG3 are most divergent in CH1 from other IgG genes and also divergent from each other. IgG1, IgG2, and IgG4 have nearly identical CH1 domains (Figs. 1, 4A). The last 19 positions are generally shared by the IgG5 and the IgG6 alleles which differ in this region from the IgG1–IgG2–IgG4 group by three to four nonconservative changes. The putative IgG1 alleles differ at only one position, the Thr/Val at position 18; this point substitution is also seen in IgG3, IgG4b, and IgG5b. There is only one difference between the putative IgG4a/IgG4b allelic variants and the IgG2a/IgG2b allelic variants. The latter involves the Gly/Ser at position 15. However, the putative IgG5 alleles differ from each other at ten positions, and the IgG6 alleles differ from each other at six positions.
IgG5a differs from IgG1a at seven positions, all of which are nonconservative substitutions. Only four of these are shared by IgG5b. IgG3 differs from IgG1a at 15 positions in CH1 of which only one (Ala/Val at position 29) is shared with both IgG5 variants but an additional five are shared with IgG5b including the Thr/Val substitution at position 18. Perhaps the CH1 domain of IgG5b and IgG3 may have arisen from a recent ancestral CH1 domain (Fig. 4A).
The upper hinge
We have divided the genetic hinge, defined as a separate exon, into an upper hinge (residues 99–105) and a middle hinge or “core” hinge from 106–114 (Nezlin 1994). The lower hinge, encoded at the beginning of the CH2 domain, encompasses residues 115–121. These divisions are indicated in Fig. 1. The upper genetic hinge of IgG2a, IgG2b, IgG1a, and IgG4a are identical. This means that IgG1 and IgG4, which are nearly sequence identical in CH1, differ allelically in their upper hinge by four out of five amino acids. IgG4a shares the upper hinge of IgG1a while IgG4b shares the upper hinge of IgG1b (Fig. 4B). Thus, IgG4b and IgG1b lose the two potential O-linked glycosylation sites at positions 100 and 102 but gain a proline at position 103. IgG3 differs from all other porcine IgGs in the upper hinge and adds a double proline, thus expressing something like a middle hinge motif that may have some of the same properties. Paired prolines are also found in the upper hinge of human IgG3, mouse IgG3 and rat IgG2c (Nezlin 1994). Interestingly, IgG5a, IgG6a, and IgG6b all share the short upper hinge, while IgG5b has a unique upper hinge. No cysteines are encoded in the upper hinge of any porcine Cγ genes that could contribute to interchain disulfide bridges or could be involved in L–H disulfide bonds as in human IgG1. As indicated above, cysteine 14 in CH1 must serve that function (Fig. 1). The Lys-Thr motif that defines a trypsin cleavage site, is present in all porcine Cγ genes except IgG1b, IgG4b, and IgG3 and of course those Cγ genes (IgG5a and IgG6a) with a short upper hinge. This could be predictive of the effect of proteases on IgGs of these putative subclasses/allelic variants (Table 4).
The middle hinge
All porcine Cγ genes carry variations of the C-P-P-C motif that characterizes the “hinge core” (Nezlin 1994). All porcine Cγ genes contain at least two cysteines presumed to be involved in H–H disulfide bonds, while IgG3 has three (Fig. 1). Recent engineering studies on human IgG1 show that middle hinge stability resulting from polyproline structures and disulfide bridges are very important in C1q binding (Dall’Acqua et al. 2006). While human IgG3 can use alternate hinge exons, all porcine IgG3 sequences we have recovered have a single hinge exon. Except for IgG3, all porcine Cγ immunoglobulins have identical core hinge sequences.
The lower hinge
The first seven amino acids of the CH2 domain (115–121) constitute the lower hinge. This is identical for both alleles of IgG1 and IgG5a which is also true for the middle but not the upper hinge of these Cγ genes. The putative alleles of IgG2 and IgG4 share with IgG3 and IgG6b the Ala at position 115, but the IgG2 alleles have a Ser at position 119, while this is a Gly for IgG4 alleles and IgG6a and IgG6b. Both IgG2 and IgG4 have a Pro at position 120, which is also shared by IgG6a but not IgG6b. IgG6b lacks this proline, and the lower hinge terminates in a hydrophilic dipeptide (Glu+ Asn). IgG3 is again most divergent with three very conserved substitutions in the lower hinge; Val → Leu, Ala → Gly, Gly → Ala. The paired leucines that form the pepsin cleavage site in human IgG1 and in lower hinge of various species is absent in all swine Cγ genes.
Figure 4b shows that when the complete genetic hinge is considered, two features stand out. First, different Cγ genes share nearly identical hinges even though they are from different putative subclasses. Second, except for those with identical genetic hinges, the porcine Cγ genes show their lowest sequence similarity (note genetic distance scale in Fig. 4b). Based on motifs in IgG of other species, the sequence differences we have described are unlikely to yield homogeneous Fab/Fc or F(Ab’)2 fragments by proteolytic digestion (Table 4). Furthermore, differences in rigidity or flexibility that are dependent on disulfide bonds and proline residues could effect complement activation (see below).
