Abstract
Serine proteases constitute the major protein granule content of cells of several hematopoietic cell lineages. A subgroup of these proteases, including the mast cell chymases, neutrophil cathepsin G, and T cell granzymes B to F and N, are in all investigated mammals encoded in one locus, the chymase locus. It is interesting to note that this locus has diversified greatly during the last 95 Myr of mammalian evolution. This divergence is exemplified by the presence of Mcpt8-related genes and multiple β-chymases in the mouse and rat, which lack direct counterparts in primates and in seven functional granzyme genes in the mouse where the human locus has only two. To study the expansion of the locus during rodent evolution and to better understand the evolutionary origin of β-chymases and the Mcpt8-family, we have performed a detailed analysis of the chymase locus of four mammalian species, i.e., human, dog, mouse, and rat. As a result, we report here a second chymase-like gene in dog, Cma2, which clusters with β-chymases in phylogenetic analyses. This finding supports a duplication of the common ancestor for α- and β-chymases before the major radiation of placental mammals, and a loss of the ancestral β-chymase gene sometime during primate evolution. Moreover, we show that in the rat, the Mcpt8-family diversified relatively recently together with sequences related to the β-chymase Mcpt2. Eight novel genes were identified in the duplication region, four of which are predicted to be functional. Duplications of rat granzyme B- and C-like sequences occurred seemingly independently within a similar time frame, but did not give rise to functional genes. Due to the duplications in rat and deletions in the carnivore/primate lineage, the rat chymase locus is approximately 15 and 9 times larger than its counterparts in dog and human, respectively. These findings illustrate the importance of gene duplications in conferring rapid changes in mammalian genomes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Proteases are central effector molecules in a large number of biological processes, such as food digestion, blood coagulation, complement activation, fertilization, tissue remodeling, and immunity (Neurath 1986). The importance of proteolysis in human biology is reflected by the fact that more than 2% of all human genes are either proteases or protease inhibitors (Puente 2005). It is interesting to note that the degradomes of rodents are even more complex. Out of the approximately 626 proteases in rat (Rattus norvegicus), 102 lack a direct counterpart in the human (Homo sapiens) genome. On the other hand, only 37 of the 561 human proteases lack a direct counterpart in the rat. This quantitative difference is mainly due to the expansion of specific protease subfamilies in the rat. Notably, most of these protease subfamilies are involved in reproductive and immunological functions (Gibbs et al. 2004; Puente and Lopez-Otin 2004).
A locus that was significantly expanded both in the mouse and rat genome is the mast cell chymase locus, which also represents the largest protease gene cluster in the rat (Puente and Lopez-Otin 2004). Thanks to the assembly of the complete dog genome (CanFam 1.0, July 2004, in Ensembl), the entire locus can now be compared between four mammalian species, i.e., mouse, rat, human, and dog. Besides mast cell chymases, the chymase locus harbors the genes of a heterogenic group of hematopoietic serine proteases (Caughey et al. 1993; Gurish et al. 1993), including the mast cell/basophil proteases of the mouse/rat mast cell protease-8 family (M/R8-family), neutrophil cathepsin G (Ctsg), and T cell granzymes B to F and N (Gzmb to f and n).
By phylogenetic analyses, the mast cell chymases fall into two groups, termed α- and β-chymases (Caughey et al. 2000; Chandrasekharan et al. 1996; Huang and Hellman 1994). It is interesting to note that the genomes of rodents, dogs, and primates seem to contain only one α-chymase gene. For example, of the multiple mast cell chymase genes that were described in mouse (Mus musculus) and rat, only mouse mast cell protease (mMCP)-5 and rat mast cell protease (rMCP)-5 are phylogenetically classified as α-chymases. In primates, a single mast cell chymase was described, and this is classified as an α-chymase (Caughey et al. 2000; Huang et al. 1991; Ide et al. 1995; Lützelschwab et al. 1997). However, these phylogenetic homologs are not always functional equivalents. For example, primate α-chymases have chymotrypsin-like substrate specificity (Hoit et al. 1995; Takai et al. 1997; Wintroub et al. 1984), whereas mMCP-5 and rMCP-5 have elastase-like specificity (Karlson et al. 2003; Solivan et al. 2002).
