Abstract
In primates and rodents, the extended FcR family is comprised of three subsets: classical FcRs, structurally diverse cell surface receptors currently designated FCRL1–FCRL6, and intracellular proteins FCRLA and FCRLB. Using bioinformatic analysis, we revealed the FcR-like genes of the same three subsets in the genome of dog, another representative of placental mammals, and in the genome of short-tailed opossum, a representative of marsupials. In contrast, a single FcR-like gene was found in the current version of the chicken genome. This in silico finding was confirmed by the gene cloning and subsequent Southern blot hybridization. The chicken FCRL gene encodes a cell surface receptor with the extracellular region composed of four Ig-like domains of the D1-, D2-, D3-, and D4-subtypes. The gene is expressed in lymphoid and non-lymphoid tissues. Phylogenetic analysis of the mammalian and chicken genes suggested that classical FcRs, FCRLA, and FCRLB emerged after the mammalian–avian split but before the eutherian–marsupial radiation. The data obtained show that the repertoire of the classical FcRs and surface FcR-like proteins in mammalian species was shaped by an extensive recombination process, which resulted in domain shuffling and species-specific gain and loss of distinct exons or entire genes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The ability to bind the constant regions of immunoglobulins is a feature of numerous and structurally diverse cell surface receptors. Among these, the family of classical Fc-receptors (FcR) that belong to the immunoglobulin superfamily (IgSF) occupies an important place (Ravetch and Kinet 1991; Daeron 1997). FcRs are involved in various immune reactions, such as phagocytosis, antibody-dependent cellular cytotoxicity, and immediate hypersensitivity. They participate in the regulation of the Ig synthesis by B lymphocytes and play an important role in antigen capture by professional antigen presenting cells.
FcRs are separated into four main classes differing by their ligand preferences, binding affinity, and signal properties: FcγRI (the high affinity receptor for IgG), FcɛRI (high affinity receptor for IgE), and low affinity receptors for IgG FcγRII and FcγRIII. The Ig-like domains of the FcR extracellular (EC) regions may be separated into three structural subtypes: D1, D2, and D3. FcγRI has all the three subtypes, whereas EC of the other FcRs are composed of D1 and D2. Studies of the human and mouse FcRs demonstrated that the family evolved in a species-specific manner. In the human genome, there is a single functional gene for FcγRI and FcɛRI, three for FcγRII (FcγRIIa, FcγRIIb, and FcγRIIc), and two for FcγRIII (FcγRIIIa and FcγRIIIb). In addition, human has two putative FcγRI pseudogenes. FcγRIIb contains immunoreceptor tyrosine-based inhibitory motif (ITIM) in its cytoplasmic (IC) region and functions as an inhibitory receptor; the other FcRs promote leukocyte activation programs. FcγRI, FcɛRI, and FcγRIIIa are expressed on the cell surface in complex with immunoreceptor tyrosine-based activating motif (ITAM)-bearing FcRγ or TcRζ subunits. FcγRIIa and FcγRIIc have ITAMs in their IC regions. FcγRIIIb lacks the typical transmembrane (TM) region, and it is anchored to the cell membrane through glycosyl-phosphatidylinositol (GPI). Until recently, four genes encoding Fc receptors were identified in mouse: one for each FcγRI, FcγRII, FcγRIII, and FcɛRI class. Based on the sequence comparisons, it has been suggested that mouse FcγRIII gene resulted from a rodent-specific recombination event that brought the EC domains of FcγRII together with the TM/IC of ancient FcγRIII (Hughes 1996). This suggestion has been supported by our identification of an additional mouse gene (CD16-2) that encodes a receptor highly similar to the human FcγRIII in both the EC and TM/IC parts (Mechetina et al. 2002a). The receptor has been further shown to bind the IgG2a and IgG2b subclasses and designated as FcγRIV (Nimmerjahn et al. 2005)
The FcR family has been recently extended by identification of two groups of the FcR-like (FCRL) genes in human and mouse (Table 1 and Fig. 1). Different names have been given to these genes by different researchers. Here, we will use the recently accepted new nomenclature for their description (Maltais et al. 2006). One group contains two genes, FCRLA and FCRLB, which have been originally described as FCRL/FREB/FcRX (Davis et al. 2002b; Facchetti et al. 2002; Mechetina et al. 2002b) and FCRL2/FREB2/FcRY (Chikaev et al. 2005; Masuda et al. 2005; Wilson and Colonna 2005). Both genes encode intracellular proteins composed of three Ig-like domains (D1D2D3), and a mucin-like domain at the COOH end. The N-terminal domain of FCRLA is truncated and shows only partial similarity to the D1-type domains. The second group of FcR-like genes mainly codes for cell surface receptors that have been described as IRTA/FcRH/IFGP/BXMAS/SPAP (Davis et al. 2001, 2004, 2005; Guselnikov et al. 2002; Hatzivassiliou et al. 2001; Miller et al. 2002; Nakayama et al. 2001; Xu et al. 2001, 2002). According to the new nomenclature (Table 1 and Fig. 1), the human and mouse genes of this group are designated FCRL1-6 and FCRLS, respectively. The proteins encoded by these genes exhibit striking diversity of domain architecture (Fig. 1). Besides three domain subtypes characteristic of classical FcRs, two additional subtypes, D4 and D5, have been identified in their EC regions. One of the mouse proteins is mosaic. It has a domain belonging to the scavenger receptor superfamily (SCRS) at its COOH-terminus and has been designated FCRLS for this reason. To distinguish the two FCRL subsets, we will refer to them as to iFCRL (intracellular) and sFCRL (surface) elsewhere.
a Schematic representation of the domain composition of the FcR, FCRL, and CD5L molecules of human, mouse, dog, opossum, and chicken. The Ig-like, SRCR-like and the mucin-like domains are depicted by circles, squares, and triangles, respectively. The domain subtypes are multicolored. Domains coded by pseudoexons are shown by semifilled circles or squares. b Schematic representation of the organization of the human, mouse, dog, opossum, and chicken FcR and FcR-like genes. The members of the family are shown by filled rectangles, the other genes by open rectangles. Orthologous genes are connected by vertical dashes. Grey horizontal lines designate parts of chromosomes. Gaps in the chromosomes are shown by double slash
The functional importance of the FcR-like proteins is still an unresolved issue. They lack Fc-binding activity (Leu et al. 2005; Polson et al. 2006). However, analysis of their expression patterns and signal potential suggests that most of the proteins are primarily implicated in regulation of immunity (Chikaev et al. 2005; Davis et al. 2001, 2002b, 2004, 2005; Ehrhardt et al. 2003, 2005; Facchetti et al. 2002; Falini et al. 2003; Guselnikov et al. 2002; Hatzivassiliou et al. 2001; Leu et al. 2005; Mechetina et al. 2002b; Miller et al. 2002; Nakayama et al. 2001; Polson et al. 2006; Xu et al. 2001).