The CH2 domain
The CH2 domain extends from 115 to 224 and contains a potential 60 amino acid intradomain disulfide loop (Cys 145 and 205) in all sequences. While the sequences of the putative subclasses differ, there are nearly no allelic differences. The allelic variant of IgG1 differs by two amino acids at the 3′ end (223, 224), and the alleles of IgG5 are identical except for the conservative Thr to Ser change at 183. All porcine IgG genes differ from IgG1a by the Gln → Arg change at position 139, and most others have a Glu → Asn or Glu → Lys change at position 142. The alleles of IgG2 differ only by the nonconservative Glu to Gly change at 178, and the IgG4 alleles are identical. The putative alleles of IgG6 stand out because of seven amino acid differences. Most unique among subclass genes is IgG3 with ten mostly nonconserved unique amino acids changes. Several (Hist → Glu at 154; Glu to Lys at 168, and Hist → Tyr at 169) are quite radical but only one (Ala → Ser at 214) involves a glycosylation site. The CH2 domain contains either two or three potential AG splice acceptors sites; this will be discussed in more detail below. The overall sequence similarity of the different Cγ sequences in CH2 (Fig. 4C) groups allelic variants as does Fig. 2, unlike when only the CH1 and the hinge domains are compared.
The CH3 domain
The domain (225–334) contains a potential 60 amino acid intradomain disulfide loop (Cys 251–311) and apart from the hinge: (a) the most subclass diagnostic domain and (b) allelic variants of putative subclasses group together in the same manner as is seen for the CH2 domain (Fig. 4D). IgG1 differs at up to 17 positions from the IgG2, IgG4, IgG6 group. This group shares many of these position changes; nine of the 15 are shared between IgG2 and IgG4, and six are also found in IgG6. Allelic differences in CH3 among the subclass Cγ genes are modest. IgG1a and IgG1b differ at five positions, IgG2 and IgG4 alleles and IgG6 alleles differ at seven positions and IgG5 alleles differ at six positions. The CH3 domains of IgG3 and IgG5 are most divergent from all other porcine Cγ genes but neither share tight sequence similarity with any of the Cγ genes reported here.
Introns are conserved and are subclass-specific
Further evidence supporting the identification of the six putative porcine IgG subclasses is based on differences in intron sequences; putative alleles of a subclass have nearly identical introns. IgG5 and IgG6 share a similar IVS1 which is shorter than the IVS1 of IgG1, IgG2, IgG3, and IgG4, while IgG3 has a unique IVS1 deletion (Fig. 1). IVS2 is almost the same length in all genomic sequences and for all but IgG3 differs by only 1–3 nt. IgG3 differs at 23 positions, i.e., 26% of the length of IVS2. IVS3 differs in length among Cγ genes; IgG3, IgG4, and IgG5 have an IVS3 that is 9 nt shorter than for IgG1, IgG2, and IgG6.
In summary, the overall sequence comparison (Figs. 1 and 2), the cladogram comparing the individual domains (Fig. 4), and the detailed analysis reveal several features of the porcine Cγ genes. First, they appear to belong to three groups; IgG1, IgG2, and IgG4 comprise one group, IgG5 and IgG6 another, and the ancestral-like IgG3 is in its own group. Second, the CH1 domain is the most conserved and very similar for IgG1, IgG2, IgG4, and IgG6 suggesting they may have duplicated from a common ancestral CH1 domain. The genetic hinge of IgG2, IgG4a, and IgG1a are identical, whereas IgG1b and IgG4b share a common hinge suggesting a type of genomic exon shuffle during diversification of porcine Cγ genes. Finally, the overall sequence of the CH2–CH3 domain complex, i.e., the Fc region, is most diagnostic of the six putative subclasses.
IgG3 is the most divergent Cγ gene of swine sharing little sequence homology with any other Cγ gene in any domain except with the CH1 of IgG5b (Fig. 4A). Most obvious is the extended genetic hinge of IgG3, the upper portion showing some similarity to that of IgG1b. Cloning of genomic DNA encoding IgG3 failed to provide evidence that the extended hinge of porcine IgG3 was due to the use of multiple hinge exons as is the case for long-hinged, human and bovine IgG3 (Lefranc and Lefranc 2001; Rabbani et al 1997). Like human IgG3, the middle hinge of IgG3 allows for additional inter-heavy chain disulfide bonding.
Alternative splicing may increase the number of expressed IgG variants
Figure 5 shows that in splicing the hinge exon to the CH2 domain, there are either two or three splice acceptor sites in the 5′ region of CH2 (ag; dependent on the putative subclass) that are situated to yield in-frame products although the alternative sites are noncanonical by their lack of a polypyrimidine tract (ppt). Nevertheless, transcripts using these sites were frequently found. It is recognized that if alternative sites are in close proximity to a canonical ppt, they can also be used (C.M. Stolzfus, personal communication). Alternatively spliced transcripts were most common in IgG5b in which the consensus splice acceptor site was frequently mutated. When 25 were sequenced, 12 spliced the hinge to an acceptor site with a canonical ppt 50 nt downstream (a position not shown in Fig. 5), but that results in a sterile transcript. Only five of the 25 transcripts used the first CH2 acceptor site, while eight used the second 12 nt downstream (Fig. 5). Both IgG5b and IgG6 transcripts have also been recovered that totally lack the CH1 domain (Fig. 5).
Porcine IgG3 is most likely to activate complement
The most attractive model describing the ionic C1q–IgG interaction involves Asp270 and Lys322 of all human IgGs (154 and 206 in swine; Fig. 1) that form salt bridges with Arg129 and Glu162 of the globular human B subunit of C1q, respectively (McCall and Easterbrook-Smith 1989; Emanuel et al. 1982; Burton et al. 1980). In addition, Tyr278, Lys326, Pro329, Pro331, Glu333 of human IgG1 (162, 210, 213, 215, 217, respectively, for swine; Fig. 1) are also involved in the interaction (Gaboriaud et al. 2003). Based on these motifs, only IgG3 is conserved at position 270 (154 in swine) where Glu represents a conserved replacement for Asp in human IgG1. Other pig IgG isotypes show major changes (His or Asn) at this critical site. While in swine the amino acid residues at 162, 206, and 215 (Tyr, Lys, and Pro) are conserved, radical changes were observed at position 210 in IgG1a, IgG1b (Val), and IgG5 (Glu) and at position 213 in IgG3 (Leu instead of Pro). There is also a radical change at position 217 in all the pig IgGs (Thr instead of Glu; Fig. 1). Our analysis also considered the hinge “spacer” effect in binding to C1q (Gaboriaud et al. 2003; Dall’Acqua et al. 2006). IgG3 has the potential for three inter-heavy chain disulfide bonds, while others have only had two cysteine residues that may form inter-heavy chain disulfide bonds (Fig. 1). Two may be insufficient to provide the rigidity and space to allow effective complement activation.
