Abstract
Previous studies initiated defining the role of host genetics in influencing the outcome of exposure to ovine progressive pneumonia virus. However, specific genes influencing host control of virus replication and disease progression have not been identified. This study, using 383 ewes of the Columbia, Polypay, and Rambouillet breeds, tested the hypothesis that host control of OPPV as measured by provirus levels in the peripheral blood associates with certain breeds and MHC class II Ovis aries (Ovar)-DRB1 expressed alleles. Rambouillet ewes were less likely to have measurable provirus levels as compared to Columbia ewes at ages 5 and 6 (P value < 0.02), and they exhibited lower provirus levels when compared to both Columbia and Polypay ewes of the same ages (P value < 0.05). The presence of DRB1*0403- or DRB1*07012-expressed alleles were significantly associated (P value = 0.019 and 0.0002, respectively) with lower OPP provirus levels but only were only found in 11% of the ewe flock. Analysis of each segregating amino acid in the β1 domain of DR β-chain revealed that amino acids Y31, T32, N37, T51, Q60, or N74 significantly associated (P value range = 0.0003–0.018) with lower OPP provirus levels, whereas amino acids H32, A38, or I67 associated (P value range = 0.013–0.043) with higher OPP provirus levels. These results suggest that Ovar-DRB1 contributes as one host genetic factor that controls OPP provirus levels, but does not fully account for the breed-specific OPP proviral differences.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ovine progressive pneumonia virus (OPPV), which is also known as maedi-visna virus (MVV) or ovine lentivirus (OvLV), infects sheep and causes life long persistent infection resulting in various degrees of arthritis, mastitis, cachexia, dypsnea, and encephalitis in some infected sheep (Narayan et al. 1988). However, OPPV disease progression is insidious in that the chronic inflammatory processes, especially in the lung and joints, take years to be clinically detectable and infected sheep are a persistent source of virus for transmission (DeBoer et al. 1979).
Pathogenesis of OPPV involves replication and persistence in the monocyte–macrophage lineage, and one measurement of infection is the presence of virus that has been reverse transcribed and integrated into the host genome as provirus (Gendelman et al. 1986; Gorrell et al. 1992). The presence of OPP provirus in alveolar macrophages associates with moderate and severe lung lesions, and the presence of 103 to 106 copies of provirus in adherent mononuclear cells associates with mild to severe lung lesions (Brodie et al. 1992; Zhang et al. 2000). In other lentivirus infections such as human immunodeficiency virus (HIV-1), provirus level is the preferred clinical marker of disease in persons being treated with anti-viral therapies (Verhofstede et al. 1994; Vitone et al. 2005).
The consistent presence of antibody to OPPV in persistently infected sheep is the basis for use of serological tests like agar gel immunodiffusion (AGID) and cELISA to detect infection (de Andres et al. 2005). However, these serological tests are limited in that they do not quantify the level of infection. The development of quantitative PCR for determination of OPPV provirus levels in peripheral blood leukocytes allows for a measurement of host viral control (Herrmann-Hoesing et al. 2007a, b).
Two reports showed lower OPPV seroprevalence in Rambouillet ewes as compared to ewes of other breeds (Gates et al. 1978; Snowder et al. 1990). Although these reports did not examine potential differences in viral exposure (dose) among these flocks, they suggest that host genetics influence the OPPV infection differences among breeds. Additionally, a study utilizing three monozygotic twin sets where one twin was experimentally infected with OPPV 85/34 strain and the other twin with OPPV 84/28 strain showed that within the same twin set, the same degree of pulmonary pathology was observed whereas between the twin sets, the extent of pathology differed (de la Concha-Bermejillo et al. 1995). These data implicate host genetics in controlling the degree of pulmonary pathology in OPPV-infected sheep.
There is abundant evidence to suggest that small ruminant lentiviruses such as OPPV are immunopathological diseases (Cheevers and McGuire 1988). Therefore, host immune response genes are viable targets for use in control of OPPV at the population level. The major histocompatibility complex (MHC) class II was evaluated as one host immune response gene that could associate with the control of OPPV. MHC class II has been identified as a putative cellular receptor for MVV and caprine arthritis encephalitis virus (CAEV); however, its main function is to present endogenous and exogenous antigens to CD4 positive T-lymphocytes cells (Dalziel et al. 1991; Hullinger 1993). CD4 positive cells, which interact with MHC class II, appear to play a major role in OPPV infection since depleting them causes reduced virus replication whereas depleting CD8 positive cells has no apparent impact on virus replication (Eriksson et al. 1999).