The remarkable diversity of the FcR-likes proteins makes the family an interesting model to study how new functions might have emerged in the evolving immune system and how species-specific generation of leukocyte receptor repertoires contributed to the species-specific immunoregulation. Here, we report the results of an in silico analysis of the FcR-family genes in the recently sequenced dog, opossum, and chicken genomes. It is demonstrated that all the three subsets of the FcR-family were maintained throughout the mammalian evolution, on the one hand, and were subject to continuous reorganization, on the other. The phylogenetic relationships of the mammalian and chicken proteins suggest also that FCRLA, FCRLB, FCRLS, and classical FcRs are mammalian-specific acquisitions.
Materials and methods
Bioinformatics tools
Nucleotide and amino acid sequences were analyzed using utilities at the NCBI (http://www3.ncbi.nlm.nih.gov), EMBL (http://www2.ebi.ac.uk), and BCM Search Launcher (http://www.hgsc.bcm.tmc.edu) web sites. Amino acid sequences were aligned with the CLUSTALW 1.7 program (Thompson et al. 1994) and shaded with the BoxShade program (http://www.ch.embnet.org). The nucleotide and amino acid sequences of known genes were retrieved from the GenBank using ENTREZ at http://www3.ncbi.nlm.nih.gov. The genomic sequences were retrieved from and analyzed at the Ensembl web site (http://www.ensembl.org; Hubbard et al. 2002) or the NCBI (http://www3.ncbi.nlm.nih.gov). Homology searches were performed using TBLASTN (Altschul et al. 1997) and TFASTA (Pearson 1990) programs. The GeneScan program (http://genes.mit.edu/GENSCAN.html; Burge and Karlin 1997) and the Webgene program package (http://l25.itba.mi.cnr.it/~webgene; Milanesi et al. 1999) were used for the automated gene structure prediction. For assessment of the manually predicted splice sites, the Splice Site Prediction by Neural Network (http://www.fruitfly.org/seq_tools/splice.html), SpliceView (http://l25.itba.mi.cnr.it/~webgene/wwwspliceview.html) and SpliceProximalCheck (http://industry.ebi.ac.uk/~thanaraj/SpliceProximalCheck.html; Thanaraj and Robinson 2000) were used. The FcR- and FCRL-surrounding genes were identified using the Ensembl utilities at http://www.ensembl.org, MapViewer at http://www3.ncbi.nlm.nih.gov/mapview and were verified by reciprocal sequence comparisons at the NCBI website using the BLASTP program. The dog and opossum genomes were sequenced and assembled at the Broad Institute (http://www.broad.mit.edu); the chicken genome was sequenced and assembled by the Genome Sequencing Center at Washington University, St Louis (http://genome.wustl.edu).
Phylogenetic Analysis
Phylogenetic analysis was performed with MEGA3 (Kumar et al. 2004) for nucleotide sequences of exons and amino acid sequences of domains after alignment with the CLUSTAL option (14). In certain cases, the CLUSTAL-generated alignments were manually corrected. Phylogenetic trees were constructed using the bootstrap and interior branch tests of the neighbor-joining (NJ) method with proportion of differences (p-distances). We also constructed unweighted pair group method with arithmetic mean (UPGMA) and minimum evolution (ME) trees, they were essentially the same as the NJ trees in the major branching patterns, and they will not be presented here. The phylogenetic trees based on amino acid and nucleotide sequences generated topologies with branching patterns similar in the major clades. However, the bootstrap values for the trees built on the basis of nucleotide sequences were higher. For this reason, only these trees are shown here.
Sequences used in analyses
GenBank accession numbers or genomic scaffold (Ensembl) numbers for sequences are as follows: human (Homo sapiens): hFcγRI (X14356), hFcγRIIA (M31932), hFcγRIIB (X52473), hFcγRIIC (X17652), hFcγRIIIA (X52645), hFcγRIIIB (X16863), hFcɛRI (X06948), hFCRLA (AF329489), hFCRLB (AY670683), hFCRL1 (AF329488), hFCRL2 (AY043465), hFCRL3 (AY043466), hFCRL4 (AF329490), hFCRL5 (AF397453), hFCRL6(AY654628), hCD5L (AF011429).
Mouse (Mus musculus): mFcγRI (M31314), mFcγRII (M16367), mCD16-2 (AF499613), mFcγRIII (M14215), mFcɛRI (J05018), mFcrla (AF329487), mFcrlb (AY513660), mFcrl1 (AF329485), mFcrl5 (AY506558), mFcrl6 (NT_039185), mFcrls (AF329486), mCD5L (AF011428).
Dog (Canis familiaris): dFcγRI (NW_139881), dFcγRII (NW_139918), dFcγRIII (NW_139918), dFcɛRI (NW_139918), dFcrla (NW_139918), dFcrlb (NW_139918), dFcrl1 (NW_139860), dFcrl2 (NW_139860), dFcrl3 (NW_139860), dFcrl4 (NW_139860), dFcrl5 (NW_139860), dFcrl6 (NW_139918), dFcrls (NW_139860), dCD5L (NW_139860). The DNA sequences from genomic database were released on January 4, 2005.
Opossum (Monodelphis domestica): oFcgRIIA (scaffold_ 19223), oFcγRIIB (scaffold_19223), oFcγRIII (scaffold_19223), oFcɛRI (scaffold_15142), oFcrla (scaffold_19223), oFcrlb (scaffold_19223), oFcrl1 (scaffold_19046), oFcrl2 (scaffold_19046), oFcrl3 (scaffold_19046), oFcrl4 (scaffold_19046), oFcrl5 (scaffold_19046), oFcrl6 (scaffold_15142), oCD5L (scaffold_19046). The scaffold designation from the Ensembl genomic database is version 0.5.
Chicken (Gallus gallus): chiFcrl_D1 (NW_084033), chiFcrl_D2 (NW_084033), chiFcrl_D3.1 (NW_084033), chiFcrl_D3.1 (NW_098409), chiFcrl_D4 (NW_098409). The DNA sequences from genomic database were released on July 29, 2004.