Specificity for Fc receptors predicted from sequence motifs
The overall structure of the human FcγRs is remarkably similar, and the critical residues involved in binding to the IgGs are highly conserved (Radaev and Sun 2001). When the pig FcγRs were compared to their human orthologues, most critical residues in FcγR that interact with human IgG1 were conserved or replaced by nonradical substitutions (amino acid residues 90, 113, 120, 129, 131–132, 155, 158; data not shown). However, some nonconserved replacements were seen (amino acid residues 134, and case of FcγRIII, 160 and 162). The interface between FcγRIII and the α-chain of Fc is dominated by hydrogen-bonding interactions, whereas hydrophobic interactions occur primarily between the FcγRIII and the β-chain of Fc.
When the lower hinge and CH2 domain of the porcine IgGs were compared to their human orthologues, we observed that all the interaction sites of the α-chain were fully conserved, although the critical interaction site for the β-chain in the lower hinge differed remarkably among the putative porcine subclasses. Only the sequence in IgG3 (positions 119, 120) is conserved (Fig. 1). While some subisotypes have nonradical substitutions that should not alter binding (IgG1 and IgG5 have Val-Ala instead of Leu-Gly), the Gly-Pro change in IgG4 and IgG6a and the Ser-Pro change in IgG2 and Gly-Asn in IgG6b , may prevent hydrophobic interaction with FcγRs and lower the affinity for the β-chain (Fig. 1). All pig IgG subisotypes except IgG3 have Glu instead of Leu at position 118 which may prevent hydrophobic interaction in this “hot spot”. The other interaction sites for the FcγR β-chain (213,216; Fig. 1) are conserved or have nonradical substitutions. For example, IgG3 has Leu instead of Pro at site 213 (Fig. 1). Based on differences in the hinge length of porcine IgG subisotypes, we speculate that IgG5a, IgG5b, and IgG6a may bind poorly to FcγRs compared to the other porcine subisotypes because their lower hinges are shorter by one to three amino acids.
A comparison of FcRn α-chains from several species, among them the pig, has been performed (Kacskovics et al. 2006) using the crystallographic analysis of the rat FcRn–heterodimeric Fc complex as a guide (Martin et al. 2001). The study found Glu117, Glu118, Glu132, Trp133, Glu137 of the rat FcRn to be highly conserved and that mutation at these sites resulted in a significant loss of binding affinity. These important residues are conserved in pig FcRn. Fig. 1 shows that all the important residues in the Fc region (Ile 137, His 194–Gln 195, His 321) are fully conserved in all porcine IgGs except IgG5a, IgG5b, and IgG6a (Fig. 1). Radical replacement at position 195 (an acidic Glu instead of the conserved uncharged polar Gln) in IgG5 and IgG6a could prevent association since this residue (Glu) faces Glu 118 in FcRn. At the slightly acidic condition at which FcRn and IgG/Fc interact, both Glu residues would be positively charged.
The bacterial-binding protein SpA establishes contact with residues in the Fc region of human IgG that are located in three separate loops of the Cγ2–Cγ3 interface (135–138, 195–196, and 319–324) (Deisenhofer 1981) (Fig. 1). These motifs also interact with FcRn. These sites are generally conserved among the putative porcine subclasses except in IgG5a, IgG5b, and IgG6a that show a radical replacement at position 195 where the acidic Glu replaces the neutral Gln (Fig. 1). This may modify interaction with SpA of these porcine isotypes. Predictions about SpG affinity proved difficult. Gln 195 in Cγ genes is believed to form a polar interaction with Glu 42 of protein G, which means that IgG5 and IgG6a that have Glu at 195 should bind with lower affinity. Glu 380 and Glu 382 (272–274 in swine, Fig. 1) are also considered “hot spots” for protein G binding, and this motif (Glu-Pro-Glu) is conserved in all porcine IgGs.
Predictions concerning specificity of porcine IgG subclass proteins for various Fc receptors and susceptibility to proteases are summarized in Table 4.
Discussion
Data presented advance our knowledge about Cγ diversity in a mammal with an emerging role in xenotransplantation (Auchincloss and Sachs 1998), cystic fibrosis (Rogers et al. 2008), the generation of humanized antibodies (Waltz 2006; Mendicino et al. 2008), an established role in developmental immunology (Butler et al. 2002, 2005, 2008b) and that is important to the world food supply.