The DR and DQ MHC Class II proteins are located on the surface of antigen-presenting cells and are composed of type 1 membrane-spanning alpha and beta chains. The MHC Class IIa region encodes for the alpha and beta chains on separate loci. Recently, a finished genomic sequence for the MHC class IIa region revealed the following order of the MHC class II loci in a Rambouillet ram: DQB2, DQA2, DQB1, DQA1, and DRB1 (Herrmann-Hoesing et al. 2008). MHC Class II Ovar-DRB1 was chosen as the immune response gene in this study because it is highly polymorphic, transcribed, and there are over 100 different DRB1 alleles reported in Genbank based upon either restriction fragment length polymorphisms (RFLP) or the deduced amino acid sequence for the β1 domain encoded by exon 2 (Dutia et al. 1994; Schwaigger et al. 1996; Jugo and Vicario 2001; Konnai et al. 2003; Herrmann et al. 2005). Others have shown specific MHC Class II Ovar-DRB1 alleles or serological types associate with resistance and/or less severe clinical signs in bacterial foot rot, experimental bovine leukemia virus infections, and gastrointestinal nematodes (see review, Dukkipati et al. 2006). Indeed, research in human leukocyte antigens (HLA) suggest that specific HLA haplotypes including HLA-DR and DP genotypes may protect against transmission and infection of specific pathogens such as human immunodeficiency virus (HIV) (Carrington and O’Brien 2003; Polycarpou et al. 2002). In this study, detection and levels of OPP provirus were evaluated among breeds, Ovar-DRB1-expressed alleles, and the deduced amino acids comprising the β1 domain of DR β-chain of an Idaho ewe flock.
Materials and methods
Animals
Four hundred and five mature ewes from a closed OPPV endemic flock at the U.S. Sheep Experiment Station, Dubois, Idaho were used in this study. Serum and peripheral blood leukocytes were isolated from 10 ml whole blood using prior methods (Herrmann-Hoesing et al. 2007a, b). The final data set included 383 ewes whereby both MHC Class II Ovar-DRB1-expressed alleles and OPP provirus levels were determined. Of these 383 ewes, 129 were Columbia, 126 were Polypay, and 128 were Rambouillet ewes. Within the 129 Columbia ewes, 41, 34, 31, and 23 were 3-, 4-, 5-, and, 6-year-olds, respectively. Within the 126 Polypay ewes, 31, 34, 35, and 26 were 3-, 4-, 5-, and 6-year-olds, respectively. Within the 128 Rambouillet ewes, 33, 33, 35, and 27 were 3-, 4-, 5-, and 6-year-olds, respectively.
DNA isolation and OPPV qPCR
DNA was isolated from PBL following manufacturer’s directions for 10 million cells using Puregene technology (Gentra Systems Inc., Minneapolis, MN). The concentration and purity of DNA was determined spectrophotometrically at 260 and 280 nm. Methods describing the OPPV qPCR have been previously reported (Herrmann-Hoesing et al. 2007a, b). This OPPV qPCR uses primers and a Taqman probe targeting the transmembrane region of envelope, which is highly conserved in North American OPPV strains.
RNA isolation and Ovar-DRB1 typing
Total RNA was extracted from 2 ml whole blood using PAXtube technology following manufacturer’s directions (Qiagen, Inc.) and was stored at −80°C until needed. Ovar-DRB1 allelic typing was conducted utilizing RT-PCR on the total RNA followed by cloning and sequencing of 20–30 DRB1 cDNA clones of 195 sheep using previously described methods and reagents (Herrmann et al. 2005). Primers used in the PCR amplification of Ovar-DRB1 first domain (β1) were DRB1A-5′-CATGGTGTGCCTGTATTTCTCC-3′ that binds to 7–28 nucleotides in GenBank accession number M93432 and DRB1B-5′-GATCACCCCAGCCTCCTCTT-3′ that binds to 504–523 nucleotides in M93432. These primers are intron spanning from exon 1 and exon 3 of Ovar-DRB1 allow for complete sequence analysis of exon 2 encoding for β1 or the peptide-binding site. New DRB1 alleles were defined once observed in at least two clones from at least two separate PCR reactions. Since there were no new alleles observed in the last 50 animals of the first 195 tested, direct sequencing of RT-PCR products from the remaining 188 animals ensued and was performed identically to the previous 195 sheep except that cDNA PCR amplified product was treated with ExoSAP-IT (USB, Cleveland, OH). The ExoSAP-IT procedure was slightly modified from manufacturer’s directions whereby 1 μl of ExoSAP was added to 7 μl of PCR product incubated at 37°C for 30 min, 80°C for 15 min, and held at 4°C for indefinite. Four microliters of ExoSAP treated PCR product in contrast to 5 μl of plasmid DNA was sequenced using the same DRB1 primers used in the initial RT-PCR and Big Dye® sequencing methods as previously reported (Herrmann et al. 2005).