Gene prediction criteria
The search for the FcR-related genes in the dog, opossum, and chicken genomes consisted of two steps. First, the sequences were analyzed using the TBASTN program (Altschul et al. 1997) for exons encoding domains similar to those found in human and mouse members of the family. The search was performed in both genomic and expression sequence tag (EST) databases. The scaffolds identified were further analyzed, using the GenScan (Burge and Karlin 1997) and/or Webgene (Milanesi et al. 1999) gene-prediction programs. Independent of the results of the automated prediction, the putative exon sequences were visually inspected for possible splice signals, frame-shift mutations, and stop codons. The constraint imposed was that the boundaries between the exons coding for the EC domains and TM region must be of phase 1. Only GT and AG dinucleotides were regarded as donor and acceptor splice signals, respectively. The predicted exons were accepted as “functional”, if they lacked frame-shift mutations or stop codons, or disrupted splice sites. The automated prediction was frequently efficient in joining exons for the EC regions, but, as a rule, failed to recognize short exons coding for the leader peptides LP, TM, and IC. For this reason, the scaffold sequences were additionally analyzed for regions homologous to the LP and TM of the human and mouse proteins, using the TFASTA algorithm (Lipman and Pearson 1985). A feature shared by the FcR-related proteins is that their LP are encoded by two exons. With the exception of FCRLA, the second LP exon is of 21 bp in all the known human and mouse FcR and FCRL genes. In the case of the FCRL1-6 proteins, FCRLB, and FcγRI, the first LP exon is invariably 31 bp long. The TM regions of activating receptors are relatively conserved and contain characteristic sequence signatures, such as, for instance, the LFAVDTGL motif in FcγRIII and FcɛRI. Furthermore, the TM and IC regions of the activating receptors are, as a rule, encoded by a single exon. The reasonable degree of homology, hydrophobicity of the encoded peptide, characteristic length and flanking splice signals of phase 1 were regarded as evidence for a particular sequence identified by TFASTA being either LP or TM exon. If the automated and visual prediction of the LP and TM exons disagreed, preference was given to visual prediction. The same approach was applied to the prediction of the IC exons. However, because of much weaker conservation of this portion of the FcR-related molecules, analysis was limited to the assessment of whether the predicted protein may possess the IC region or the tyrosine-based motifs of the YxxL/I/V type. The efficiency of this approach was confirmed by the comparison of the predicted sequences with the available EST sequences. An important guideline for tracing gene relationships among various species is conservation of the gene synteny throughout phylogeny. For this reason, genes closely linked to the FcR-related genes in the examined genomes were identified using the gene prediction programs followed by reciprocal sequence comparisons. Alignment of amino acid sequences of the predicted Ig-like domains is shown in supplementary Figs. 12 and 13 in Appendix 1.
Southern blot analysis
Genomic DNA from chicken blood cells was isolated as described by Sambrook et al. (1989) and digested to completion with the restriction endonucleases EcoRI and HindIII. The digested DNA (10 μg/lane) was separated on 0.5% agarose gel and transferred onto Zeta-probe nylon membranes (BioRad Laboratories, USA) by the vacuum blotting technique in 0.4 M NaOH. The blotted membranes were fixed by baking in a vacuum oven at 80°C for 30 min. The blotted membranes were prehybridized in a solution consisting of 6× SSC, 5× Denhardt’s, 0.5% SDS, and 20 μg/ml salmon sperm DNA at 60°C for 1 h, and hybridized with the 32P-labeled probes in a solution consisting of 6× SSC, 5× Denhardt’s, 0.5% SDS, 10% dextran sulfate, and 20 μg/ml salmon sperm DNA at 60°C for 12 h. The probes were two purified PCR fragments coding for the D3- type domain (255 bp) and D4- type domain (250 bp) of chiFCRL. Primer sequences for PCR amplification of the first probes were 5′-CAGTGCTGAACGTAGCAAG-3′ and 5′-CACCCGAGGACTCCATTTCT-3′, for PCR amplification of the second probes were 5′-GTGTCCCTGGAGGTTTGGC-3′ and 5′-CTACAGTGACCTGCACCTTC-3′. The filters were washed two times with 2× SSC/1% SDS at 55°C for 30 min and autoradiographed at −70°C.
Reverse transcriptase-polymerase chain reaction (RT-PCR) assay
Total RNA was extracted from chicken tissues as described by Chomczynski and Sacchi (1987). The first-strand cDNA was synthesized from 2 μg of total RNA using SuperScript II RNase H- Reverse Transcriptase (Gibco-BRL) according to the manufacturer’s instructions. The following primers were used to amplify the chicken Fcrl cDNAs: forward D1 primer corresponding to the chicken D1-type domain (5′-CGTCCAGCAACTCCGTCAC-3′), forward D3 primer corresponding to the chicken D3-type domain (5′-CCAGTGCTGAACGTAGCAAG-3′), and reverse D4 primer corresponding to the chicken D4-type domain (5′-TACTGCCCGTTGTCACTCTG-3′). The samples were denatured at 94°C for 5 min followed by amplifications for 40 cycles (94°C for 30 s, 62°C for 20 s, 72°C for 45 s). The negative controls contained all the reagents except cDNA or all the reagents except one of the two primers. The cDNA samples were additionally checked using PCR for β-actin. Primer sequences for PCR amplification of the β-actin fragment were 5′-CGCGAGAAGATGACCCAGATC-3′ and 5′-TTGCTGATCCACATCTGCTGG-3′. The PCR products were cloned into the pBluescript KS vector (Stratagene) and sequenced to verify PCR specificity.
Results and discussion
The dog FcR-related genes
The search for the FcR-related genes revealed 13 putative members of the FcR family in the dog genome (Fig. 1). Three genes mapped by the sequencing consortium to chromosome 38 code for classical FcRs, such as dFcγRII, dFcγRIII, and dFcɛRI. The dFcγRI gene was found on the genomic sequence assigned to chromosome 17 by the sequencing consortium. The gene prediction analysis suggests that dFcγRI, dFcγRIII, and dFcɛRI encode activating receptors. In each gene, exons for the EC domains are followed by a single exon coding for the joined TM and IC region. Furthermore, the TM regions of these receptors contain characteristic charged residues. In contrast, the TM and IC portions of dFcγRII are encoded by separate exons. One of the two predicted IC exons encodes an amino acid sequence with typical ITIM, suggesting that dFcγRII, like human FcγRIIb and mouse FcγRII, is an inhibitory receptor. The current analysis of the dog genome provided no evidence for lineage-specific duplication of any of the four genes for classical FcRs. Chromosome 23 contains an orphan exon structurally similar to the one for the dFcγRI TM/IC region (57% identity at the amino acid level). However, we did not find any other ORFs in the close proximity to this exon. The EST database search revealed partial cDNA sequences for the dFcγRIII gene. Dog cDNA for FcγRI has been recently cloned (Nakamura et al. 2003). The sequence data for dFcγRI and dFcγRIII are in good agreement with those predicted from genomic analysis.
Like the human and mouse FCRLA and FCRLB genes, their dog counterparts are closely linked to the dFcγRII gene on chromosome 38 (Fig. 1). The deduced amino acid sequences of these genes show a high degree of homology to the human and mouse proteins. The structural similarity is particularly high in the case of dFCRLB whose D1- and D2-type domains have 97 and 95% residue identities, respectively, with the counterpart human FCRLB domains. In the C-terminal mucin-like domains of dFCRLA and dFCRLB, the predicted a-helical motif, comprised of two–three di-leucine repeats that are interspersed with charged residues, is strongly conserved (see the supplementary Fig. 7 in Appendix). The predicted structure of the dFCRLA gene was supported by a partial cDNA from the EST databases. Like in the human genome, the cluster that contains genes for low affinity dFcRs, dFCRLA, and dFCRLB is flanked by the HSPA6, DUSP12, and ATF6 genes (Fig. 1).