Our initial studies on the Cγ genes of swine utilized genomic blots and cDNA sequences to reveal a diverse Cγ repertoire (Kacskovics et al 1994). Here we show that many of the porcine Cγ genes share similar or identical exons. IgG1, IgG2, IgG4, and perhaps IgG6 appear to have obtained their CH1 exon by recent duplication of an ancestral CH1 exon that is genomically rearranged to the same hinge as seen in IgG1a, the IgG2 alleles and IgG4a whereas IgG1b and IgG6 were rearranged to two different hinge exons. The hinge used by IgG1b is identical to that found in IgG4b while the IgG6 hinge is shared with IgG5a (Fig. 4B) so that IgG4a is essentially “an IgG6 with an IgG1a hinge,” whereas IgG4b is an “IgG6 with an IgG1b hinge.” Given the alternative splicing observed in this study and discussed below, we initially entertained the idea that the shared exons found among the 11 Cγ genes were “exon chimeras” resulting from transcriptional/splicing events or PCR artifacts. Since we recovered all 11 from genomic DNA together with their introns, the former is unlikely (Fig. 1). However, heavy chain “hybrids” are known, especially after the advent of hybridoma technology (Mihaesco et al. 1988; Kunkel et al. 1969; Birshstein et al. 1980; Colle et al. 1987). The argument that the Cγ genes described are PCR cloning artifacts is inconsistent with our previous study on PCR-generated VDJ chimeras in the same species (Sinkora et al. 2000). PCR chimeras can be generated in vitro when only two to five different templates are present and are rarely recovered when large numbers of different templates are present as in a normal cDNA (Sinkora et al. 2000). Most important is that two such chimeras seldom have the same sequence, i.e., the polymerase does not “jump” at the same point. Considering the >9,000 cDNA and ~ 3,000 DNA clones recovered and that the >270 that were sequenced from different variants had identical sequences (Table 2), it is likely that the IgG subclass/allotypic diversity we have described is genuine. The possibility that the sharing of genomic exons represents some form of AID-dependent templated somatic mutation, i.e. “gene conversion” is inconsistent with what is known about this process in chicken (Ratcliffe 2006) in which: (a) each mosaic product has a unique sequence, whereas the 184 cDNA and 88 DNA Cγ genes that were sequenced (Table 2) yielded the same seven sequences that we ascribed to subclasses and allelic variants, (b) the mosaic is confined to a 10- to 300-bp region of a single exon not mixing of exons and (c) templated somatic mutation among Ig genes has never been described for constant region genes. Authentic gene conversion is a genomic event in yeast and distinct from the vernacular use of the term in immunology textbooks (Janeway et al 2005). Therefore, it is conceivable that some form of this process combined with gene duplication over the 50 million years of evolution in family Suidae could explain the origin of the porcine subclass genes, specifically the “exon shuffle” aspect. Without further information about the IgG subclasses of other contemporary members of family Suidae and/or DNA sequences recovered from fossils, we are more comfortable with attributing the origin of the porcine Cγ genes to gene duplication that is well documented among the hundreds of members of the Ig supergene family (Hunkapillar and Hood 1989; Williams 1987) with the added feature of “exon shuffle” (Fig. 4A,B) without assigning the latter to a particular mechanism. Differences between exons of actual or putative allelic variants are very small, and we believe are explained by single point mutations in the genome that occurred during or after duplication and exon shuffle.
Our analysis of the Cγ genes of swine and other mammals is consistent with earlier studies suggesting that diversification of IgG occurred after speciation (Kehoe and Capra 1974). Figure 6 shows that the various Cγ genes of swine group together and are not intermixed with those of other mammals. This pattern is seen throughout the cladogram for the IgGs of other species. The overall pattern in Fig. 6 is generally consistent with accepted taxonomic schemes in that the Artiodactyls (swine, cattle, camel) group together while rabbits are grouped with rodents. Noteworthy is that porcine IgG3 appears to be most similar in sequence to the stem IgG of all mammals. Data of Fig. 6 also show that even when intron sequences are not included as in Fig. 2, the putative alleles of the putative swine IgG subclasses group together. Figure 6 also shows that “name homologs” of IgG subclasses in other mammals do not share an evolutionary homolog indicating the danger of extrapolating function from a subclass in one species to one of the same name in another.
Despite their apparent species-specific divergence, IgG subclass proteins in different mammals appear to perform similar functions, and certain functions are associated with different subclasses in a number of these mammals (Burnett et al. 1989; Lefranc and Lefranc 2001; Butler 1983; Clark 1997). As a first step in determining which porcine IgG subclasses have analogous behavior to those in other mammals, we analyzed the sequences of the various porcine Cγ genes to identify motifs that might predict their effector activities. Since our predications are based only on sequence motifs, they await confirmation when purified subclass proteins become available for this species. In any case, our predictions show that all porcine IgGs except IgG5 and IgG6a should be able to bind Fcγ receptors, but some degree of affinity differences are likely (Table 4). FcRn protects IgG from degradation and is responsible for the long serum half-life of certain subclasses of IgG as well as mediating transepithelial transport (Roopenian and Akilesh 2007). Based on sequence motifs, we predict that IgG5 and IgG6a would have the shortest plasma half-life and would be least likely to be transported through epithelial cells such as the acinar epithelial cells of the mammary gland (Butler 1974; Brambell 1971). Since SpA and SpG bind IgG to the same or similar motifs as FcRn (Deisenhofer 1981; Nezlin and Ghetie 2004), all porcine IgG isotypes should bind to SpA and SpG although with different affinity.
C1q shows marked differences in its binding to the CH2 domain of different human IgG subclasses (McCall and Easterbrook-Smith 1989; Duncan and Winter 1988; Idusogie et al. 2000). Furthermore, the Fab/Fc orientation, controlled at least partly by the hinge region, is a critical factor in recognition of C1q (Gaboriaud et al. 2003) as is the importance of the hinge as a spacer (Dall’Acqua et al. 2006). Therefore we predict that pig IgG3 has all the necessary features to activate complement (Table 4). While differences in the ability of the porcine IgG subclasses in complement activation have been suggested, previous investigators identified the IgG subclasses involved using commercial mAbs of untested specificity (Crawley and Wilkie 2003). Our predictions concerning the function of IgG3 in complement activation, affinty for FcγRs (Table 4), and its ancestral nature (Fig. 6) is interesting because IgG3 comprises >60% of total IgG transcripts in the ileal Peyers patches (IPP) and mesenteric lymph nodes of late term and newborn fetuses but expression of IgG3 in the IPP precipitously drops in the first 4–6 weeks postpartum (Butler and Wertz 2006). We therefore speculated that IgG3, like those from marginal zone B cells (Lopez-Carvalho et al 2005) may be an important part of the antigen-independent preadaptive response in newborn piglets and thus important in the innate and pre-adaptive immune response of swine.
