Abstract
Reduced numbers and function of invariant NKT (iNKT) cells partially contribute to type 1 diabetes (T1D) development in NOD mice. Previous linkage analysis identified a genetic locus on chromosome 2 controlling numbers of thymic iNKT cells. Interestingly, this locus resides within the Idd13 region that distinguishes NOD mice from the closely genetically related, but strongly T1D-resistant NOR strain. Thus, we tested if a genetic variant that confers T1D resistance in NOR mice may do so by enhancing iNKT cell numbers. iNKT cells were enumerated by an α-GalCer analog loaded CD1d tetramer in NOD and NOR mice as well as in NOD stocks carrying NOR-derived congenic regions on chromosome 1, 2, or 4. Significantly, more thymic and splenic iNKT cells were present in NOR than NOD mice. The NOR-derived Idd13 region on chromosome 2 contributed the most significant effect on increasing iNKT cell numbers. Subcongenic analyses indicated that at least two genes within the Idd13 region regulate iNKT cell numbers. These results further define the genetic basis for numerical iNKT cell defects contributing to T1D development in NOD mice.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Invariant NKT (iNKT) cells possessing important immune-regulatory capacity are distinct from conventional CD4 and CD8 T cells. Mouse iNKT cells utilize a Vα14Jα18 TCR chain preferentially paired with a Vβ8, Vβ7, or Vβ2 chain and recognize lipid antigens presented by MHC class I-like CD1d molecules (Godfrey et al. 2000; Kronenberg 2005). iNKT cells develop in the thymus into CD4+ and CD4−CD8− double-negative (DN) subsets (Benlagha et al. 2002; Egawa et al. 2005; Gapin et al. 2001; Pellicci et al. 2002). While both subsets are capable of producing a large quantity of immune-regulatory cytokines immediately upon TCR stimulation, CD4+ and DN iNKT cells appear to be functionally distinct (Wilson and Delovitch 2003). In several cases, CD4+ iNKT cells were shown to exert tolerogenic activity (Chen et al. 2006; Nakamura et al. 2003; Roelofs-Haarhuis et al. 2004). On the other hand, DN iNKT cells may preferentially possess the ability to promote immunological effector responses (Crowe et al. 2005). The development of autoimmune type 1 diabetes (T1D) in NOD mice appears to partly result from the numerical and/or functional defects in iNKT cells that characterize this strain (reviewed in Wilson and Delovitch 2003). This conclusion is based on the finding that NOD mice were protected from T1D by administration of the iNKT cell super-angonist α-galactosylceramide (α-GalCer; Hong et al. 2001; Naumov et al. 2001; Sharif et al. 2001; Wang et al. 2001). In addition, it has been reported that CD1d-deficient NOD mice lacking iNKT cells exhibit accelerated T1D (Shi et al. 2001; Wang et al. 2001). Moreover, adoptively transferred iNKT cells prevent T1D development in NOD mice (Hammond et al. 1998; Lehuen et al. 1998). Thus, it seems possible that some subset of the multiple susceptibility (Idd) genes contributing to T1D development in NOD mice may mechanistically do so by limiting the generation or survival of iNKT cells.
The NOR strain is closely related to NOD, but remains T1D-free due to contributions from genes of C57BL/6 (B6) or DBA/2 origin that comprise ∼12% of its genome (Prochazka et al. 1992). It has been reported that NOD and NOR mice harbor equivalent numbers of total iNKT cells in the thymus, spleen, and liver (Matsuki et al. 2003). However, a genetic linkage analysis of backcross progeny from an outcross of B6 mice with a NOD.Nkrp1 b chromosome 6 congenic stock indicated one of the two most significant genetic loci controlling numbers of thymic iNKT cells mapped to chromosome 2 between the markers D2Mit490 and D2Mit280 (Esteban et al. 2003). This genetic interval lies within the previously defined Idd13 locus from NOR mice containing at least two genes originally derived from the B6 strain that contribute to T1D resistance (Serreze et al. 1998). Allelic variants of β2-microglobulin (β2m) represent a known Idd13 region gene that contributes to T1D susceptibility or resistance by inducing conformational differences in MHC class I molecules, which, in turn, influences their ability to positively select autoreactive pathogenic CD8 T cells (Hamilton-Williams et al. 2001). Dimerizing with different β2m variants could also theoretically influence the ability of CD1d molecules to mediate the selection or activation of iNKT cells. Thus, we decided to use a congenic strain approach to test if polymorphic genes respectively contributing to T1D susceptibility and resistance in NOD and NOR mice might do so by altering the generation or maintenance of iNKT cells.