The β-chymases, on the other hand, often have several family members. Up till now, they were described in rodents only. In mouse, four β-chymases were identified, i.e., mMCP-1, -2, -4, and -9 (Huang et al. 1991; Hunt et al. 1997; Newlands et al. 1987; Serafin et al. 1990; Trong et al. 1989). Similarly, five β-chymases were described in the rat, i.e., rMCP-1 to -4 and rat vascular chymase (VCh), a serine protease expressed in mast cell lines and vascular smooth muscle cells (Benfey et al. 1987; Guo et al. 2001; Ide et al. 1995; Kido et al. 1986; Le Trong et al. 1987; Lützelschwab et al. 1997). It was suggested that the presence of ancestors for both α- and β-chymase genes is ancient and that β genes were lost at some point during primate evolution (Caughey et al. 2000; Huang and Hellman 1994). As no β-chymase was described in any non-rodent species, it is however unclear at which point during the mammalian evolution this loss has occurred.
Moreover, it is also unclear what evolutionary events have given rise to the M/R8-family. We have previously identified one member of this family with basophil-specific expression in the mouse, mMCP-8 (Lützelschwab et al. 1998; Poorafshar et al. 2000), and three family members in the rat, rMCP-8 to -10, which are all expressed in mucosal mast cells (MMC) (Lützelschwab et al. 1997). However, members of this family were not described in any other species.
In addition, the number of Gzm genes differs between mammalian species, ranging from two in human (Gzm b and h) to eight in mouse (Gzmb, c, d, e, f, g, l, and n; reviewed in Grossman et al. 2003). Of these eight genes in the mouse, only Gzml appears to be a pseudogene.
The presence of the M/R8-family genes and multiple β-chymase and granzyme genes in rodents indicates a massive lineage-specific expansion of the chymase locus. In contrast, only a single Ctsg gene is found in the chymase locus of all investigated mammals. This gene always appears to be located near the center of the chymase locus (Fig. 1).
Schematic comparison of the chymase locus in human, dog, mouse and rat. a Chromosomal localization of the chymase locus in human, dog, mouse and rat. Arrowheads indicate the position of the chymase locus on the ideograms. Grey bars next to the respective ideograms linked by black lines indicate regions of conserved synteny. b Comparative analysis of the genetic organization of the human, dog, mouse and rat chymase locus. Bars indicate gene positions; arrows indicate transcriptional orientation. Intervals between genes are drawn to scale. Numbers signify distance between genes in kb. Solid lines connect orthologues present in all four species. Dashed lines connect orthologues present in several, but not all four shown species. Transcripts have been described for all denoted genes except for dog Cma2. The total dimension of the chymase locus is approximately 73 kb in dog, 129 kb in human, 324 kb in mouse and 1.1 Mb in rat
We wanted to understand what evolutionary mechanisms have created these diverse protease repertoires in mammals and how the genetic differences translate into cellular function. Therefore, as a first step, we conducted an extensive bioinformatic analysis of the chymase loci of mouse, rat, dog, and human. Subsequently, a complete picture of the proteases encoded in the respective chymase loci can help us to analyze functional homologies and to understand the role these proteases play in the immune systems of different mammals.
Materials and methods
Chromosomal localization
The chromosomes harboring the chymase locus were identified by nucleotide–nucleotide basic local alignment search tool (BLAST) searches in the Ensembl and National Center for Biotechnology Information (NCBI) databases with the sequences listed below. The chromosomal localization of the chymase locus in human, dog, mouse, and rat was determined by analyzing the respective chromosomes in Ensembl (http://www.ensembl.org).