Splice variants within the Ig supergene family are well known including swine. The allotypic variants of porcine IgA differ because of a mutated splice acceptor site in CH2 resulting in a four amino acid hinge deletion (Brown et al. 1995) and in in vitro studies, Cμ1 can be spliced to either Cδ1 or Hδ (Zhao et al. 2003). Alternative porcine Cγ transcripts are not unusual since all but five of the 25 IgG5b transcripts we sequenced were alternatively spliced. Although it remains to be shown whether any are expressed, there are examples of truncated heavy chain proteins such as in human heavy chain disease in which the Cγ chain lacks the CH1 domain (Franklin et al. 1964; Osserman and Takatsuki 1964). The 5 S protein described 40 years ago from newborn piglets (Franek and Riha 1964) may be the product of an IgG5b alternative transcription (Fig. 5). Because the putative cysteine involved in the L–H disulfide bond is found at position 14 in the CH1 domain, porcine splice variants lacking a CH1 domain may encode “heavy chain only antibodies” as in camelids (Hamers-Castermann et al. 1993) unless the light chain is noncovalently associated as in the case of human IgA2 (Grey et al. 1968; Marquart and Deisenhofer 1982) and some heavy chain disease proteins (Franklin et al. 1964; Osserman and Takatsuki 1964).
Conclusions regarding allotypic variants are based on: (a) relative occurrence (Tables 2 and 3), (b) genomic sequences in which introns sequences are included (Fig. 2), (c) the absence of some variants in genomic DNA, and (d) the sequence similarity of their CH2–CH3 domain complex, i.e., Fc. Confirmation of our conclusions awaits locus maps and breeding studies.
The complexity of the IgG system of swine and the inability to physico-chemically purify porcine IgG subclass proteins, challenges the process of preparing subclass-specific mAbs. The use of vectors that allow in vitro expression of each variant will be needed to provide purified subclass proteins to: (a) test the specificity of existing mAb, (b) prepare the next generation of mAbs and, (c) test the functional predictions of Table 4. Since the Fc regions (CH2–CH3) of the porcine Cγ genes are subclass-specific regardless of putative allotype differences (Fig. 4C,D), they offer the best targets for the preparation of subclass-specific mAbs.
Domesticated farm animals have been randomly inbred and in the case of NIH minipigs, selectively inbred to establish class I homozygous lines suitable for transplantation research (Auchincloss and Sachs 1998). This process has also impacted Ig genetics. We have previously shown that NIH minipigs and Meishans from a single herd were homozygous for IgAa (Navarro et al. 2000a). This now appears only to be true for one strain of Meishans, i.e., it is founder related (Table 3). Most outbred swine are IgG1a/IgG1b and IgAa/IgAb heterozygotes (Butler and Wertz 2006; Navarro et al. 2000a), and the gene deficiencies we observed were only observed in swine homozygous for IgG1a/IgAa (Table 3). Considering the >270 Cγ gene sequences recovered form DNA that were examined, it is unlikely that random sampling would have missed any variant of IgG2 or IgG4 in the animals selected. However, data presented in Table 3 are only an example of Cγ gene deletions; extensive population studies are needed to determine if this phenomenon is widespread. Deletion of complete CH genes in humans is known albeit uncommon, and individuals often present with recurrent infections or other abnormalities (Migone et al. 1984; Terada et al. 2001; Pan and Hammarstrom 2000). However, CH gene and haplotype deletions in healthy individuals have also been reported (Lefranc et al. 1982; Olsson et al. 1993). Since our predictive analysis of complement, FcγR and FcRn binding did not identify any unique function for porcine IgG2 and IgG4, their loss may confer no selective disadvantage.
The data presented represent a major advance in the characterization of IgG diversity in swine, a model to explain the evolutionary diversification of IgG subclasses in all mammals and the predictive role of different IgG subclasses in the porcine immune system. The studies also highlight the potential difficulties in the preparation of subclass-specific mAbs and the danger of extrapolating subclass function from one species to another. We hope this study will encourage further immunogenetic, breeding and functional studies to test the predictions arising from this work.