Analysis of PCR sequencing of Ovar-DRB1
In order to analyze a polymorphic gene like Ovar-DRB1 through direct PCR sequencing, a 32 × 32 DRB1 expressed allele matrix was generated. With this matrix, most, if not all of the possible 2 DRB1 allele combinations would be represented. PCR products that were directly sequenced in 188 sheep were initially aligned against the cloned and sequenced 32 DRB1 expressed sequences. The two alleles with the highest percent nucleotide identity to the unknown sample were further analyzed using the matrix. For example, if a PCR DRB1 cDNA product had the highest nucleotide sequence identity to DRB1*0206- and DRB1*1101-expressed alleles, then the 31 possible two-allele combinations for DRB1*0206 and the 31 possible two-allele combinations for DRB1*1101 would be aligned against the PCR DRB1 cDNA product. A PCR DRB1 cDNA product required 100% nucleotide identity to one allele (if homozygous) or to two-allele combinations in order to identify the genotype for that animal.
PHASE verification of haplotypes
Matrix-based method of haplotype determination was verified using PHASE v2.0 (Stephens and Donnelly 2003; Stephens et al. 2001). The option for phase-known haplotype entry in PHASE was used for the 195 sheep with haplotypes known from clones, and the 188 other animals were entered as phase unknown. Phase haplotype predictions on the 188 remaining animals were verified by cloning and sequencing if PHASE could not identify the alleles present based upon phase-known animals.
Phylogenetic analysis of Ovar-DRB1
Nucleotide and amino acid alignments were performed using DNAStar Lasergene v. 7.0 integrated software programs. Initial nucleotide sequences were assembled/edited in SeqMan, transferred to Editseq, and aligned using Clustal W in MegAlign. Phylogenetic analysis was conducted using Phylip (Phylogeny Inference Package) version 3.67.
Statistical analyses
Two types of analyses were performed on provirus levels data. First, logistic regression models from the logistic procedure of SAS v9.1 (SAS Institute, Cary, NC) were used to examine probability of positive (nonzero) provirus results. The odds ratios for significant associations were converted to risk ratios according to a previously published method (Zhang and Yu 1998). The second set of analyses involved using general linear models from the GLM procedure of SAS v9.1 to examine variation in logarithm10 transformed provirus levels. For each type of analysis, an initial model included breed, a linear covariate of age, and an interaction term. The interaction was further investigated by analyzing sheep of each age separately in a model including breed. DRB1 genotype data were analyzed on a presence/absence basis to account for the codominant inheritance of MHC Class II molecules, and a minimum of 5% allele frequency in the overall sample of 383 sheep was required for testing individual allelic associations. The presence of each of the 32 DRB1 expressed alleles, the presence of each segregating amino acid encoded within nucleotides 1–273 of DRB1 exon 2, and each silent nucleotide difference amongst the 32 DRB1 expressed alleles were analyzed for associations with the presence/absence of provirus and provirus levels. Both logistic analysis of infection probability and general linear model analysis of logarithm transformed provirus levels were used for testing allelic association. In each case, the association model included breed, a linear covariate of age, a breed-by-age interaction, and an indicator variable for the presence/absence of the allele. All reported P values are nominal and are not corrected for multiple testing.
Results
Breeds in a ewe flock were evaluated for differences in the frequency of animals that had detectable OPP provirus in peripheral blood. The interaction of breed and age was significant, indicating that the breeds differed in odds of detectable provirus over the observed age range. There were no significant differences among breeds at age 3 in terms of detectable OPP provirus (data not shown), but there were significantly fewer OPP provirus positive Rambouillet ewes than Columbia ewes at 5 years (P value = 0.016; Risk Ratio (RR) 2.80; 95% confidence interval (CI) 1.39–4.10) and at 6 years of age (P value = 0.0048; RR 10.03; 95% CI 1.97–19.30). There were significantly fewer Polypay ewes with detectable provirus compared to either Rambouillet ewes (P value = 0.0070; RR 1.71; 95% CI 1.05–2.22) or Columbias (P value = 0.0070; RR 2.01; 95% CI 1.19–2.67) at age 4. However Polypay ewes did not significantly differ in the frequency of animals with detectable OPP provirus compared to Rambouillet ewes or Columbia ewes at 3, 5, or 6 years of age.
Furthermore, the mean OPP provirus levels (log10 env copies/microgram DNA) were examined for each age and breed in the 245 sheep with detectable OPP provirus to investigate host ability to control OPPV. The interaction of breed and age was significant, indicating that breeds differed in OPPV provirus levels over the observed age range. The Rambouillet ewes at ages 3 and 4 had generally lower mean OPP provirus levels than the Columbia ewes at the same ages, but it was not significant (P values at 0.053 and 0.083 for 3 and 4 years of age, respectively, see Fig. 1). However, the Rambouillet ewes at ages 5 and 6 had significantly lower OPP provirus levels than the Columbia ewes at 5 (P value = 0.045) and 6 (P value = 0.013) years of age or the Polypay ewes at 5 (P value = 0.0030) and 6 (P value = 0.0009) years of age (Fig. 1).