Materials and methods
Mice and reagents
NOD/LtDvs mice and the closely related NOR/Lt strain (Prochazka et al. 1992) are maintained by brother–sister mating at The Jackson Laboratory (Bar Harbor, ME). Stocks of NOD background mice carrying NOR-derived congenic intervals of chromosome 1 NOD.NOR-(D1Mit532-D1Mit8)/DvsJ (designated NOD.Chr1 NOR hereafter) or 4 NOD.NOR-(D4Mit31-D4Mit310)/DvsJ (designated NOD.Chr4 NOR hereafter) have been described previously (Reifsnyder et al. 2005). It should be noted that in the latter of these two stocks, the NOR-derived chromosome 4 congenic interval was found to extend more distally than previously reported (Reifsnyder et al. 2005) to also encompass the marker D4Mit310. NOD.NOR-(D2Mit63-D2Mit48)/LtJ (designated NOD.Chr2A NOR hereafter), NOD.NOR-(D2Mit63-D2Mit224)/LtJ (designated NOD.Chr2B NOR hereafter), and NOD.NOR-(D2Mit256-D2Mit307)/LtJ (designated NOD.Chr2C NOR hereafter) congenic mice have also been described (Serreze et al. 1998). The designation of these NOR-derived chromosome 2 congenic intervals have been modified from that originally reported (respectively termed NOD.D2Mit490-Mit144 NOR, NOD.H3a-Il1 NOR, and NOD.Il1-Pcna NOR in Serreze et al. 1998) because of updated knowledge of marker positions. Mice used for all experiments were age-matched (6–8 weeks old) females. Monoclonal antibodies used for flow cytometry were fluorochrome-conjugated anti-B220 (RA3-6B2), anti-CD4 (RM4-5), anti-TCR Vβ (H57-597), anti-TCRβ2 (B20.6), anti-TCRβ7 (TR310), anti-TCRβ8 (F23.1). These mAbs were purchased from BD Bioscience (San Diego, CA). CD1d tetramers loaded with an α-GalCer analog (PBS57) were provided by the NIH Tetramer Facility.
Flow cytometry
Single cell suspensions were prepared from the spleen and thymus. Red blood cells were removed. Cells were treated with Fc block (anti-CD16/CD32, clone 2.4G2) at room temperature for 10 min followed by staining with the indicated antibodies and CD1d tetramer at 4°C for 30 min. Stained cells were washed and analyzed on a FACSCalibur or a Cytek-upgraded 5-color FACScan flow cytometer (Becton Dickinson) using the CellQuest software. Propidium iodide was used to gate out dead cells.
Statistical analysis
All statistical comparisons between two groups were performed using the non-parametric Wilcoxon rank sum test.
Results
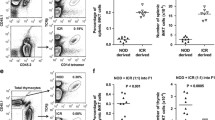
An α-GalCer analog (PBS57) loaded CD1d tetramer was used to quantify iNKT cells. Two subsets of iNKT cells (CD4+ and DN) were further distinguished based on CD4 expression. As shown in Fig. 1a and b, the frequency as well as the absolute numbers of total thymic iNKT cells were significantly greater in NOR than NOD mice. Furthermore, numbers of both CD4+ and DN thymic iNKT cells were higher in NOR than NOD mice. Similarly, compared to NOD mice, the NOR strain also exhibited an increase in the proportion and numbers of total iNKT cells in the spleen (Fig. 1c and d). In contrast to the thymus, only the CD4+ subset of iNKT cells was increased in the spleens of NOR mice. There was a previous report (Matsuki et al. 2003) that iNKT cell numbers do not differ in NOD and NOR mice. One possible explanation for the discrepancy between our results and those reported by Matsuki et al. (2003) is that they identified iNKT cells by co-staining with a CD1d tetramer and anti-TCR Vβ8.1-2. While iNKT cells are most frequently characterized by TCR Vβ8 expression, they can also utilize Vβ2 or Vβ7 elements (Godfrey et al. 2000; Kronenberg 2005). Hence, we determined if iNKT cells from NOD and NOR mice differentially utilize TCR Vβ2, Vβ7, or Vβ8 elements. To do this, we used CD1d tetramers in conjunction with antibodies specific for TCR Vβ2, Vβ7, or Vβ8 elements. As shown in Fig. 2, Vβ2+ iNKT cells were more prevalent in NOR than NOD mice. The numbers of Vβ7+ and Vβ8+ iNKT cells were equivalent in NOR and NOD mice. The Vβ2, Vβ7, and Vβ8 chain usage accounted for about 80 and 72% of total splenic iNKT cells in NOD and NOR mice, respectively (data not shown). Thus, we agree with Matsuki et. al. that the numbers of TCR Vβ8 expressing iNKT cells do not differ in NOD and NOR mice. However, our results indicate that an increase in the TCR Vβ2-positive subset and others expressing non-Vβ7 and Vβ8 elements account for the significantly higher numbers of total iNKT cells in NOR than NOD mice.