References
Auchincloss H Jr, Sachs DH (1998) Xenogeneic transplantation. Annu Rev Immunol 16:433–470
Bianchi ATJ, Schotten JW, Jongenelen IMCA, Koch G (1990) The use of monoclonal antibodies in an enzyme immunospot assay to detect isotype-specific antibody secreting cells in pigs and chickens. Vet Immunol Immunopathol 24:125–134
Birshstein BK, Campbell R, Greenberg ML (1980) A γ2b–γ2a hybrid immunoglobulin heavy chain produced by a variant of the MPC11 mouse myeloma cell line. Biochem 19:1730–1737
Brambell FWR (1971) The transmission of passive immunity from mother to young. Frontiers of Biology Series. North Holland, Amsterdam
Brown WR, Kacskovics I, Amendt B, Shinde R, Blackmore N, Rothschild M, Butler JE (1995) The hinge deletion variant of porcine IgA results from a mutation at the splice acceptor site in the first Cα intron. J Immunol 154:3836–3842
Burnett RC, Hanly WC, Zhai S-K, Knight KL (1989) The IgA heavy chain gene family in rabbit: cloning and sequence analyses of 13 Cα genes. EMBO J 8:4041–4047
Burton DR, Boyd J, Brampton AD, Easterbrook-Smith SB, Emanuel J, Novotny EJ, Rademacher TW, van Schravendijk MR, Sternberg MJ, Dwek RA (1980) The Clq receptor site on immunoglobulin G. Nature 288:338–344
Butler JE (1969) Bovine immunoglobulins. J Dairy Sci 52:1895–1909
Butler JE (1974) Immunoglobulins of the mammary secretions. In: Larson BL, Smith V (eds) Lactation, a comprehensive treatise. Academic Press, New York Vol. III, Chapter V:217–55
Butler JE (1983) Bovine immunoglobulins: An augmented review. Vet Immunol Immunopathol 4:43–152
Butler JE (1997) Immunoglobulin gene organization and the mechanism of repertoire development. Scand J Immunol 45:455–462
Butler JE (2006) Preface: Why I agreed to do this. In: Butler JE, guest ed. Antibody repertoire development. Dev Comp Immunol 30:1–17
Butler JE, Wertz N (2006) Antibody repertoire development in fetal and neonatal pigs. XVII. IgG subclass transcription in newborns revisited with emphasis on new IgG3. J Immunol 177:5480–5489
Butler JE, Sun J, Weber P, Francis D (2000) Antibody repertoire development in fetal and neonatal piglets. III. Colonization of the gastrointestinal tracts results in preferential diversification of the pre-immune mucosal B-cell repertoire. Immunol Br 100:119–130
Butler JE, Sun J, Weber P, Ford SP, Rehakova Z, Sinkora J, Lager KM (2001) Antibody repertoire development in fetal and neonatal piglets. IV. Switch recombination, primary in fetal thymus occurs independent of environmental antigen and is only weakly associated with repertoire diversification. J Immunol 167:3239–3249
Butler JE, Weber P, Sinkora M, Baker D, Schoenherr A, Mayer B, Francis D (2002) Antibody repertoire development in fetal and neonatal piglets. VIII. Colonization is required for newborn piglets to make serum antibodies to T-dependent and type 2 T-independent antigens. J Immunol 169:6822–6830
Butler JE, Francis D, Freeling J, Weber P, Sun J, Krieg AM (2005) Antibody repertoire development in fetal and neonatal piglets. IX. Three PAMPs act synergistically to allow germfree piglets to respond to TI-2 and TD antigens. J Immunol 175:6772–6785
Butler JE, Lemke CD, Weber P, Sinkora M, Lager KM (2007) Antibody repertoire development in fetal and neonatal piglets. XIX. Undiversified B cells with hydrophobic HCDR3s preferentially proliferate in PRRS. J Immunol 178:6320–6331
Butler JE, Weber P, Wertz N, Lager KM (2008a) Porcine reproductive and respiratory syndrome virus (PRRSV) subverts development of adaptive immunity by proliferation of germline-encoded B cells with hydrophobic HCDR3s. J Immunol 180:2347–2356
Butler JE, Lager KM, Splichal I, Francis D, Kacskovics I, Sinkora M, Wertz N, Sun J, Zhao Y, Brown WR, DeWald R, Dierks S, Muyldermanns S, Lunney JK, McCray PB, Rogers CS, Welsh MJ, Navarro P, Klobasa F, Habe F, Ramsoondar J (2008b) The piglet as a model for B cell and immune system development. Vet Immunol Immunopathol (in press).
Clark MR (1997) IgG effector mechanisms. Chem Immunol 65:88–110
Colle JH, Perret R, Truffa-Bachi P (1987) Hybrid polypeptide heavy chains produced by two hybridoma lines. Mol Immunol 24:39–46
Crawley A, Wilkie BN (2003) Porcine Ig isotypes: function and molecular characteristics. Vaccine 21:2911–2922
Dall’Acqua WF, Cook KE, Damschroder MD, Woos RM, Wu H (2006) Modulation of the effector functions of a human IgG1 through engineering of its hinge. J Immunol 177:1129–1138
Deisenhofer J (1981) Crystallographic refinement and atomic models of a human Fc fragment and its complex with fragment B of protein A from Staphylococcus aureus at 2.9- and 2.8-A resolution. Biochem 20:2361–2370
Duncan AR, Winter G (1988) The binding site for C1q on IgG. Nature 332:738–740
Emanuel EJ, Brampton AD, Burton DR, Dwek RA (1982) Formation of complement subcomponent C1q–immunoglobulin G complex. Thermodynamic and chemical-modification studies. Biochem J 205:361–372
Franek F, Riha I (1964) Purification and structural characterization of 5 S γ-globulin of newborn pigs. Immunochemistry 1:49–63