Mapping the chymase locus in human, mouse, rat, and dog
The chymase locus in human (H. sapiens), mouse (M. musculus), and rat (R. norvegicus) was mapped using the following mRNA sequences from the NCBI database: H. sapiens CMA1 (NM_001836), CTSG (NM_001911), GZMH (NM_033423), and GZMB (NM_004131); M. musculus Mcpt5 (NM_010780), Mcpt9 (NM_010782), Mcpt1 (NM_008570), Mcpt2 (NM_008571), Mcpt4 (NM_010779), Mcpt8 (NM_008572), cathepsin-G (X78544), Gzme (NM_010373), Gzmd (NM_010372), Gzmg (NM_010375), Gzmn (NM_153052), Gzmf (NM_010374), Gzmc (NM_010371), and Gzmb (NM_013542); and R. norvegicus Cma1 (NM_013092), RMCP-4 (U67907), RMCP-3 (U67888), VCH (AF063851), RMCP-8 (U67911), RMCP-10 (partial cds; U67913), RMCP-9 (partial cds; U67912), Mcpt2 (NM_172044), RMCP-1 (U67915), Ctsg_predicted (XM_214205), LOC290259 (XM_214196), Rnkp7 (NM_153466), LOC290262 (XM_224224), Gzmc (NM_134332), and Gzmb (NM_138517). The chymase locus of the dog (Canis familiaris) was mapped using the following sequences from the Ensembl database: ENSCAFT00000019746 (Cma1), GENSCAN00000053517 (Cma2), ENSCAFT00000019749 (sim. Ctsg), ENSCAFT00000019767 (sim. Gzmh), and ENSCAFT00000019785 (sim. Gzmb). The dog sequences were named according to the orthologs suggested by Ensembl, taking into account their positions in the phylogenetic analyses shown in Fig. 2.
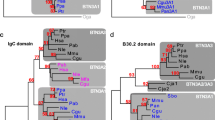
Phylogenetic analyses of all known and novel sequences in the chymase locus in human, dog, mouse, and rat. Numbers represent bootstrap values out of 1,000 trees. The prefixes h, d, r, and m refer to sequences from human, dog, rat, and mouse, respectively. Sequences without reported protein expression are shaded. For “d sim. CtsG” and “r CtsG,” protein fragments were reported. (corr.) Corrected. a Phylogenetic tree based on neighbor joining analysis of exon sequences corresponding to mature proteins. Numbers Ia through IIb signify nodes utilized for the calculation of evolutionary distances (Materials and methods). b Phylogenetic tree based on neighbor-joining analysis of mature protein sequences
Using the above sequences, we carried out BLASTN searches in the respective genomes (NCBI and Ensembl, May 2005). Our BLASTN searches in the rat genome yielded the novel sequences Mcpt2-rs1, Mcpt2-rs2a, Mcpt2-rs2b, Mcpt2-rs2c, Mcpt8-rs1, Mcpt8-rs2, Mcpt8-rs3, Mcpt8-rs4, Gzmc-rs1, Gzmc-rs2, and Gzmc-rs3. These sequences were named according to the rules of the Rat Genome and Nomenclature Committee (http://rgnc.gen.gu.se). Moreover, several incomplete sequences with similarity to previously known genes were identified in the rat chymase locus. These incomplete sequences were not given names, but are included in Fig. 4.
Exon–intron boundaries of novel genes were determined in comparison with the canonical sequence for GT-AG introns (Breathnach and Chambon 1981; Burset et al. 2000) and by sequence similarity with previously known related genes.