Six of the seven dog genes belonging to the sFCRL subset were mapped to chromosome 7 (Fig. 1). Five genes, structurally similar to the human FCRL1, FCRL2, FCRL3, FCRL4, and FCRL5 genes, are ordered as in the human genome, and form a tight cluster. Compared to man, dog possesses an additional dFCRLS gene, coding for a mosaic protein similar to the mouse FCRLS. This protein is presumably secreted. Its EC region is composed of the D2, D3, D4, D5 Ig-like domains and of the C-terminal SRCR-domain. A gene for CD5L/SPα/AIM, a secreted member of the SRCR superfamily, comprises three exons for the SRCR-like domains and lies between the dFCRL1 and dFCRLS genes. The SRCR domain of dog FCRLS is more closely related to its counterpart in mFCRLS (74% residue identity) than to the N-terminal domain of human, mouse, and dog CD5L (61–64% identity). We have previously suggested that the mFCRLS gene might have arisen by an intergenic recombination between CD5L and one of the sFCRL genes (Guselnikov et al. 2002). Furthermore, the sequence comparisons suggested that this recombination might have predated radiation of primates and rodents. The presence of the FCRLS gene in the dog genome strongly supports this suggestion.
The exon organization of the dFCRL1, dFCRL2, and dFCRL4 genes is similar to that of the human FCRL1, FCRL2, and FCRL4 genes, and accordingly, the predicted dog proteins do not differ in domain architecture from their human counterparts. The predicted dFCRL3 and dFCRL5 genes encode proteins with unique domain architecture. Like the hFCRL3 gene, dFCRL3 has exons for the D1, D2, D3, D4 and two D5-type domains. However, the D1-, D3-, and D4-exons have in-frame stop codons, while the 5′splice site of the first D5 exon appears to be aberrant. This gene contains well-conserved LP exons with the characteristic length of 31 and 21 bp, an exon for typical TM, and at least two exons encoding the tyrosine-based motifs highly similar to those in the human FCRL3 IC region (not shown). It remains unclear whether this gene is functional. If so, the encoded protein may have a D2D5 architecture. The dFCRL5 gene encodes a protein consisting of eight domains, of which five belong to the D5 type. In human FCRL5, six D5-type domains may be further divided into two subtypes, D5a and D5b, with three D5b followed by three D5a domains (Miller et al. 2002; Davis et al. 2002a). In the predicted dog FCRL5, a single D5b-type domain is followed by four D5a domains. In the EST collections, partial cDNAs for dFCRL1, dFCRL2, and dFCRLS were found. The human and mouse FCRL1–FCRL5 clusters are flanked by the KIRREL and ETV3 genes. The ETV gene was found at a distance of 230 kb from dFCRL5. However, the KIRREL gene is on chromosome 38 not far away from the FcɛRI gene.
FCRL6 is the most distant member of the sFCRL subset. In the human genome, the FCRL6 gene is located on chromosome 1, between FceRI and a cluster of genes for FCRLA, FCRLB, and low affinity FcRs (Fig. 1). Judging from the sequence comparisons, FCRL6 is most similar to rat gp42, an NK cell-specific protein described more than two decades ago (Imboden et al. 1989). However, in contrast to hFCRL6, the EC region in gp42 consists of only two domains (D3D5), lacks IC, and attaches to the cell membrane via GPI anchor. Davis et al. (2002a) have previously reported the presence of the gp42-like gene in the mouse genome. However, the gene has not been described in detail. It remained unclear whether hFCRL6, gp42, and mouse FCRL6 are true orthologs. To clarify the relationship of these genes, we searched the mouse genomic sequences for the gp42-like gene. It was found on mouse chromosome 1 as a part of the linkage group flanked by the DUSP23 and SLAMf8 genes (Fig. 1). The gene order is the same as in the human genome. Analysis of the available rat genomic sequences showed that the rat gp42 gene is also linked to the DUSP23 and CRP gene (not shown). It may be inferred that the human FCRL6 and rodent FCRL6/gp42 are diverged orthologs. Mouse FCRL6 and rat gp42 share about 80% identical residues in the EC and TM regions. In contrast to the rat protein, mouse FCRL6/gp42 appears to have the IC region (R. Davis, personal communication). Analysis of the dog genomic sequences demonstrated that the dFCRL6 gene is located between the DUSP23 and SLAMf8 genes, like in the human and mouse genomes. Again, like the mouse FCRL6 and rat gp42, the encoded dFCRL6 appears to be a two-domain receptor (D3D5). However, upstream of the exon for the D3-type domain, the dog gene has an aberrant exon for the D2-type domain, suggesting thereby that the common ancestor of primates, rodents, and carnivores perhaps had a gene for a three-domain protein with EC made up of the D2-, D3-, and D5-type domains.
The opossum FcR-related genes
Primates, rodents, and carnivores belong to placental mammals or eutherians. Opossum belongs to marsupials, a sister group that is believed to have diverged from placental mammals about 130 million years ago. Marsupials possess typical IgG and IgE (Vernersson et al. 2002). The current analysis of the opossum genome revealed that this species possesses genes for all the three main groups of the FcR family (Fig. 1). There are at least four genes for classical FcRs in the opossum genome. According to sequence comparisons and synteny data, opossum has FcɛRI, FcγRIII, and two genes for the FcγRII-like proteins. The FcγRII-like genes have apparently resulted from a recent duplication event, as their deduced amino acid sequences share up to 90% identical residues. Both genes appear to code for inhibitory receptors. They have distinct exons for the TM and IC regions, and in each locus, one of the IC exons encodes a polypeptide bearing a strongly conserved ITIM-like motif. In the FcγRIII and FcɛRI genes, exons that code for joined TM and IC regions were revealed. Furthermore, the TM regions of the predicted FcγRIII and FcɛRI contain the conserved Fxx(D/N)TxL motif characteristic of their counterparts in placental mammals.
We did not find a counterpart of the FcγRI gene in the opossum genome. One of the predicted opossum genes encodes a receptor with the EC region composed of the D1, D2, and D3-type domains, like that in FcγRI (Fig. 1). However, this gene occupies a position between DUSP23 and SLAMf8 genes, like the FCRL6 gene in the placental mammal genomes. The gene structure analysis suggested that, unlike the FcγRI gene of placental mammals, the TM and IC regions of the opossum FCRL6 are encoded by distinct exons.
According to the Ensembl mapping, the organizational order of the opossum genes for classical FcRs, FCRLA, and FCRLB is very similar to that in the placental mammals. oFCRLB is closely linked to the oDUSP12 gene and FcgRIII to the HSPA6-like gene. FCRLA and FCRLB proved to be the most conserved members of the family. Each of the genes codes for a four-domain protein with a mucin-like domain at the C-end. This domain is rich in proline, serine, and threonine residues and contains a characteristic a-helical motif. A distinctive feature of human, mouse, and dog FCRLA is a short N-terminal domain resembling the D1-type domains at the C-end only. The predicted N-terminal domain of opossum FCRLA is longer, and it shares about 25–26% identical residues with the D1-type domains of FcgRI and FCRLB. However, the domain has a single cysteine residue.