Comparison of iNKT cell numbers between NOD and NOR mice. Thymocytes (a and b) and splenocytes (c and d) were stained with a cocktail containing an α-GalCer analog loaded CD1d tetramer and TCRβ chain plus CD4 specific antibodies. iNKT cells were identified by co-staining with the tetramer and the TCRβ chain antibody. Representative plots are shown in the left panels of a and c. The numbers (mean±SEM, n = 8–11) depict the percentages of iNKT cells among the total cell population. Expression of CD4 was further analyzed on iNKT cells as shown in the right panels of a and c. The numbers (mean±SEM, n = 8–11) depict the percentages of the CD4+ subset among total iNKT cells. The absolute numbers of total iNKT cells and individual subsets in the thymus and spleen are shown in b and d, respectively (mean±SEM, n = 8–11). *p < 0.01; **p < 0.001 when compared to the same iNKT cell population in NOD mice
Analyses of TCR Vβ chain usage by iNKT cells in NOD and NOR mice. Splenocytes from NOD and NOR mice were stained with CD1d tetramers as well as antibodies that recognize the indicated TCR β chain elements or B220. B220+ cells were electronically gated out. a Representative FACS profiles of TCR Vβ2, Vβ7, or Vβ8 expressing iNKT cells. The value in each plot depicts the frequency of iNKT cells expressing the indicated Vβ chain among total splenocytes (mean±SEM, n = 9). b Absolute numbers of iNKT cells utilizing TCR Vβ2, Vβ7, or Vβ8 elements (mean±SEM, n = 9). *p < 0.05 when compared to the same iNKT cell population in NOD mice
We had previously generated NOD background stocks congenic for NOR-derived intervals of chromosomes 1, 2, or 4 (here designated NOD.Chr1 NOR, NOD.Chr2A NOR, NOD.Chr4 NOR) respectively containing Idd5, Idd13, or Idd9/11 region resistance alleles (Reifsnyder et al. 2005; Serreze et al. 1998). These congenic stocks allowed us to test the hypothesis that genetic loci contributing to T1D resistance in the NOR strain co-localize with those allowing them to generate higher numbers of iNKT cells than in NOD mice. Indeed, one quantitative trait locus (Nkt2) regulating iNKT cell numbers was previously mapped in linkage analyses to lie within the NOR-derived chromosome 2 interval transferred to the NOD strain (Esteban et al. 2003). Therefore, thymic and splenic iNKT cells were compared between NOD mice and the individual congenic stocks.
Consistent with the location of Nkt2, the numbers of thymic iNKT cells were dramatically higher in the NOD.Chr2A NOR congenic stock than in NOD mice (Fig. 3a). The significant increase of total thymic iNKT cells observed in NOD.Chr2A NOR mice was contributed by both the CD4+ and DN subsets (Fig. 3a). Compared to standard NOD mice, the NOD.Chr1 NOR congenic stock also had marginally increased numbers of thymic iNKT cells, including both CD4+ and DN subsets (Fig. 3b). However, it should be noted that the congenic region in the NOD.Chr1 NOR stock does not overlap with the previously described more distal Nkt1 locus that contains Slamf1 and Slamf6 as candidate genes (Jordan et al. 2007). In contrast, numbers of thymic iNKT cells were equivalent in NOD.Chr4 NOR and NOD mice (Fig. 3c).