Phylogenetic analyses
The nucleotide sequences were compared, translated into amino acid (aa) sequences and analyzed with the DNASTAR software (DNASTAR, Madison, USA). Retrieved exon sequences from previously known genes were translated into aa sequences and checked for identity with the following respective NCBI entries: human (H. sapiens) P23946 (Cma1), P08311 (Ctsg), NP_219491 (Gzmh), and NP_004122 (Gzmb); dog (C. familiaris) P21842 (Cma1); mouse (M. musculus) P21844 (Mcpt5), P11034 (Mcpt1), O35164 (Mcpt9), P15119 (Mcpt2), P21812 (Mcpt4), P43430 (Mcpt8), CAA55290 (Ctsg), P08884 (Gzme), P11033 (Gzmd), P13366 (Gzmg), BAB68562 (Gzmn), P08883 (Gzmf), P08882 (Gzmc), and NP_038570 (Gzmb); and rat (R. norvegicus) NP_037224 (Mcpt5), AAB48260 (Mcpt4), XP_214189 (Mcpt3), AAC16657 (VCh), AAB48264 (Mcpt8), Q06606 (Mcpt10), AAB48265 (incomplete, Mcpt9), P00770 (Mcpt2), AAB48268 (Mcpt1), AAB05241 (Nkpt7), NP_599159 (Gzmc), and NP_612526 (Gzmb).
For dog XP_547751 (Gzmh), the aa encoded by the predicted exon 1 differ from our analysis, but the mature proteins are identical. Rat XP_214205 (Ctsg) contains an additional 26 aa compared to our analysis, and XP_224224, the predicted protein to LOC290262, is 17 aa shorter than our prediction.
For phylogenetic analyses, sequences of immature proteins, mature proteins, or the nucleotide sequences corresponding to them, respectively, were used with similar results. In cases where NCBI entries of hypothetical proteins differed from our analysis, or where no NCBI entries were available, aa sequences translated from the predicted exons were used. Where necessary, frameshift or stop mutations were corrected to obtain sensible protein sequences for phylogenetic analysis. Phylogenetic analyses were performed with CLUSTALX (Thompson et al. 1997) as described previously (Vernersson et al. 2004) and with PHYLIP version 3.5c (Felsenstein 1989), yielding similar results. Neighbor-joining and parsimony analyses produced essentially identical topologies. Data shown were obtained with sequences of mature proteins or the corresponding nucleotide sequences using CLUSTALX and neighbor-joining.
The following mRNA and protein sequences provided an outgroup: H. sapiens β-tryptase (M37488, NP_003285), R. norvegicus RMCP-6 (U67909, P50343), and R. norvegicus RMCP-7 (U67910, P27435).
Cladograms were depicted using TreeView Version 1.6.6 (2000, Roderic D. M. Page) or njplot (Manolo Gouy, Lyon).
Sequence analysis of the R2/R8 duplication region
The R2/R8 duplication region was analyzed by overlapping pairwise alignments and Nucleic Acid Dot Plots (http://arbl.cvmbs.colostate.edu/molkit/dnadot). (AC)N repeats, (CAA)N repeats, and parts of L1_RN were identified using the Censor server repeat screening (http://www.girinst.org/Censor/) (Jurka et al. 1996).
Calculation of dN/dS values
As a measurement of evolutionary pressure, the portion of nonsynonymous vs synonymous base pair substitutions within the R2- and R8-family, respectively, was calculated using PAML software (http://abacus.gene.ucl.ac.uk/software/paml.html) (Yang 1997). Results shown are based on dN±SE and dS±SE values obtained by the method of Yang et al. (2000). The final values for dN/dS±SE were calculated in Microsoft Excel 10 for Mac Service Release 1.
Calculation of evolutionary distance
Evolutionary distances were calculated for the M/R8 and M1/R2 gene families separately. Reference values of percent nucleotide substitutions that were accumulated after the time of the mouse–rat split were obtained by calculating the pairwise percent nucleotide substitutions between all family members in rat and their respective ortholog in mouse, and by determining the average (Fig. 2, nodes Ia and Ib). The divergence time between the mouse and rat was assumed to be 18±6 Myr (Gibbs et al. 2004). We compared exon nucleotide sequences and intron nucleotide sequences separately. For exon nucleotide comparisons, only presumably functional genes were considered. For all comparisons, insertions and deletions (indels) comprising several nucleotides were counted as one substitution. Pairwise comparisons were then also performed for rat sequences only, both within the R8- and R2-family (Fig. 2, nodes IIa and IIb, respectively), and for the Mcpt2-rs2-subfamily (Fig. 2, node IIIa). Only sequence pairs crossing the respective node were considered. In addition, the parts of the truncated sequences s2a, s2b, s2c, and s2d that are homologous to exons and introns of the complete members of the R2-family were compared to the corresponding parts of mMcpt1 and to each other. Due to the truncations, this subgroup is not included in the phylogenetic analyses described above. However, nucleotide sequence similarity suggests that s2a branched out before the divergence of s2b, s2c, and s2d, and we therefore only considered pairs of the latter three with s2a in our analysis.