The opossum sFCRL cluster contains five genes spanning more than 350 kb. All appear to encode cell surface receptors with the EC regions composed of D1D2D3D4 (oFCRL1 and oFCRL4), D1D2D3D4D5 (oFCRL2 and oFCRL3) and D1D2D3D4D5D5 (oFCRL5) (Fig. 1). The cluster includes also the CD5L-like gene, located between oFCRL1 and oFCRL2 genes. As in placental mammals, the opossum CD5L-like gene was predicted to contain three exons for the SRCR domains. However, the 5′-exon contains two frame-shift mutations. The oFCRL1 and oFCRL2 genes neighboring the CD5L gene contain exons for the TM regions and appear to encode the cell surface receptors. No additional exons for the SRCR-like domains were found in this gene cluster, suggesting thereby that opossum lacks the FCRLS-like gene. The ETV3 and KIRREL genes that flank this cluster in the human and mouse genomes were found on the other scaffolds in the opossum genomic sequence.
The chicken FcR-related genes
In contrast to mammals, birds are known to have the IgY isotype, the closest relative of IgG and IgE (Warr et al. 1995). The IgY-binding entities have been described on the surface of chicken leukocytes (Ewald et al. 1976), erythrocytes (Manghi et al. 1987), and on the yolk sac (Tressler and Roth 1987). The yolk sac IgY receptor has been recently characterized as a phospholipase A2 receptor homolog (West et al. 2004). Thus, it was of particular interest to examine the FcR family genes in the chicken genome. Ten EST cDNAs coding for polypeptides structurally similar to mammalian members of the FcR family were found. These appeared to be alternatively spliced transcripts originated from the same gene or distinct highly similar genes. Search in the chicken genomic sequences resulted in identification of only three short contigs that proved to be unordered pieces of the genome. Of these, one carried three exons for the D1, D2, and D3-type domains; the second had exons for the D3 and D4 domains. An exon for a joined TM region and IC region was revealed in the third contig. The D3-exons from the two contigs differed in two nucleotides only. Moreover, the 5′-noncoding regions flanking the D3 exons in these contigs showed a single nucleotide substitution suggesting that the contigs may be overlapped.
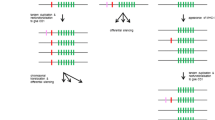
To examine complexity of the chicken FcR locus, we used Southern blot analysis. The EcoRI or HindIII digests of individual chicken genomic DNA were hybridized under mild conditions with the D3- or D4-specific probes. The probes were prepared using PCR of chicken DNA. The probes revealed a single hybridizing fragment in both digests (Fig. 2a). We also tested distribution of FCRL mRNA in chicken tissues. RT-PCR using primers matching the D1 and D4 exons detected two to three transcripts in both lymphoid and non-lymphoid tissues (Fig. 2b). Cloning and sequencing of these fragments showed that they represent alternatively spliced transcripts of the same gene. The largest fragment included the D1, D2, D3, and D4 exons. The intermediate was aberrant because of a frame-shifting deletion of the main part of the D2 exon. The corresponding transcript was apparently generated through a cryptic splice site in the D2 exon. The small fragment lacked the entire exon for the D2 domain but otherwise was identical to the full-length transcript. These results indicate that chicken has a single FCRL gene coding for a transmembrane receptor with the EC portion composed of the D1, D2, D3, and D4 domains (Fig. 1). The protein has a short IC region devoid of tyrosine residues. The TM region contains an arginine residue at the N-terminus. It remains to be examined if the protein may mediate activating signals through association with an ITAM-bearing signal subunit and if it is able to interact with chicken IgY.
Experimental study of the chicken FCRL gene. a Southern blot hybridization. The DNA was isolated from individual chicken blood cells, digested with EcoRI and HindIII, electrophoresed in 0.5% agarose, blotted, and hybridized with the 32P-labeled probes. The probes were two purified PCR fragments for the D3- type domain (255 bp) and D4- type domain (250 bp) of chiFcrl. b RT-PCR analysis of the chicken FCRL gene expression in tissues using primers matching the D1 and D4 exons. Arrow indicates a fragment corresponding to a full length transcript
No evidence for the presence of chicken genes similar to the classical FcR, FCRLA, and FCRLB genes was found in the genomic and EST databases. The small size of the chicken FcR family was quite unexpected in view of our recent finding that more primitive amphibians have up to 75 FcR-related genes (Guselnikov et al. 2004 and our unpublished data). It has been reported that the chicken genome is smaller than the mammalian and teleostean (Hillier et al. 2004). Gene number has been estimated as 20,000–23,000 in chicken. Many gene families were found to be underrepresented compared to mammals. The presence of a single FcR-related gene in chicken may be thus explained by lineage-specific gene loss.
An alternative explanation implies gaps in the genome assembly. To verify this with reference to the FcR-related genes, we analyzed the genomic sequences for genes closely linked to the members of the FcR family in mammals, such as CRP, CD5L, DUSP12, ATF6, DUSP23, ETV3, NESG1, and KIRREL (Fig. 1). The DUSP12 and ATF6 genes that are linked to the FCRLB gene in the mammalian genomes are on chromosomes 1 and 8 in the chicken genome, respectively. The genes neighboring DUSP12 in the chicken genome (ATP6V1A and Mac3) reside in chromosome 3 in the human genome. One of the chicken ATF6 neighbors (OLFML2B) is closely linked to ATF6 in human. Thus, the entire syntenic group, which in mammals includes genes for the low affinity classical FcRs, FCRLA, and FCRLB, appears to be dispersed throughout the chicken genome.
The FCRL6-neighboring DUSP23 gene was revealed in the disordered piece of the chicken genome together with the CRP-like gene. Another disordered fragment of the chicken genome was found to contain the NESG1-like gene, the counterparts of another FCRL6 neighbor in the mammalian genomes. Orthologs of the ETV3 and KIRREL genes that flank the main FCRL1–FCRL5 cluster in mammals were revealed in short fragments scattered throughout the chicken genome. Thus, most of the genes closely linked to the human and mouse FcR-related genes may be found in the available chicken genomic sequences. Although the exact order of many of these genes cannot, as yet, be established, it appears unlikely that the absence of the FcR-related genes in the chicken genome is due to gaps in the current assembly. Furthermore, it should be noted that cDNAs for classical FcRs and/or FcR-like proteins are very abundant in the mammalian and amphibian EST collections because of high expression level and broad cellular distribution of the corresponding mRNAs. The chicken EST collection was estimated to include up to 460,000 clones, and it is thought to cover most of the chicken genes (Smith et al. 2004). The absence of EST cDNAs for the classical FcRs is thus a further argument in favor of the suggestion that the FcR-family has become dramatically reduced in this particular species.
Phylogenetic analysis
The organization of the mammalian FcR-family genes and their exon composition clearly indicate that the FcR, iFCRL, and sFCRL subsets emerged before the radiation of marsupials and placental mammals. The orthology of the FCRLA and FCRLB genes of all the four mammalian species is unequivocal. The sequence comparisons, however, are insufficient for making inferences about the relationships among the various FcR and sFCRL proteins in the four species. To gain a deeper insight into the evolution of the family, we constructed a series of phylogenetic trees for separate or joined EC exon/domain subtypes using the neighbor-joining (NJ) method (Kumar et al. 2004).