Franklin EC, Lowenstein J, Bigelow B, Meltzer M (1964) Heavy chain disease—a new disorder of serum γ-globulins. Am J Med 37:332–350
Gaboriaud C, Juanhuix J, Gruez A, Lacroix M, Darnault C, Pignol D, Verger D, Fontecilla-Camps JC, Arlaud GJ (2003) The crystal structure of the globular head of complement protein C1q provides a basis for its versatile recognition properties. J Biol Chem 278:46974–46982
Grey HM, Abel CA, Jount WJ, Kunkel HG (1968) A subclass of human γA-globulins (γA2) which lacks the disulfide bonds linking heavy and light chains. J Exp Med 128:1223–1236
Hamers-Castermann C, Atarhouch T, Muyldermans S, Robinson G, Hamers C, Songa EB, Bendahman N, Hamers R (1993) Naturally occurring antibodies devoid of light chains. Nature 363:446–448
Hunkapillar T, Hood L (1989) Diversity of the immunoglobulin gene superfamily. Adv Immunol 44:1–63
Idusogie EE, Presta LG, Gazzano-Santoro H, Totpal K, Wong PY, Ultsch M, Meng YG, Mulkerrin MG (2000) Mapping of the C1q binding site on rituxan, a chimeric antibody with a human IgG1 Fc. J Immunol 164:4178–4184
Janeway CA Jr, Travers P, Walport M, Shlomchik MJ (2005) Immunobiology Garland Press, N.Y. 823 pp
Kacskovics I, Sun J, Butler JE (1994) Five subclasses of swine IgG identified from the cDNA sequences of a single animal. J Immunol 153:3566–3573
Kacskovics I, Mayer B, Kis Z, Frenyo LV, Zhao Y, Muyldermans S, Hammarstrom L (2006) Cloning and characterization of the dromedary (Camelus dromedarius) neonatal Fc receptor (drFcRn). Dev Comp Immunol 30:1203–1215
Kaltreider HB, Johnson JS (1972) Porcine immunoglobulins. I. Identification of subclasses and preparation of specific antisera. J Immunol 109:992–998
Kehoe JM, Capra JD (1974) Nature and significance of immunoglobulin subclasses. N Y State J Med 74:489–491
Kilian M, Russell MW (2004) Microbial evasion of IgA function. In: Mestecky J, Lamm ME, Strober W, Bienenstock J, McGhee JR, Mayer L (eds) Mucosal immunology. Elsevier/Academic, Amsterdam, pp 291–303
Kunkel HG, Natvig JB, Jostin FG (1969) A “lepore” type of hybrid γ globulin. Proc Natl Acad Sci USA 62:144–149
Lefranc M-P, Lefranc G (2001) The immunoglobulin facts book. Academic, NY
Lefranc M-P, Lefranc G, Rabbitts TH (1982) Inherited deletion of immunoglobulin heavy chain constant region genes in normal individuals. Nature 300:760–762
Lemke CD, Haynes JS, Spaete R, Adolphson D, Vorwald A, Lager KM, Butler JE (2004) Lymphoid hyperplasia resulting in immune dysregulation is caused by PRRSV infection in pigs. J Immunol 172:1916–1925
Lopez-Carvalho T, Foote J, Kearney JF (2005) Marginal zone B cells in lymphocyte activation and regulation. Curr Opin Immunol 17:244–250
Marquart M, Deisenhofer J (1982) The three-dimensional structure of antibodies. Immunol Today 3:160–166
Martin WL, West AP Jr, Gan L, Bjorkman PJ (2001) Crystal structure at 2.8 A of an FcRn/heterodimeric Fc complex: mechanism of pH-dependent binding. Mol Cell 7:867–877
McAleer J, Weber P, Sun J, Butler JE (2005) Antibody repertoire development in fetal and neonatal piglets. XI. The thymic B cell repertoire develops independently from that in blood and mesenteric lymph nodes. Immunology 114:171–183
McCall MN, Easterbrook-Smith SB (1989) Comparison of the role of tyrosine residues in human IgG and rabbit IgG in binding of complement subcomponent C1q. Biochem J 257:845–851
Mendicino M, Ramsoondar J, Phelps C, Vaught T, Ball S, Dai Y, LeRoith T, Monahan J, Chen S, Dandro A, Boone J, Jobst P, Vance A, Wertz N, Polejaeva I, Butler J, Ayares D, Wells K (2009) Targeted disruption of the porcine immunoglobulin heavy chain locus produces a B cell null phenotype. Nature Biotechnology (pending)
Mestecky J, Moro I, Kerr MA, Woof JM (2004) Mucosal immunoglobulins. In: Mestecky J, Lamm ME, Strober W, Bienenstock J, McGhee JR, Mayers L (eds) Mucosal immunology. Elsevier/Academic, Burlington, MA, pp 153–181
Metzger JJ, Fougereau M (1967) Characterization of two subclasses of γG immunoglobulin in swine. CR Hebd Seances Acad Sci Ser P Sci Nat 265:724–727
Migone N, Oliviero S, de Lange G, Delacroix DL, Boschis D, Altruda F, Silengo L, DeMarchi M, Carbonara AO (1984) Multiple gene deletions within the human immunoglobulin heavy chain cluster. Proc Natl Acad Sci USA 81:5811–5815
Mihaesco E, Gendron M-C, Congy N, Frangione B (1988) Protein ROU, a human IgA hybrid. J Immunol 140:1236–1238
Navarro P, Christenson R, Ekhardt G, Lunney JK, Rothschild M, Bosworth B, Lemke J, Butler JE (2000a) Genetic differences in the frequency of the hinge variants of porcine IgA is breed dependent. Vet Immunol Immunopathol 73:287–295
Navarro P, Christenson R, Weber P, Rothschild M, Erhardt G, Lemky J, Butler JE (2000b) Porcine IgA allotypes are not equally transcribed or expressed in heterozygous swine. Mol Immunol 37:653–664
Nezlin R (1994) Immunoglobulin structure and function. In: van Oss CJ, van Regenmortel MHV (eds) Immunochemistry. Marcel Dekker, New York, pp 3–45