For all subfamilies, the respective average of percent substitutions between the rat paralogs was divided with the corresponding mouse–rat reference value. The average of all ratios within a group was calculated and the significance of differences between intron and exon comparisons was tested with Student’s t test.
Results
Comparative analysis of the chymase locus in human, dog, mouse, and rat
The chymase locus is situated on chromosomes 14q11.2, 8, 14C1/2, and 15 p12/13 in human, dog, mouse, and rat, respectively (Fig. 1a). Conserved synteny spanning over large regions is found pairwise between human/dog and mouse/rat. However, only a small region, containing the chymase locus, displays conserved synteny between all four species. Thus, large-scale chromosomal rearrangements have occurred in the course of speciation, which result in different chromosomal surroundings for the chymase locus in rodents vs primates and carnivores.
The chymase locus itself has also undergone major changes during mammalian evolution, both in size and absolute number of functional genes. For example, this locus is approximately 15 times larger in rat (1.1 Mb) than in dog (73 kb) (Fig. 1b), and the number of functional genes is only 4 in human and at least 14 in mouse. However, a common feature for all four species is that the α-chymase (Cma1 or Mcpt5) gene borders the locus on one flank and the Gzmb gene on the other flank. All four genomes also contain a single Ctsg gene near the middle of the locus.
The four genes in the human chymase locus are Cma1, Ctsg, and two granzyme genes Gzmb and Gzmh (Figs. 1b and 2). All these genes have the same transcriptional orientation (Fig. 1b). The region between Cma1/Mcpt5 and Ctsg will in the following be referred to as “Mcpt region,” and the region between Ctsg and Gzmb as “Gzm region” (see Figs. 1b and 2).
The structure of the dog chymase locus is almost identical to human, except for the presence of an additional gene in the Mcpt region (Caughey 2005), the Cma2 gene. This novel gene, which lies in the same orientation as the other genes in the locus, clusters with β-chymases in phylogenetic analyses of exon nucleotides, but with α-chymases when considering protein sequences (Fig. 2). Both statements hold true with neighbor-joining and parsimony methods. The dog Cma2 gene is the most β-chymase-like gene so far described in a non-rodent species. However, a deletion of a single base pair in codon 139 has lead to a frame shift and a premature stop in the reading frame. This has most likely inactivated the gene and transformed it into a pseudogene. However, the deduced (corrected) dog Cma2 protein has a substrate-binding region (Perona and Craik 1995) with greatest similarity to that of the β-chymases mMCP-4 and rMCP-1 (Fig. 3), which indicates that it had a β-chymase like function before it was transformed into a pseudogene.