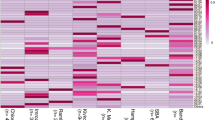
In addition, Ovar-DRB1 was tested as a possible host factor influencing control of OPP provirus. In continuation of an earlier study showing a total of 15 Ovar-DRB1 expressed alleles in an ewe–lamb flock of 32 originating from Idaho (Herrmann et al. 2005), Ovar-DRB1 expressed alleles were cloned and sequenced from 195 out of 383 ewes of the same Idaho flock and 18 additional DRB1 expressed alleles were identified (DQ659114–DQ659135, EF380349–EF380350, EF566981, Table 1). These DRB1 expressed alleles include 66 nucleotides of exon 1 and 273 nucleotides of exon 2. Using the allele information from cloning and sequencing (see “Materials and methods”), Ovar-DRB1 expressed alleles were identified in an additional 188 ewes from direct sequencing of cDNA products that were PCR amplified using DRB1 primers. Collectively, a total of 33 DRB1 expressed alleles have been identified in a total of 424 sheep from Idaho, 32 of which were found in the 383 ewes of this study. An alignment of the deduced amino acid sequence encoded by exon 2 or the β1 domain of the beta chain is shown in Fig. 2. Of the additional 18 expressed DRB1 alleles identified in this study, DRB1*0144, DRB1*02032, DRB1*0803, DRB1*1102 are new DRB1 expressed alleles in terms of Genbank accessions, and all but one new DRB1 expressed allele (DRB1*1102) was found in two or more ewes. DRB1*02032 has a synonymous nucleotide change in exon 1 as compared to DRB1*0203 and has been named according to nomenclature methods in order to distinguish between the two for this association study (Konnai et al. 2003).
In order to evaluate the diversity of the DRB1 expressed alleles identified in the ewe flock and to evaluate whether DRB1 phylogeny groups DRB1 alleles according to breed, both deduced amino acid sequence identities and phylogenetic analysis were conducted. Deduced amino acid sequence identities in the β1 domain of the 33 Ovar-DRB1 expressed alleles ranged from 72.5 to 98.9% between any two alleles. A phylogenetic tree was constructed for the 33 Ovar-DRB1 expressed alleles using Phylip (Phylogeny Inference Package) version 3.67 with the criterion set at distance with neighbor joining and 1,000 bootstraps (Fig. 3). A similar tree resulted when the criteria were set to parsimony with heuristic and 1,000 bootstraps (data not shown). Both of the phylogenetic analyses revealed that the DRB1 alleles in these sheep had three main branches signified by A, B, and C; however, the three main branches did not correspond to the three specific breeds (Fig. 3).
Phylogenetic analyses of the 33 Ovar-DRB1 expressed alleles including nucleotides 1 through 273 of exon 2 using Phylip (Phylogeny Inference Package) version 3.67 with parameters set at distance with neighbor joining with 1,000 bootstraps. Three major branches of Ovar-DRB1 expressed alleles were observed in this data set, which are indicated by circles and the letters A, B, and C
Our previous data showed that certain Ovar-DRB1 expressed alleles were identified in specific breeds of sheep (Herrmann et al. 2005). Therefore, total and breed-specific DRB1 allelic frequencies were calculated for the 32 DRB1 expressed alleles of 383 sheep within this study (Table 2). Thirty-seven ewes (9.6%) in the population were homozygous for DRB1 where 2.9%, 1.8%, and 1.3% of the population were homozygous for DRB1*02032, DRB1*1202, and DRB1*0206, respectively. In terms of breed-specific alleles, DRB1*GS1 was only found in Columbia ewes, and DRB1*0803 and DRB1*j were only found in Rambouillet ewes. There were no DRB1 alleles found exclusively in Polypay ewes.
Since OPP provirus levels significantly associated with breed, the possible association between specific DRB1 expressed alleles and detection or levels of OPP provirus was investigated. Initially, each of the 32 MHC Class II Ovar-DRB1 expressed alleles was examined for association with detectable OPP provirus. However, none of the MHC Class II Ovar-DRB1 expressed alleles significantly associated (P value < 0.05) with either undetectable or detectable provirus. In addition, each MHC Class II Ovar-DRB1 expressed allele was examined for its restrictiveness or permissiveness for OPPV, resulting in lower or higher provirus levels, respectively, as compared to ewes without this Ovar-DRB1 expressed allele. In this analysis, DRB1*0403 and DRB1*07012 appeared more restrictive of OPPV than the average DRB1 allele as seen by significant associations with lower provirus levels (P value = 0.019 and 0.0002, respectively). The corresponding effect sizes were 0.4 and 1.01 for DRB1*0403 and DRB1*07012, respectively, where the corresponding effect size is defined as the difference in log10 mean provirus levels between animals with this DRB1 expressed allele and animals without this DRB1 expressed allele. However, none of the Ovar-DRB1 expressed alleles significantly associated with higher provirus levels.