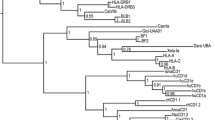
The phylogenetic analysis of the joined D1D2 exons showed separation of the mammalian genes into three main groups corresponding to FcRs, FCRL1-5 and FCRLB (Fig. 3). In the tree, the chicken D1D2 does not associate with any of these groups, suggesting that the subdivision of the FcR-family might have occurred in the mammalian lineage after its split from the avian. Based on the branching pattern of the classical FcR exons, it may be deduced that the common ancestor of placental mammals had a single gene for each functional class. These genes persisted in carnivores, whereas primates and rodents acquired additional through lineage-specific gene duplications. The common ancestor of mammals had at least three genes coding for the ancestral FcɛRI, FcγRII, and FcγRIII. The opossum FcγRII and FcγRIII form a cluster separated from that formed by eutherian FcγRII and FcγRIII. The topology was the same when we used amino acid sequences or applied the minimal evolution (ME) method for the tree construction. However, it should be noted that FcγRII and FcγRIII have opposite signal properties and their TM/IC regions are quite different. For this reason, it appears unlikely that the low affinity FcRs have independently subdivided into FcgRII and FcgRIII in marsupials and placental mammals. Instead, the branching pattern of the FcgRII/III EC domains may be explained by their lineage-specific exchange in the course of marsupial phylogenesis. Such an exchange is apparent in the case of rodent low affinity FcRs (Hughes 1996; Mechetina et al. 2002a). Indeed, phylogenetic analysis clearly demonstrates the monophyletic origin of the FcγRIII TM regions in mammals (supplementary Fig. 11 in Appendix 1).
NJ tree of the human (h), mouse (m), dog (d), opossum (o), and chicken (chi) exon sequences coding for the D1 and D2 domain subtypes of the FcR and FCRL molecules. This tree and those shown below and in the electronic supplementary figures, were constructed using the MEGA3 package with p-distances for nucleotide sequences sites. The numbers on the trees represent values for the bootstrap and interior branch tests after 250 replicates. Only values above 50 are shown. Dashes indicate values below the cutoffs
The absence of the FcγRI-like gene in the current version of the opossum genome may be accounted for (1) gaps in the assembly; (2) loss of the gene in marsupials; and (3) lineage-specific acquisition of the gene by placental mammals. The latter possibility is unlikely because phylogenetic analysis of the D1D2 exons shows that FcγRIII and FcγRII of all mammalian species are significantly closer to each other than to FcγRI D1D2 (Fig. 3). However, further data are needed to resolve this issue.
We omitted the FCRLA genes from the D1 + D2 trees because they did not contain the typical D1 exon. Construction of the trees for the D2 and D3 exons did not provide statistical support for close relationships of FCRLA and FCRLB (supplementary Figs. 9 and 10 in Appendix 1). Nevertheless, the presence of the exons for the conserved mucin-like domain in these two genes and close linkage in all the mammalian species, strongly argue in favor of their monophyletic origin.
The sFCRL subset is the most peculiar part of the FcR family. Each of the four species has its own number of genes and domain architecture of the encoded proteins is highly diverse. The branching patterns of the human and dog D1–D4 type exons support the suggestion that the dog FCRL1–FCRL6 genes are orthologous to human FCRL1–FCRL6 (Figs. 4 and 5 and supplementary Figs. 8, 9, and 10 in Appendix 1). The D2 exon and the putative D1, D3, and D4 pseudoexons of dFCRL3 associate with the corresponding exons of the human FCRL3. Dog FCRL1 and FCRL6 are orthologs of the human and mouse FCRL1 and FCRL6, respectively. In the case of FCRL6, statistically supported association is observed for the D5-type exons of the three species genes (Fig. 5) and for the dFCRL6 and hFCRL6 D2- and D3-type exons (supplementary Figs. 9 and 10 in Appendix 1). Prominent association is also observed for the D4 exons of human, mouse, and dog FCRL1 (Fig. 4). The availability of dog sequences makes it possible to resolve relationships among mFCRL5 and the other members of the family. The mFCRL5 gene contains an exon for the D4 domain, and in this respect, is similar to the human and dog FCRL3 genes. However, phylogenetic analysis demonstrates that the D1 and D2 exons of the mouse gene are closely related to the D1 and D2 exons of hFCRL5 and dFCRL5 (supplementary Figs. 8 and 9 in Appendix 1). Furthermore, the D5 exon of mFCRL5 is associated with the exons of the D5b subtype found only in hFCRL5 and dFCRL5 (Fig. 5). Finally, the TM and IC regions of mFCRL5 are most similar to those of hFCRL5 (supplementary Fig. 11 in Appendix 1 and data not shown). Thus, despite the presence of the D4 exon in the mFCRL5, this gene may be regarded as the diverged ortholog of the human and dog FCRL5. Hence, it may be concluded that rodents have lost three genes, namely, FCRL2, FCRL3, and FCRL4, after their separation from other mammals.
Dog FCRLS appears to be the ortholog of mouse FCRLS. Not only these two genes share exon for the SRCR domain, but also two of their four EC exons show statistically supported association (Figs. 4 and 5 and supplementary Figs. 9 and 10 in Appendix 1). The presence of the Ig-SRCR gene in the dog and mouse genomes suggests that this gene has been present in the common ancestor of placental mammals. Interestingly, we did not find its counterpart in the opossum genome. Aiming to reconstruct the phylogenetic history of this gene, we analyzed relationships among the FCRLS SRCR exons and the exons of the closely linked CD5L genes. In the tree constructed, the FCRLS SRCR domains are significantly closer to the first domain of the mouse, dog, and human CD5L than to that of opossum CD5L (Fig. 6). This branching pattern supports the suggestion that the FCRLS gene might have emerged after the split of marsupials and eutherians. As the human and mouse CD5L are closer to each other than to the dog CD5L, it appears likely that the FCRLS gene has been lost later in the primate lineage.
Importantly, different exon subtypes frequently exhibit different branching patterns in the trees. Thus, D4-type exons of dFCRL4 and dFCRLS are more closely related to each other than to those of hFCRL4 or mFCRLS (Fig. 4); close association may be seen between the D1-type exons of dFCRL4 and dFCRL5 and between the D1 exons of hFCRL4 and hFCRL5 (supplementary Fig. 8 in Appendix 1); the D5 exon of dFCRLS associates with D5.2–D5.4 exons of dFCRL5, while is weakly related to D5 of mFCRLS (Fig. 5). The most plausible explanation of these differences in the tree topologies is that exon shuffling occurred among the various members of the family during species-specific evolution. The same explanation is applicable to the fact that with the rare above exceptions, the D5-type exons of the sFCRL genes are poorly resolved. For instance, the D5 exons of the human genes tend to associate with each other and the same pattern can be seen for most of the dog D5 exons. Although the bootstrap values to support this association are low, the D5 branching pattern contrasts with the clear resolution of the D1-, D2-, and D4-type domains and suggests that the D5 exons were partially homogenized during species-specific evolution of the sFCRL gene subset.