Alignment of substrate-binding regions in α- and β-chymases. The aa sequences in the substrate-binding regions of all known α- and β-chymases in human, dog, mouse and rat are aligned to the deduced aa sequence of d Cma2. Horizontal dark grey bars on top mark previously reported substrate-binding regions. The catalytic S195 is shaded in orange. Numbers correspond to chymotrypsin numbering. Position 210, which is distinctive for the considered α- and β-chymases, and the common residues in the substrate-binding regions of mMCP-4, rMCP-1 and d Cma2 (P) are shaded in light grey. (P): predicted
Detailed genetic structure and possible duplication units in the rat chymase locus. Genes with previously reported protein expression are assigned with bold; novel sequences with plain text. Highlighted names signify novel sequences with predicted protein expression. Dashed boxes represent probable pseudogenes. Sequences shown in the same color are >85% identical. Black arrows show gene orientation. Distances between genes are drawn to scale. Numbers denote distances in kilobase. Tables below the maps show which exons were identified at the respective location on the map. + denotes the presence of a full-length exon, (+) the presence of part of an exon. Colored arrows below the tables indicate possible units in duplication events
In mouse and rat, multiple β-chymases were described. In the mouse, the genes for the four known β-chymases mMCP-1, mMCP-2, mMCP-4, and mMCP-9 are all situated in the Mcpt region (Fig. 1b) (Huang et al. 1991; Hunt et al. 1997; Newlands et al. 1987; Serafin et al. 1990; Trong et al. 1989). Based on nucleotide and protein sequence similarity analyses (data not shown), these closely related sequences are most likely the product of multiple gene duplication events. Two of the genes, Mcpt1 and Mcpt2, lie in opposite orientation to all other sequences in the locus, indicating that a gene inversion has taken place. The mouse chymase locus also contains an additional gene, Mcpt8. Mcpt8 is a member of a more cathepsin G-related/granzyme-related gene family, the M/R8-family, which so far was identified in rodents only (Lützelschwab et al. 1997, 1998). To date, its evolutionary origin remains elusive.
The Gzm region in the mouse chymase locus has also undergone several duplication events, leading to the presence of multiple functional granzyme genes, i.e., Gzme, Gzmd, Gzmg, Gzmn, Gzmf, Gzmc, and the pseudogene Gzml (reviewed in Grossman et al. 2003). Thus, the murine chymase locus has most likely gained in size mainly by duplication of ancestral β-chymase and granzyme genes.
Similarly, the rat chymase locus also appears to have undergone gene duplication and inversion events, which have lead to a massive increase in its size. As for the mouse, several β-chymases were described, i.e., rMCP-1, -2, -3, -4, and rat VCh (Benfey et al. 1987; Guo et al. 2001; Ide et al. 1995; Kido et al. 1986; Le Trong et al. 1987; Lützelschwab et al. 1997). All of these β-chymase genes, except Mcpt1, lie in opposite orientation to the genes that have orthologs in human and dog, i.e., Mcpt5/Cma1, Ctsg, and Gzmb. Moreover, the M/R8-family also has undergone an expansion in the rat. Three expressed family members were identified previously, i.e., Mcpt8, -9, and -10 (Lützelschwab et al. 1997). The Gzm region is also greatly expanded in the rat chymase locus. However, in addition to Gzmb, only two functional genes, Nkpt7 and Gzmc, are reported from this region.
Although the rat chymase locus is more than three times larger than its mouse counterpart, it harbors approximately the same number of reported functional genes (13 in the rat vs 14 in the mouse). To understand how the rat chymase locus has gained its size and to possibly identify novel genes, we have performed a detailed analysis of this locus.
Detailed genetic structure and organization of the rat chymase locus
The rat genome was screened by BLASTN searches with all previously annotated and predicted gene sequences within the rat chymase locus (as extracted from GenBank by February 2005) (Fig. 4). Novel sequences with highest similarity to the chymase genes were found within the Mcpt region only, whereas those with higher similarity to Nkpt7, Gzmb, or Gzmc were confined to the Gzm region. Phylogenetic analysis also places the known and novel sequences into well-defined groups within the borders of the Mcpt- and Gzm region, respectively (Fig. 2). Thus, no duplication events involving genes from both regions seem to have occurred. Instead, these regions seem to have expanded independently. Moreover, sequences with highest similarity to Mcpt3 or -4 (Fig. 4, green) were found exclusively in a 130-kb region near Cma1/Mcpt5. Similarly, in the Gzm region, sequences with highest similarity to Nkpt7 (brown) were found only within the 80-kb region neighboring Ctsg.