Comparison of iNKT cell numbers between standard NOD mice and those carrying NOR-derived congenic intervals on chromosomes 1, 2, or 4. Total and subsets of iNKT cells in the thymus (a–c) and spleen (d–f) were identified as described in Fig. 1. In each experiment, NOD controls and an individual congenic stock was compared simultaneously. The results are presented as mean±SEM (n = 9 to 11 per group). *p < 0.01; **p < 0.001 when compared to the same iNKT cell population in NOD mice
Similar to the case in the thymus, both the CD4+ and DN subsets contributed to the significantly higher numbers of splenic iNKT cells in NOD.Chr2A NOR than NOD mice (Fig. 3d). The slight increase of thymic iNKT cells in the NOD.Chr1 NOR congenic stock was also carried over to the spleen (Fig. 3e). However, the higher numbers of splenic iNKT cells seen in the NOD.Chr1 NOR stock compared to standard NOD mice were solely due to the difference in the CD4+ subset. As observed in the thymus, NOD.Chr4 NOR and NOD mice had comparable numbers of splenic iNKT cells (Fig. 3f). Collectively, these results indicate that strongly acting genes on chromosomes 2, and possibly weaker contributors on chromosome 1, allow for the development of higher numbers of immunoregulatory thymic and splenic iNKT cells in NOR than NOD mice. In turn, these differing numbers of iNKT cells may contribute to the T1D susceptibility and resistance, respectively characterizing NOD and NOR mice.
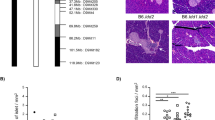
As shown in Fig. 4a, the NOR-derived chromosome 2 congenic interval that in the above studies exerted the strongest effect in elevating iNKT cell numbers beyond the level seen in standard NOD mice was relatively large (38.1 Mb; flanking markers D2Mit63 and D2Mit48). Therefore, the location of the NOR-derived chromosome 2 gene(s) controlling iNKT cell numbers was further refined by evaluating this trait in two other subcongenic stocks we had previously produced (Serreze et al. 1998). One of these strains (NOD.Chr2B NOR) contains a shorter 11.3 Mb NOR-derived chromosome 2 congenic interval delineated by the flanking markers D2Mit63 and D2Mit224 (Fig. 4a). The other (NOD.Chr2C NOR) carries a 4.9-Mb region derived from NOR chromosome 2 delineated by the flanking markers D2Mit256 and D2Mit307 (Fig. 4a). Both the NOD.Chr2B NOR and NOD.Chr2C NOR stocks exhibited significantly more total and individual subsets of iNKT cells in the thymus (Fig. 4b). This could be due to a gene common to both the NOD.Chr2B NOR and NOD.Chr2C NOR stocks, as the NOR-derived chromosome 2 congenic intervals they carry overlap slightly. It is also possible two distinct genes individually contribute to the elevated numbers of thymic iNKT cells in the NOD.Chr2B NOR and NOD.Chr2C NOR stocks, as the levels in both remain lower than that induced by the longer 2A congenic interval (compare Figs. 3 and 4). In the spleen, numbers of total as well as CD4+ and DN iNKT cells in the NOD.Chr2A NOR and NOD.Chr2B NOR stocks were characterized by similar elevations compared to standard NOD mice (compare Figs. 3d and 4c). While achieving statistical significance, total numbers of splenic iNKT cells, resulting solely from a change in the CD4+ compartment, were only slightly elevated in the NOD.Chr2C NOR stock compared to standard NOD mice (Fig. 4c). Thus, a gene(s) in the region of chromosome 2 defined by the 2A and 2B, but distinct from the 2C congenic region, most strongly regulates the differential peripheral levels of iNKT cells in NOD and NOR mice.