Based on the gene localization and significant bootstrap support of the clustering of the D2 exon, oFCRL6 may be regarded as the ortholog of the eutherian FCRL6 genes. The combined D1 and D2 exons of oFCRL6 obviously belong to the sFCRL group (Fig. 3). Thus, the similar domain architecture in oFCRL6 and FcgRI may be due to molecular convergence rather than to common origin. The presence of the exon for the conventional TM domain without charged residues in the opossum FCRL6 gene further supports this view. Relationships among five other opossum FCRL genes with those of placental mammals are not clear. The trees generated suggest monophyletic origin of oFCRL1, oFCRL2, and oFCRL3, on one hand, and oFCRL4 and oFCRL5, on the other.
The chicken FCRL gene encodes a protein similar in domain architecture to human and dog FCRL4. However, phylogenetic analysis provided no support for close relationships between these proteins. Like in the case of D1 + D2 (Fig. 3), no statistically significant association of the chicken gene with any of the mammalian FcR-related genes was observed in the analysis of the separate D1, D2, D3, and D4 exons (Fig. 4 and supplementary Figs. 8, 9, and 10 in Appendix 1). These data are consistent with the suggestion that separation of the FcR-family into FcRs, sFCRL, and iFCRL subgroups might have occurred after the mammalian–avian split. Furthermore, these results are consistent with our assessment that the FcR-family has been substantially reduced in the chicken genome.
Concluding remarks
The analysis of the gene structure and phylogenetic relationships of the FcR-related genes in four mammalian representatives and chicken clearly demonstrated that the common ancestor of mammals possessed all the three main subsets of the family. Of these, the iFCRL subset persisted without significant changes throughout mammalian phylogeny. The radiation of mammals was, however, accompanied by species-specific duplication of the genes for classical FcRs. As a result, the dog genome acquired the minimum set of genes, one for each FcR class. The rodent and primate genomes got additional genes. The duplications of the FcR genes in primates and rodents were apparently accompanied by exon shuffling (Hughes 1996; Mechetina et al. 2002b). However, the domain architecture remained unaltered in the newly created FcR proteins.
Compared to FcRs, the species-specific evolution of the sFCRL genes resulted in dramatic differences between the mammalian lineages not only in gene number, but in domain architecture of the encoded proteins as well. Besides gene/exon deletions and duplications, multiple inter- and intragenic recombination events clearly contributed to the subfamily diversification. The maintenance of these proteins in various species, along with their extensive structural variability, may indicate that their ligands have also evolved in a species-specific manner. What these ligands are remains to be determined. Except for mFCRLS, all the members of the sFCRL subset are expressed in lymphoid cells. Human FCRL1–FCRL5 and mouse FCRL1 and FCRL5, are B cell-specific receptors (Davis et al. 2005). FCRL6 is expressed by CD8 T and NK cells in human (Ershova et al. 2004) and by B and NK cells in mouse (R. Davis, personal communication). The expression patterns suggest that these diverse receptors may contribute to the species-specific features of function and/or development of the effector lymphocyte subsets.
Our recent experimental and genome mining data demonstrated that FcR-related genes are abundant in the amphibian Xenopus (Guselnikov et al. 2004 and unpublished data). Like in the chicken genome, no evidence for the presence of the FcR-, FCRLA- and FCRLB-like genes in the Xenopus genome was obtained. With this in mind and also taking in consideration that IgE and IgG, the main ligands of classical FcRs, are mammalian-specific Ig isotypes (Vernersson et al. 2002; Warr et al. 1995), we hypothesize that the sFCRL group is the most ancient part of the FcR family, whereas classical FcRs and FCRLA and FCRLB, are relatively recent derivatives that arose during mammalian phylogenesis.
References
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Burge C, Karlin S (1997) Prediction of complete gene structures in human genomic DNA. J Mol Biol 268:78–94
Chikaev NA, Bykova EA, Najakshin AM, Mechetina LV, Volkova OY, Peklo MM, Shevelev AY, Vlasik TN, Roesch A, Vogt T, Taranin AV (2005) Cloning and characterization of the human FCRL2 gene. Genomics 85:264–272
Chomczynski P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction. Anal Biochem 162:156–159
Daeron M (1997) Fc receptor biology. Annu Rev Immunol 5:203–234
Davis RS, Wang YH, Kubagawa H, Cooper MD (2001) Identification of a family of Fc receptor homologs with preferential B cell expression. Proc Natl Acad Sci USA 98:9772–9777
Davis RS, Dennis G Jr, Odom MR, Gibson AW, Kimberly RP, Burrows PD, Cooper MD (2002a) Fc receptor homologs: newest members of a remarkably diverse Fc receptor gene family. Immunol Rev 190:123–136
Davis RS, Li H, Chen CC, Wang YH, Cooper MD, Burrows PD (2002b) Definition of an Fc receptor-related gene (FcRX) expressed in human and mouse B cells. Int Immunol 14:1075–1083
Davis RS, Stephan RP, Chen C-C, Dennis G Jr, Cooper MD (2004) Differential B cell expression of mouse Fc receptor homologs. Int Immunol 16:1343–1353
Davis RS, Ehrhardt GR, Leu CM, Hirano M, Cooper MD (2005) An extended family of Fc receptor relatives. Eur J Immunol 35:674–680
Ehrhardt GR, Davis RS, Hsu JT, Leu CM, Ehrhardt A, Cooper MD (2003) The inhibitory potential of Fc receptor homolog 4 on memory B cells. Proc Natl Acad Sci USA 100:13489–13494
Ehrhardt GR, Hsu JT, Gartland L, Leu CM, Zhang S, Davis RS, Cooper MD (2005) Expression of the immunoregulatory molecule FcRH4 defines a distinctive tissue-based population of memory B cells. J Exp Med 202:783–791
Ershova SA, Peklo MM, Shevelev AY, Najakshim AM, Mechetina LV, Volkova OY, Chikaev NA, Vlasik TN, Taranin AV (2004) IFGPG: a novel human member of the IFGP/IRTA/FcRH family that is selectively expressed by CD8 T and NK cells. Clin Invest Med Proceedings of the 12th International Congress of Immunology, July 18–23, 2004, Montreal, Canada 27(4):10C
Ewald S, Freedman L, Sanders BG (1976) EA rosette-forming lymphoid cells in chickens: specificity of the Fc receptor and its relationship to other surface antigens. Immunology 31:847–854
Facchetti F, Cella M, Festa S, Fremont DH, Colonna M (2002) An unusual Fc receptor-related protein expressed in human centroblasts. Proc Natl Acad Sci USA 99:3776–3781