To further investigate the role of DRB1, we examined segregating amino acids as biological markers that could have functional involvement in antigen presentation. There were 30 variable amino acid positions out of 91 total in the β1 domain of the Ovar DR β-chain (Fig. 2). The amino acid variants may reflect the ability of the β1 domain of the DR β-chain to bind to OPPV, to bind to the T-cell receptor, and/or bind and present an assortment of OPPV peptides to CD4+ T-lymphocytes. Therefore, we also evaluated each polymorphic amino acid in the β1 domain of DR β-chain for restrictiveness or permissiveness of OPPV as reflected in lower or higher provirus levels compared to other β1 domain amino acids found in this ewe flock. Grouping by amino acid residues encoded represents a form of cladistic analysis to take advantage of the evolutionary relationships among underlying haplotypes to construct statistical tests for evidence of relationship to proviral levels. Table 3 shows that the presence of Y31, T32, or N37 significantly associated with lower provirus levels (P value = 0.0003), and these amino acids were found in expressed alleles DRB1*0702 and DRB1*07012. In addition, the presence of T51 or Q60 also significantly associated with lower provirus levels (P value = 0.014), and these amino acids were found in animals with expressed alleles DRB1*0702, DRB1*07012, and DRB1*0803. However, the effect was not as great and the P values were larger for T51 or Q60 as compared to Y31, T32, or N37, and this indicates that amino acids T51 and Q60 are not as strongly associated with lower provirus levels. In addition, the presence of N74 significantly associated with lower provirus levels (P value = 0.018), and this amino acid was identified in the following DRB1 expressed alleles: DRB1*0109, DRB1*0132, DRB1*0403, DRB1*0404, and DRB1*0412.
The presence of specific amino acids in the β1 domain of MHC Class II DR β-chain also associated with higher provirus levels (see Table 3 under Higher OPP Provirus Levels). The presence of H32 significantly associated with higher provirus levels (P value = 0.043), and this amino acid was found in DRB1*0104, DRB1*0141, DRB1*0144, DRB1*0301, DRB1*0803, and DRB1*1301. In addition, the presence of A38 significantly associated with higher provirus levels (P value = 0.043), and this amino acid was found in DRB1*0114, DRB1*0116, DRB1*0141, DRB1*0144, DRB1*0324, and DRB1*0803. And finally, the presence of I67 significantly associated with higher provirus levels (P value = 0.013), and this amino acid was found in DRB1*01072, DRB1*0114, DRB1*0116, DRB1*0141, DRB1*0144, DRB1*0201, DRB1*02032, DRB1*0803, and DRB1*GS1. Interestingly, DRB1*0803 was the only allele that contained amino acids that significantly associated with both lower (T51 and Q60) and higher provirus levels (H32, H38, and I67) and was only found in the Rambouillet breed.
To determine if there were any nucleotides that encoded for synonymous amino acids and could be used as a genetic marker, the presence of specific nucleotides in exon 2 of DRB1 were also evaluated for association with lower or higher provirus levels. The presence of G32, C99, or A207 significantly associated with lower provirus levels (P value = 0.0003, 0.0006, and 0.0004, respectively), and these nucleotides were found in DRB1*07012 and DRB1*0702 or DRB1*07012 alone. The presence of a C63 or T174 also significantly associated with lower provirus levels (P values = 0.025 and 0.028, respectively). Like the amino acids T51 and Q60 in DRB1*0803, it appears that nucleotides C63 and T174 in DRB1*0803 helped to decrease the effect size and increase P values. Only the presence of one nucleotide, G207, significantly associated with higher provirus levels (P value = 0.0004), and this particular nucleotide was found in all DRB1 alleles except DRB1*07012.
Discussion
Quantitation of virus that has integrated into the host genome in the form of provirus provides insight into host control of infection that expands upon previous studies involving serological measures. Lentiviruses are persistent in nature whereby there has been no evidence of elimination of provirus once it has integrated into the host genome. One in two sheep of open-range U.S. flocks are infected with OPPV, and 92% of U.S. sheep producers do not test for OPPV (APHIS 2003). Therefore, the persistence of OPPV in U.S. flocks forces the host to regulate rather than eliminate OPPV. It is possible that lowering OPP provirus levels in the host may cause less clinical disease, may result in better production, and may reduce the total viral reservoir available to mutate into more virulent strains. Further studies need to be conducted to determine whether OPP provirus level is a good host genetic tool for controlling OPPV clinical disease and future outbreaks.