Comparison of iNKT cell numbers between NOD- and NOR-derived chromosome 2 subcongenic mice. a Genetic maps of NOD.Chr2A NOR, NOD.Chr2B NOR and NOD.Chr2C NOR congenics. The positions (Mb) of the markers are based on NCBI Build 36 and are obtained from Mouse Genome Informatics (http://www.informatics.jax.org/). The distance between markers is not drawn to scale. Total and subsets of iNKT cells in the thymus (b) and spleen (c) were identified as described in Fig. 1. The results are presented as mean±SEM (n = 9–13 per group). *p < 0.05; **p < 0.01; ***p < 0.001 when compared to the same iNKT cell population in NOD mice
Discussion
In the current study, we analyzed iNKT cell numbers in NOD and NOR mice as well as NOD background stocks carrying NOR-derived congenic intervals on chromosomes 1, 2, or 4. The collective results shown here indicate that the NOD.Chr2A NOR stock is characterized by more than one gene on chromosome 2 that polymorphically differ from the variants found in standard NOD mice and allow for enhanced development and/or homeostasis of iNKT cells in the former strain. However, at least in the periphery, a NOR-derived gene(s) also found in the NOD.Chr2B NOR stock most strongly enhances iNKT cell levels. One candidate gene within the shorter congenic region in the NOD.Chr2B NOR stock is β2m. The β2m a versus β2m b alleles, respectively characterizing NOD and NOR mice, have been shown to regulate T1D susceptibility by differentially enabling the MHC class I molecules shared by these two strains to support the positive selection of pathogenic CD8 T cells (Hamilton-Williams et al. 2001). Hence, iNKT cell numbers were also assessed in previously described stocks of normally β2m-deficient NOD mice in which a transgenic rescue approach was used to restore either β2m a or β2m b expression (Hamilton-Williams et al. 2001). These analyses revealed that the T1D-protective β2m b allele did not increase iNKT cell numbers to a greater extent than the disease permissive β2m a variant (Dr. Alan Baxter, Dr. Robyn Slattery, Townsville and Melbourne Australia, personal communication). However, previous studies could not rule out the possibility that in addition to β2m b, other genetic components within the NOR-derived interval in the NOD.Chr2B NOR stock also contribute to T1D resistance. The current results suggest that if in addition to β2m b, the NOR-derived congenic interval in the NOD.Chr2B NOR stock does contain another T1D resistance gene(s), it may mediate disease protection by elevating iNKT cell numbers.
In summary, we showed that fewer immunoregulatory iNKT cells reside in T1D-susceptible NOD mice than in the closely genetically related, but disease-resistant NOR strain. At least two genes within a 38.1-Mb interval on chromosome 2 primarily contribute to the increased numbers of iNKT cells in NOR mice. Significantly, this region of chromosome 2 in the NOR strain has also been previously shown to contain at least two genes contributing to T1D resistance (Serreze et al. 1998). The current results support the possibility that one mechanism of T1D resistance mediated by genes on NOR chromosome 2 is to allow for the generation of greater numbers of immunoregulatory iNKT cells than in disease susceptible NOD mice.
References
Benlagha K, Kyin T, Beavis A, Teyton L, Bendelac A (2002) A thymic precursor to the NK T cell lineage. Science 296:553–555
Chen YG, Chen J, Osborne MA, Chapman HD, Besra GS, Porcelli SA, Leiter EH, Wilson SB, Serreze DV (2006) CD38 is required for the peripheral survival of immunotolerogenic CD4+ invariant NK T cells in nonobese diabetic mice. J Immunol 177:2939–2947
Crowe NY, Coquet JM, Berzins SP, Kyparissoudis K, Keating R, Pellicci DG, Hayakawa Y, Godfrey DI, Smyth MJ (2005) Differential antitumor immunity mediated by NKT cell subsets in vivo. J Exp Med 202:1279–1288
Egawa T, Eberl G, Taniuchi I, Benlagha K, Geissmann F, Hennighausen L, Bendelac A, Littman DR (2005) Genetic evidence supporting selection of the Valpha14i NKT cell lineage from double-positive thymocyte precursors. Immunity 22:705–716
Esteban LM, Tsoutsman T, Jordan MA, Roach D, Poulton LD, Brooks A, Naidenko OV, Sidobre S, Godfrey DI, Baxter AG (2003) Genetic control of NKT cell numbers maps to major diabetes and lupus loci. J Immunol 171:2873–2878
Gapin L, Matsuda JL, Surh CD, Kronenberg M (2001) NKT cells derive from double-positive thymocytes that are positively selected by CD1d. Nat Immunol 2:971–978