Nezlin R, Ghetie V (2004) Interactions of immunoglobulins outside the antigen-binding site. Adv Immunol 82:155–215
Olsson PG, Rabbani H, Hammarstrom L, Smith CIE (1993) Novel human immunoglobulin heavy chain constant region gene deletion haplotypes characterized by pulse-filed electrophoresis. Clin Exp Immunol 94:84–90
Osserman EF, Takatsuki K (1964) Clinical and immunochemical studies of four cases of heavy (Hγ2) chain disease. Am J Med 37:351–373
Pan Q, Hammarstrom L (2000) Molecular basis of IgG subclass deficiency. Immunol Rev 178:99–110
Plaut AG, Gilbert JV, Artenstern MA, Capra JD (1975) Neisseria gonorrhoeae and Neisseria meningitidis: extracellular enzyme cleaves human immunoglobulin A. Science 193:1103–1105
Rabbani H, Brown WR, Butler JE, Hammarström L (1997) Polymorphism of the IgHG3 gene in cattle. Immunogenetic 46:326–331
Radaev S, Sun PD (2001) Recognition of IgG by Fcgamma receptor. The role of Fc glycosylation and the binding of peptide inhibitors. J Biol Chem 276:16478–16483
Radaev S, Motyka S, Fridman WH, Sautes-Fridman C, Sun PD (2001) The structure of a human type III Fc gamma receptor in complex with Fc. J Biol Chem 276:16469–16477
Rapacz J, Hasler-Rapacz J (1982) Immunogenetic studies on polymorphism, postnatal passive acquisition and development of immunoglobulin gamma (IgG) in swine. In: Proc 2nd Int. Congress Gen and Appl Livestock Production. Vol III. Editoral Garsi, Madrid
Ratcliffe MJH (2006) Antibodies, immunoglobulin genes and the bursa of Fabricius in chicken B cell development. Dev Comp Immunol 30:101–118
Ravetch JV, Kinet J-P (1991) Fc receptors. Annu Rev Immunol 9:457–492
Rogers CS, Stoltz DA, Meyerholz DK, Ostedgaards LS, Rokhlina T, Taft PJ, Rogan MP, Pezuzulo AA, Karp PH, Itani OA, Kabel AC, Wohlford-Lenane CL, Davis GJ, Hanfland RA, Smith TL, Samuel M, Wax D, Murphy CN, Rieke A, Whitworth K, Uc A, Starner TD, Brogden KA, Shilyansky J, McCray PB Jr, Zabner J, Prather RS, Welsh MJ (2008) Disruption of the CFTR gene produces a model of cystic fibrosis in newborn pigs. Science 321:1837–1841
Roopenian DC, Akilesh S (2007) FcRn: the neonatal Fc receptor comes of age. Nat Rev Immunol 7:715–725
Sauer-Eriksson AE, Kleywegt GJ, Uhlen M, Jones TA (1995) Crustal structure of the C2 fragment of streptococcal protein G in complex with the Fc domain of human IgG. Structure 3:265–278
Sinkora M, Sun J, Butler JE (2000) Antibody repertoire development in fetal and neonatal piglets. V. VDJ gene chimeras resembling gene conversion products are generated at high frequency by PCR in vitro. Mol Immunol 37:1025–1034
Spieker-Polet H, Yam P-C, Knight KL (1993) Differential expression of 13 IgA heavy chain genes in rabbit lymphoid tissues. J Immunol 150:5457–5465
Sun J, Butler JE (1996) Molecular characteristics of VDJ transcripts from a newborn piglet. Immunol Br 88:331–339
Sun J, Hayward C, Shinde R, Christenson R, Ford SP, Butler JE (1998) Antibody repertoire development in fetal and neonatal piglets. I. Four VH genes account for 80% of VH usage during 84 days of fetal life. J Immunol 161:5070–5078
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) Software Version 4.0. Mol Biol Evol
Terada T, Kneko H, Li AL, Kasahara K, Ibe M, Yokota S, Kondo N (2001) Analysis of Ig subclass deficiency: first reported case of IgG2, IgG4 and IgA deficiency caused by deletion of C alpha1, PSI C gamma, C gamma 2, C gamma 4 and C epsilon in a Mongoloid patient. J Allergy Clin Immunol 108:602–606
Vincent AL, Lager KM, Ma W, Lekcharoensuk P, Gramer MR, Loiacona C, Richt JA (2006) Evaluation of hemagglutinin subtypes 1 swine influenza viruses from the United States. Vet Microbiol 118:212–222
Waltz E (2006) Polyclonal antibodies step out of the shadow. Nat Biotechnol 24:1181
Williams AF (1987) A year in the life of the immunoglobulin superfamily. Immunol Today 8:298–303
Zhao Y, Pan-Hammarstrom Q, Kacskovics I, Hammarstrom L (2003) The porcine Ig δ gene: Unique chimeric splicing of the first constant region domain in its heavy chain transcripts. J Immunol 171:1312–1318
Zhao Y, Pan-Hammarstrom Q, Yu S, Wertz N, Zhang X, Li N, Butler JE, Hammarstrom L (2006) Identification of IgF, a hinge-region containing Ig class and IgD in Xenopus tropicalis. Proc Natl Acad Sci USA 103:12087–12092
Acknowledgements
This research supported by Cooperative Agreement IOWR 2003-02669 with the USDA-ARS, The University of Iowa Carver Trust and Grants 05-015 and 06-043 from the National Porkboard and grant OTKA T049015 from the Hungarian Academy of Sciences.
Author information
Authors and Affiliations
Corresponding author
Additional information
An erratum to this article can be found at http://dx.doi.org/10.1007/s00251-009-0356-0
Rights and permissions
About this article
Cite this article
Butler, J.E., Wertz, N., Deschacht, N. et al. Porcine IgG: structure, genetics, and evolution. Immunogenetics 61, 209–230 (2009). https://doi.org/10.1007/s00251-008-0336-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-008-0336-9