There were breed differences in detectable provirus under the extensive management conditions and with the OPPV strains present in this Idaho flock. Specifically, Rambouillet ewes were half as likely to be positive at age 5, and even less likely at age 6 compared to Columbia ewes. This is consistent with previous studies that showed Rambouillet sheep to have the lowest seroprevalence as compared to the Columbia, Polypay, Finnsheep-cross breeds (Gates et al. 1978; Snowder et al. 1990). The quantitative differences in provirus levels also showed that under these conditions, Rambouillet sheep were better able to control virus once infected than the other breeds, particularly at older ages (see Fig. 1). The relative enhanced ability to control virus of Rambouillet ewes may be related to separate underlying genetic and subsequent biological factors, for example some mechanisms may prevent infection while others aid in virus control after infection occurs. Furthermore, the biological mechanisms underlying the breed differences may be at work even at younger ages, but it is possible that they may take time to accumulate sufficient impact on measurable provirus levels to become statistically detectable at older ages. Regardless, the breed differences imply a significant genetic component in host control of OPP infection.
Since MHC class II is the putative cellular receptor for OPP virus and is highly variable, one would hypothesize that specific Ovar-DRB1 expressed alleles may result in differential host control of OPP provirus levels due to the efficiency of virus binding to different DR β-chains on monocytes/macrophages lineage cells. Alternatively, specific MHC class II DRB1 expressed alleles may contribute to subsequent beneficial or detrimental humoral and regulatory immune responses to OPPV which could differ in the degree permissiveness/restrictiveness of viral replication. In this study, none of the specific DRB1-expressed alleles or deduced amino acids comprising the β1 domain of the DR β-chain significantly associated with the presence or absence of provirus. Therefore, the breed and age specific differences observed with regard to the presence of provirus indicate that there are other yet unidentified host genes influencing the host’s capacity to prevent OPPV infection.
In terms of controlling OPPV after infection, the presence of DRB1*0403, and DRB1*07012 significantly associated with lower provirus levels; but these ewes account for only 11% of the population in this ewe flock. And, none of the named DRB1 alleles were permissive for higher provirus levels. However, there were three amino acids (H32, A38, I67) encoded in the β1 domain of the DR β chain that significantly associated with viral permissiveness (higher proviral levels), and six amino acids (Y31, T32, N37, T51, Q60, N74) that significantly associated with viral restriction (lower provirus levels). The effect sizes were generally lower and P values higher for the amino acids that associated with higher levels (H32, A38, and I67) as compared to the amino acids that associated with lower provirus levels. Several amino acid variants had higher frequencies than full-length named alleles, which may increase the strength of the statistical comparisons for association with amino acid residues. While it is possible that other genetic factors in tight linkage disequilibrium with DRB1 could contribute to the observed allelic associations, these results suggest involvement of DRB1 named alleles and specific amino acid residues in binding to and/or dictating the type and magnitude of the CD4 T-lymphocyte immune response to OPPV.
Interestingly, DRB1*0803 is the only allele that encodes amino acids that had significant associations with both lower (T51 and Q60) and higher provirus levels (H32, A38, and I67). The fact that amino acids of the same allele can associate with both higher and lower provirus may reflect the combination of amino acids along the β1 domain that bind different OPPV strains during receptor–ligand interactions or presentation to CD4 positive T-lymphocytes. Although we did not specifically evaluate viral strains in this study, they could also be playing a role in the association of DRB1 with OPPV control.
Compared to the DRB1 codons that yielded non-synonymous deduced amino acids, there were less DRB1 codons encoding synonymous amino acids that associated significantly with OPP provirus levels. This is not surprising in the context of MHC class II DRB1 since there is enormous pathogen and environmental evolutionary pressure on MHC genes to cause functional changes. The nucleotides encoding a synonymous amino acid that associated with lower OPP provirus levels were found in one of three DRB1 alleles: DRB1*07012, DRB1*0702, and DRB1*0803. The only nucleotide (G207) encoding a synonymous amino acid that associated with higher provirus levels was found in all DRB1 alleles except for DRB1*07012. All together, this indicates that nucleotide A207 in DRB1*07012 would be the only useful marker for identifying animals with the potential of having lower OPP provirus levels.
In summary, this study shows Ovar-DRB1 to be one genetic component in host control of OPP; however, there are many other unaccounted host genetic factors that could play roles in initial OPPV infection and the control OPPV. MHC class II could still play a role in control of OPPV as observed by the fact that MHC class II expression positively correlates with the severity histopathological lesions in the brains of maedi-visna infected sheep (Bergsteinsdottir et al. 1998). In addition, an increase in MHC class II positive cells from the bronchoalveolar lavage fluid of maedi-visna infected sheep as compared to specific pathogen free controls has been observed (Lujan et al. 1993). Our observations in this study indicate that specific MHC class II DRB1 genetics do not play a large role in the control of OPPV. These findings indicate the need for further experiments to address how and which MHC class II acts as a receptor for OPPV and to address whether expression levels of MHC class II positively correlate with the severity of histopathological lesions in lung, mammary gland, and carpal synovial joints.