Godfrey DI, Hammond KJ, Poulton LD, Smyth MJ, Baxter AG (2000) NKT cells: facts, functions and fallacies. Immunol Today 21:573–583
Hamilton-Williams EE, Serreze DV, Charlton B, Johnson EA, Marron MP, Mullbacher A, Slattery RM (2001) Transgenic rescue implicates beta2-microglobulin as a diabetes susceptibility gene in nonobese diabetic (NOD) mice. Proc Natl Acad Sci USA 98:11533–11538
Hammond KJ, Poulton LD, Palmisano LJ, Silveira PA, Godfrey DI, Baxter AG (1998) alpha/beta-T cell receptor (TCR)+CD4-CD8- (NKT) thymocytes prevent insulin-dependent diabetes mellitus in nonobese diabetic (NOD)/Lt mice by the influence of interleukin (IL)-4 and/or IL-10. J Exp Med 187:1047–1056
Hong S, Wilson MT, Serizawa I, Wu L, Singh N, Naidenko OV, Miura T, Haba T, Scherer DC, Wei J et al (2001) The natural killer T-cell ligand alpha-galactosylceramide prevents autoimmune diabetes in non-obese diabetic mice. Nat Med 7:1052–1056
Jordan MA, Fletcher JM, Pellicci D, Baxter AG (2007) Slamf1, the NKT cell control gene Nkt1. J Immunol 178:1618–1627
Kronenberg M (2005) Toward an understanding of NKT cell biology: progress and paradoxes. Annu Rev Immunol 23:877–900
Lehuen A, Lantz O, Beaudoin L, Laloux V, Carnaud C, Bendelac A, Bach JF, Monteiro RC (1998) Overexpression of natural killer T cells protects Valpha14–Jalpha281 transgenic nonobese diabetic mice against diabetes. J Exp Med 188:1831–1839
Matsuki N, Stanic AK, Embers ME, Van Kaer L, Morel L, Joyce S (2003) Genetic dissection of V alpha 14J alpha 18 natural T cell number and function in autoimmune-prone mice. J Immunol 170:5429–5437
Nakamura T, Sonoda KH, Faunce DE, Gumperz J, Yamamura T, Miyake S, Stein-Streilein J (2003) CD4+ NKT cells, but not conventional CD4+ T cells, are required to generate efferent CD8+ T regulatory cells following antigen inoculation in an immune-privileged site. J Immunol 171:1266–1271
Naumov YN, Bahjat KS, Gausling R, Abraham R, Exley MA, Koezuka Y, Balk SB, Strominger JL, Clare-Salzer M, Wilson SB (2001) Activation of CD1d-restricted T cells protects NOD mice from developing diabetes by regulating dendritic cell subsets. Proc Natl Acad Sci USA 98:13838–13843
Pellicci DG, Hammond KJ, Uldrich AP, Baxter AG, Smyth MJ, Godfrey DI (2002) A natural killer T (NKT) cell developmental pathway involving a thymus-dependent NK1.1(−)CD4(+) CD1d-dependent precursor stage. J Exp Med 195:835–844
Prochazka M, Serreze DV, Frankel WN, Leiter EH (1992) NOR/Lt mice: MHC-matched diabetes-resistant control strain for NOD mice. Diabetes 41:98–106
Reifsnyder PC, Li R, Silveira PA, Churchill G, Serreze DV, Leiter EH (2005) Conditioning the genome identifies additional diabetes resistance loci in type I diabetes resistant NOR/Lt mice. Genes Immun 6:528–538
Roelofs-Haarhuis K, Wu X, Gleichmann E (2004) Oral tolerance to nickel requires CD4+ invariant NKT cells for the infectious spread of tolerance and the induction of specific regulatory T cells. J Immunol 173:1043–1050
Serreze DV, Bridgett M, Chapman HD, Chen E, Richard SD, Leiter EH (1998) Subcongenic analysis of the Idd13 locus in NOD/Lt mice: evidence for several susceptibility genes including a possible diabetogenic role for beta 2-microglobulin. J Immunol 160:1472–1478
Sharif S, Arreaza GA, Zucker P, Mi QS, Sondhi J, Naidenko OV, Kronenberg M, Koezuka Y, Delovitch TL, Gombert JM et al (2001) Activation of natural killer T cells by alpha-galactosylceramide treatment prevents the onset and recurrence of autoimmune type 1 diabetes. Nat Med 7:1057–1062
Shi FD, Flodstrom M, Balasa B, Kim SH, Van Gunst K, Strominger JL, Wilson SB, Sarvetnick N (2001) Germ line deletion of the CD1 locus exacerbates diabetes in the NOD mouse. Proc Natl Acad Sci USA 98:6777–6782
Wang B, Geng YB, Wang CR (2001) CD1-restricted NK T cells protect nonobese diabetic mice from developing diabetes. J Exp Med 194:313–320
Wilson SB, Delovitch TL (2003) Janus-like role of regulatory iNKT cells in autoimmune disease and tumour immunity. Nat Rev Immunol 3:211–222
Acknowledgements
This work was supported by National Institutes of Health grants DK46266 and DK51090; Cancer Center Support Grant CA34196; as well as by grants from the Juvenile Diabetes Research Foundation International.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chen, YG., Driver, J.P., Silveira, P.A. et al. Subcongenic analysis of genetic basis for impaired development of invariant NKT cells in NOD mice. Immunogenetics 59, 705–712 (2007). https://doi.org/10.1007/s00251-007-0236-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-007-0236-4