Most of the recovered sequences are not functional genes because they lack at least one exon. The least complete, with only one or two remaining exons (out of five exons for the complete genes), were novel Cma1/Mcpt5 and Nkpt7-related sequences. In the entire Gzm region, only three of the novel Gzmc-related sequences contained all five exons (Gzmc-rs1, -rs2, and -rs3). However, all three novel sequences contain two to three indels and/or point mutations and thereby encode prematurely terminating proteins. None of the predicted proteins contains all three aa of the catalytic triad [His57 (H57), Asp102, and Ser195 (S195)]. Thus, the main part of the Gzm region consists of apparently nonproductive duplications. The three novel granzyme-related sequences share approx. 95% nucleotide sequence identity with Gzmc (Table 1a), suggesting a quite recent duplication. Several blocks of Gzmc- and Gzmb-related sequences are found in the Gzm region, indicating that duplications probably occurred in units enclosing both a Gzmc-related and a Gzmb-related ancestor (Fig. 4).
In the rat Mcpt region, there are numerous sequences with similarity to Mcpt8 or to Mcpt2 (Fig. 2). For both families, we report four novel sequences containing all five exons (Mcpt2-rs1, -rs2a, -rs2b, -rs2c and Mcpt8-rs1, -rs2, -rs3, -rs4). Phylogenetic analysis (Fig. 2 and Table 1b) suggests that the rat sequences in the M1/R2-family have evolved in two rounds of duplication. The first duplications gave rise to the ancestors of VCh, Mcpt2, and the novel Mcpt2-rs1, which to date share between 86.9 and 89.1% nucleotide sequence identity. Around the same time, the ancestor of Mcpt2-rs2a, -rs2b, and -rs2c also seems to have appeared as a separate gene. However, the descendants in this subgroup are ∼97% identical, and the Mcpt2-rs2-subfamily therefore probably did not expand until quite recently.
In addition, the Mcpt2-rs2 subgroup also includes a set of four novel sequences, which all lack exon 1 and part of intron 1, here called s2a (for short Mcpt2-related), s2b, s2c, and s2d (Fig. 4). These truncated sequences are >94% identical to each other and to the other members of the Mcpt2-rs2 subgroup (Table 1b) and therefore probably have been part of the more recent expansion of the Mcpt2-rs2-subfamily.
The members of the Mcpt2-rs2 subgroup and the truncated s2a, s2b, s2c, and s2d share a similar degree of identity as the Mcpt8-related sequences (Table 1), and they are situated in the chymase locus interleaved with them (Fig. 4). This suggests that this part of the locus has evolved by duplication of units including a member of each family.
To test this hypothesis, we calculated the evolutionary distance of each family relative to the divergence of mouse and rat (Materials and methods), which took place approximately 18 Myr ago. Indeed, the R8-family, R2a/b/c-subfamily, and s2a/b/c/d-subfamily seem to have diverged approximately at the same time, 4 to 5 Myr ago, when considering percent nucleotide substitutions in introns. It is interesting to note that a significantly higher percentage of substitutions is however found in the R8-family exons, indicating that these genes are under positive evolutionary pressure. The same is observed for the sequences of the presumed functional members of the R2-family (rVch, rMcpt2, rMcpt2-rs2a, and rMcpt2-rs2c; data not shown).
Evolution of the R2/R8 duplication region in the rat
The high degree of nucleotide sequence identity in exon regions between members of the R8- and Mcpt2-rs2-family, respectively, has hampered the analysis of their evolutionary relations. To overcome this problem, we also including introns and intergenic regions within the entire ∼250-kb region in the analysis (data not shown, but see Fig. 4, R2/R8 duplication region). The Mcpt1 gene does not seem to have taken part in the more recent evolution of this region and is therefore not included in this analysis.