References
APHIS-Veterinary Services-Centers for Epidemiology and Animal Health December (2003) posting date. Ovine Progressive Pneumonia: Awareness, Management, and Seroprevalence. Info Sheet. [Online.] http://www.aphis.usda.gov/vs/ceah/ncahs/nahms/sheep/sheep01/OPP.pdf
Bergsteinsdottir D, Arnadottir S, Torsteinsdottir S, Agnarsdottir G, Andresdottir V, Pettursson G et al (1998) Constitutive and visna induced expression of class I and II major histocompatibility complex antigens in the central nervous system of sheep and their role in the pathogenesis of visna lesions. Neuropathol Appl Neurobiol 24:224–232 doi:10.1046/j.1365-2990.1998.00100.x
Brodie SJ, Marcom KA, Pearson LD, Anderson BC, de la Concha-Bermejillo A, Ellis JA et al (1992) Effects of virus load in the pathogenesis of lentivirus-induced lymphoid interstitial pneumonia. J Infect Dis 166:531–541
Carrington M, O’Brien SJ (2003) The influence of HLA genotype on AIDS. Annu Rev Med 54:535–551 doi:10.1146/annurev.med.54.101601.152346
Cheevers WP, McGuire TC (1988) The Lentiviruses: Maedi/visna, Caprine Arthritis Encephalitis, and Equine Infectious Anemia. In: Maramorosch K, Murphy FA, Shatkin AJ (eds) Advances in Virus Research. vol. 34. Academic, San Diego, CA, pp 189–215
Dalziel RG, Hopkins J, Watt NJ, Dutia BM, Clarke HA, McConnell I (1991) Identification of a putative cellular receptor for the lentivirus visna virus. J Gen Virol 72:1905–1911 doi:10.1099/0022-1317-72-8-1905
De Andres D, Klein D, Watt NJ, Berriatua E, Torsteinsdottir S, Blacklaws BA et al (2005) Diagnostic tests for small ruminant lentiviruses. Vet Microbiol 107:49–62 doi:10.1016/j.vetmic.2005.01.012
De Boer GF, Terpstra C, Houwers DJ (1979) Studies in epidemiology of maedi/visna in sheep. Res Vet Sci 26:202–208
de la Concha-Bermejillo A, Brodie SJ, Magnus-Corral S, Bowen RA, DeMartini JC (1995) Pathologic and serologic responses of isogeneic twin lambs to phenotypically disinct lentiviruses. J Acquir Immune Defic Syndr Hum Retrovirol 8:116–123 doi:10.1097/00042560-199502000-00002
Dukkipati VSR, Blair HT, Garrick DJ, Murray A (2006) ‘Ovar-Mhc’-Ovine major histocompatibility complex: role in genetic resistance to diseases. N Z Vet J 54:153–160
Dutia BM, McConnell I, Ballingall KT, Keating P, Hopkins J (1994) Evidence for the expression of two distinct MHC class II DRB like molecules in the sheep. Anim Genet 25:235–241
Eriksson K, McInnes E, Ryan S, Tonks P, McConnell I, Blacklaws B (1999) CD4(+) T-cells are required for the establishment of maedi-visna virus infection in macrophages but not dendritic cells in vivo. Virology 258:355–364 doi:10.1006/viro.1999.9711
Gates NL, Winward LD, Gorham JR, Shen DT (1978) Serologic survey of prevalence of ovine progressive pneumonia in Idaho range sheep. J Am Vet Med Assoc 173:1575–1577
Gendelman HE, Narayan O, Kennedy-Stoskopf S, Kennedy PG, Ghotbi Z, Clements JE et al (1986) Tropism of sheep lentiviruses for monocytes: susceptibility to infection and virus gene expression increase during maturation of monocytes to macrophages. J Virol 58:67–74
Gorrell MD, Brandon MR, Sheffer D, Adams RJ, Narayan O (1992) Ovine lentivirus is macrophage tropic and does not replicate productively in T lymphocytes. J Virol 66:2679–2688
Herrmann LM, Brown WC, Lewis GS, Knowles DP (2005) Identification and phylogenetic analysis of 15 MHC Class II DRB1 beta1 expressed alleles in a ewe–lamb flock. Immunogenetics 57:855–863 doi:10.1007/s00251-005-0050-9
Herrmann-Hoesing LM, Palmer G, Knowles DP (2007a) Evidence of proviral clearance following postpartum transmission of a ovine lentivirus. Virology 362:226–234 doi:10.1016/j.virol.2006.12.021
Herrmann-Hoesing LM, White SN, Lewis GS, Mousel MR, Knowles DP (2007b) Development and validation of an ovine progressive pneumonia virus (OPPV) quantitative PCR. Vaccine Clin Immunol 14:1274–1278 doi:10.1128/CVI.00095-07