Falini B, Tiacci E, Pucciarini A, Bigerna B, Kurth J, Hatzivassiliou G, Droetto S, Galletti BV, Gambacorta M, Orazi A, Pasqualucci L, Miller I, Kuppers R, Dalla-Favera R, Cattoretti G (2003) Expression of the IRTA1 receptor identifies intraepithelial and subepithelial marginal zone B cells of the mucosa-associated lymphoid tissue (MALT). Blood 102:3684–3692
Guselnikov SV, Ershova SA, Mechetina LV, Najakshin AM, Volkova OY, Alabyev BY, Taranin AV (2002) A family of highly diverse human and mouse genes structurally links leukocyte FcR, gp42 and PECAM-1. Immunogenetics 54:87–95
Guselnikov SV, Erilova AY, Najakshin AM, Cohen N, Rober J, Taranin AV (2004) FcR-like genes in the amphibian Xenopus: species-specific expansion, structure diversity, and developmental expression. Clin Invest Med. Proceedings of the 12th International Congress of Immunology, July 18–23, 2004, Montreal, Canada 27(4):82C
Hatzivassiliou G, Miller I, Takizawa J, Palanisamy N, Rao PH, Iida S, Tagawa S, Taniwaki M, Russo J, Neri A, Cattoretti G, Clynes R, Mendelsohn C, Chaganti RS, Dalla-Favera R (2001) IRTA1 and IRTA2, novel immunoglobulin superfamily receptors expressed in B cells and involved in chromosome 1q21 abnormalities in B cell malignancy. Immunity 14:277–289
Hillier LW et al (2004) Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432:695–716
Hubbard T, Barker D, Birney E, Cameron G, Chen Y, Clark L, Cox T, Cuff J, Curwen V, Down T, Durbin R, Eyras E, Gilbert J, Hammond M, Huminiecki L, Kasprzyk A, Lehvaslaiho H, Lijnzaad P, Melsopp C, Mongin E, Pettett R, Pocock M, Potter S, Rust A, Schmidt E, Searle S, Slater G, Smith J, Spooner W, Stabenau A, Stalker J, Stupka E, Ureta-Vidal A, Vastrik I, Clamp M (2002) The Ensembl genome database project. Nucleic Acids Res 30:38–41
Hughes AL (1996) Gene duplication and recombination in the evolution of mammalian Fc receptors. J Mol Evol 43:4–10
Imboden JB, Eriksson EC, McCutcheon M, Reynolds CW, Seaman WE (1989) Identification and characterization of a cell-surface molecule that is selectively induced on rat lymphokine-activated killer cells. J Immunol 143:3100–3103
Kumar S, Tamura K, Nei M (2004) MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163
Leu CM, Davis RS, Gartland LA, Fine WD, Cooper MD (2005) FcRH1: an activation coreceptor on human B cells. Blood 105:1121–1126
Maltais LJ, Lovering RC, Taranin AV, Colonna M, Ravetch JV, Dalla-Favera R, Burrows PD, Cooper MD, Davis RS (2006) New nomenclature for Fc receptor-like molecules. Nat Immunol 7:431–432
Manghi MA, Venturiello SM, Gutierrez MI, Etchevierrigaray M, Margni RA (1987) Isolation and partial characterization of biologically active Fc receptor of chicken red cells. Biochim Biophys Acta 923:381–388
Masuda K, Davis RS, Maruyama T, Zhang J, He T, Cooper MD, O-Wang J, Burrows PD (2005) FcRY, an Fc receptor related gene differentially expressed during B lymphocyte development and activation. Gene 363:32–40
Mechetina LV, Najakshin AM, Alabyev BY, Chikaev NA, Taranin AV (2002a) Identification of CD16-2, a novel mouse receptor homologous to CD16/Fc gamma RIII. Immunogenetics 54:463–468
Mechetina LV, Najakshin AM, Volkova OY, Guselnikov SV, Faizulin RZ, Alabyev BY, Chikaev NA, Vinogradova MS, Taranin AV (2002b) FCRL, a novel member of the leukocyte Fc receptor family possesses unique structural features. Eur J Immunol 32:87–96
Milanesi L, D’Angelo D, Rogozin IB (1999) GeneBuilder: interactive in silico prediction of gene structure. Bioinformatics 15:612–621
Miller I, Hatzivassiliou G, Cattoretti G, Mendelsohn C, Dalla-Favera R (2002) IRTAs: a new family of immunoglobulin-like receptors differentially expressed in B cells. Blood 99:2662–2669
Nakamura R, Sato Y, Takagi K, Sasaki N, Sawada J, Kitani S, Teshima R (2003) Presence and primary sequence of a high-affinity IgG receptor on canine mastocytoma (CM-MC) cells. Immunogenetics 55:271–274
Nakayama Y, Weissman SM, Bothwell AL (2001) BXMAS1 identifies a cluster of homologous genes differentially expressed in B cells. Biochem Biophys Res Commun 285:830–837
Nimmerjahn F, Bruhns P, Horiuchi K, Ravetch JV (2005) FcgammaRIV: a novel FcR with distinct IgG subclass specificity. Immunity 23:41–51
Pearson WR (1990) Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol 183:63–98
Polson AG, Zheng B, Elkins K, Chang W, Du C, Dowd P, Yen L, Tan C, Hongo JA, Koeppen H, Ebens A (2006) Expression pattern of the human FcRH/IRTA receptors in normal tissue and in B-chronic lymphocytic leukemia. Int Immunol 18:1363–1373
Ravetch JV, Kinet J-P (1991) Fc receptors. Annu Rev Immunol 9:457–492
Sambrook T, Fritsch EF, Maniatis T (eds) (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, Plainview, NY
Smith J, Speed D, Law AS, Glass EJ, Burt DW (2004) In-silico identification of chicken immune-related genes. Immunogenetics 56:122–33
Thanaraj TA, Robinson AJ (2000) Prediction of exact boundaries of exons. Brief Bioinform 1:343–356
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Tressler RL, Roth TF (1987) IgG receptors on the embryonic chick yolk sac. J Biol Chem 262:15406–15412
Vernersson M, Aveskogh M, Munday B, Hellman L (2002) Evidence for an early appearance of modern post-switch immunoglobulin isotypes in mammalian evolution (II); cloning of IgE, IgG1 and IgG2 from a monotreme, the duck-billed platypus, Ornithorhynchus anatinus. Eur J Immunol 32:2145–2155
Warr GW, Magor KE, Higgins DA (1995) IgY: clues to the origins of modern antibodies. Immunol Today 16:392–398
West AP Jr, Herr AB, Bjorkman PJ (2004) The chicken yolk sac IgY receptor, a functional equivalent of the mammalian MHC-related Fc receptor, is a phospholipase A2 receptor homolog. Immunity 20:601–610
Wilson TJ, Colonna M (2005) A new Fc receptor homolog, FREB2, found in germinal center B cells. Genes Immun 6:341–346
Xu MJ, Zhao R, Zhao ZJ (2001) Molecular cloning and characterization of SPAP1, an inhibitory receptor. Biochem Biophys Res Commun 280:768–775
Xu MJ, Zhao R, Cao H, Zhao ZJ (2002) SPAP2, an Ig family receptor containing both ITIMs and ITAMs. Biochem Biophys Res Commun 293:1037–1046
Acknowledgments
This work was in part supported by the Russian Foundation for Basic Research grant 05-04-49268 (A.M.N) and the RAS Program “Origin and evolution of life” (A.V.T). The authors are grateful to Mrs. A. Fadeeva for the help in preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link of the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Fayngerts, S.A., Najakshin, A.M. & Taranin, A.V. Species-specific evolution of the FcR family in endothermic vertebrates. Immunogenetics 59, 493–506 (2007). https://doi.org/10.1007/s00251-007-0208-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-007-0208-8