Herrmann-Hoesing LM, White SN, Lewis GS, Mousel MR, Knowles DP (2008) Genetic analysis of Ovis aries (Ovar) MHC Class IIa loci from a Rambouillet ram. Immunogenetics 60:167–176 doi:10.1007/s00251-008-0275-5
Hullinger G (1993) Ph.D. thesis. Washington State University, Pullman WA. A GP135 Cellular Receptor Mediates Infection of Caprine Synovial Membrane Cells by the Lentivirus Caprine Arthritis-Encephalitis Virus
Jugo BM, Vicario A (2001) Lymphocyte antigens in sheep: linkage to the MHC class II DRB1 gene. Eur J Immunogenet 28:451–458 doi:10.1046/j.1365-2370.2001.00242.x
Konnai S, Nagaoka Y, Takeshima S, Onuma M, Aida Y (2003) Sequences and diversity of 17 new Ovar-DRB1 alleles from three breeds of sheep. Eur J Immunogenet 30:275–282 doi:10.1046/j.1365-2370.2003.00399.x
Lujan L, Begara I, Collie DD, Watt NJ (1993) Phenotypic analysis of cells in bronchoalveolar fluid and peripheral blood of maedi visna-infected sheep. Clin Exp Immunol 91:272–276
Narayan O, Kennedy-Stoskof S, Zink MC (1988) Lentivirus–host interactions: lessons from visna and caprine arthritis-encephalitis viruses. Ann Neurol 23S:S95–S100 doi:10.1002/ana.410230725
Polycarpou A, Ntais C, Korber BT, Elrich HA, Winchester R, Krogstad P, Ariel Project et al (2002) Association between maternal and infant class I and II alleles and of their concordance with the risk of perinatal HIV type 1 transmission. AIDS Res Hum Retroviruses 18:741–746 doi:10.1089/08892220260139477
Schwaigger FW, Maddox J, Ballingall K, Buitkamp J, Crawford AM, Dutia BM et al (1996) The Ovine Major Histocompatibility Complex, p.121–176. In: Schook LB, Lamont SJ (eds) The major histocompatibility complex region of domestic animal species. CRC, Florida
Snowder GD, Gates NL, Glimp HA, Gorham JR (1990) Prevalence and effect of subclinical ovine progressive pneumonia virus infection on ewe wool and lamb production. J Am Vet Med Assoc 197:475–479
Stephens M, Donnelly P (2003) A comparison of Bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet 73:1162–1169 doi:10.1086/379378
Stephens M, Smith N, Donnelly P (2001) A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 68:978–989 doi:10.1086/319501
Stern LJ, Brown JH, Jardetzky TS, Gorga JC, Urban RG, Strominger JL et al (1994) Crystal structure of the human class II MHC protein HLA-DR1 complexed with an influenza virus peptide. Nature 368:215–221 doi:10.1038/368215a0
Verhofstede C, Reniers S, Van Wanzeele F, Plum J (1994) Evaluation of proviral copy number and plasma RNA level as early indicators of progression in HIV-1 infection: correlation with virological and immunological markers of disease. AIDS 8:1421–1427 doi:10.1097/00002030-199410000-00008
Vitone F, Gibellini D, Schiavone P, Re MC (2005) Quantitative DNA proviral detection in HIV-1 patients treated with antiretroviral therapy. J Clin Virol 33:194–200
Zhang J, Yu KF (1998) What’s the relative risk? A method of correcting the odds ratio in cohort studies of common outcomes. JAMA 280:1690–1691 doi:10.1001/jama.280.19.1690
Zhang Z, Watt NJ, Hopkins J, Harkiss G, Woodall CJ (2000) Quantitative analysis of maedi-visna virus DNA load in peripheral blood monocytes and alveolar macrophages. J Virol Methods 86:13–20 doi:10.1016/S0166-0934(99)00169-X
Acknowledgments
We thank Becky Tallmadge for the critical reading of this manuscript. We thank Liam Broughton, Nic Durfee, Nancy Kumpula-McWhirter, Lupita Gonzales, Tom Kellom, and the farm crew staff at USSES for technical assistance. This research is funded by USDA-ARS CWA No. 5348-32000-025-00D.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Herrmann-Hoesing, L.M., White, S.N., Mousel, M.R. et al. Ovine progressive pneumonia provirus levels associate with breed and Ovar-DRB1. Immunogenetics 60, 749–758 (2008). https://doi.org/10.1007/s00251-008-0328-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-008-0328-9