The entire locus is mainly composed of sequences with homology to Mcpt2 (Mcpt2, Mcpt2-rs2a, -rs2b, -rs2c, and the truncated sequences s2a, s2b, s2c, and s2d) or Mcpt8 (Mcpt8, -9, -10, Mcpt8-rs1, -rs2, -rs3, and -rs4; Fig. 4). These sequences are interspersed with different parts of the rat-specific repeat L1_RN (>92% nucleotide sequence identity). Although the L1_RN repeat itself was subject to duplications, inversions, and new insertions in this region, it is not clear whether this was a driving force in the duplication process. Numerous other repeats, including L1 elements, are also found in the human and dog chymase locus (data not shown). However, the presence of these repeats has not resulted in a comparable expansion of these chymase loci. Throughout the region, several sequences with high similarity (>85% nucleotide sequence identity) to exon 5 of Cma1/Mcpt5 were also identified (Fig. 4). Even noncoding parts in the R2/R8 duplication region are highly homologous to each other (>80% identity). Only a part in the 3′-flanking region of s2d consists of sequences not found elsewhere in the locus.
The sequence similarity of all Mcpt2-like genes stretches over an ∼8-kb region 5′ of the ATG (5′-flanking region), and all of these genes, except Mcpt2, also share a similar ∼11-kb 3′-flanking region. The Mcpt8-like genes have only a short 5′-flanking region (<1 kb) in common, but all except Mcpt8-rs2 and Mcpt9 share a somewhat longer 3′-flanking region (∼ 3 kb). The s2-like sequences, like the complete Mcpt2-like genes, have a rather long (∼ 8.5 kb) 3′-flanking region in common. The flanking regions of both gene families are modified by insertions, deletions, and replacements involving the rat-specific repeat L1_RN (data not shown).
These extended sequence similarities and the almost complete absence of nonalignable sequence within the region, confirm the hypothesis that members of the R2- and R8-family evolved via duplications of a common ancestral unit, probably of the form Mcpt2→Mcpt8→s2.
Analysis of the rMCP-2- and rMCP-8-family at the protein level
Analysis of the coding regions of the novel members of the rMCP-2-family reveals that Mcpt2-rs1 probably is a pseudogene. The open reading frame terminates prematurely after aa position 68 (Fig. 5a), i.e., shortly after the first of the three aa forming the catalytic triad, H57. The open reading frame of Mcpt2-rs2b also terminates prematurely at aa position 190 and thereby lacks the third of the catalytic residues, S195. However, the other members of this subfamily, Mcpt2-rs2a and -rs2c, are most likely functional genes. The deduced protein sequences contain all three aa of the catalytic triad (H57, D102, and S195) and all six conserved cysteine residues common to this family. The assumption that Mcpt2-rs2a and -2c are functional genes whereas Mcpt2-rs2b is a pseudogene is further supported by the finding that the former seem to be exposed to conservative evolutionary pressure (dN/dS<1), whereas the latter is not (Table 2a). Note that in the substrate-binding region of Mcpt2-rs2a and -rs2c (Perona and Craik 1995), two substitutions replace positively charged or neutral aa with negatively charged aa (E192 and E220), which might confer distinct substrate specificity.
Alignments of known and predicted aa sequences. Numbers on top of the alignments correspond to chymotrypsin numbering. Horizontal grey bars on top mark substrate-binding regions. The catalytic residues H57, D102 and S195 are shaded in orange. Cysteine residues are shaded in blue, and deleterious mutations in red. Dashed lines correspond to exon borders. Solid lines mark the start of mature proteins. (P): predicted, corr.: corrected. a Alignment of known and predicted aa sequences belonging to the rMCP-1/2 family and their homologues in mouse, mMCP-4 and mMCP-1. Arrows mark residues 192 and 220 in the substrate-binding region and H210, which is common to β-chymases. b Alignment of known and predicted aa sequences belonging to the R8-family and their homologue in mouse, mMCP-8. The arrow marks Q210, which is common to α-chymases and the M/R8-family
Regarding the M/R8-family, the deduced aa sequences for Mcpt8-rs1 and Mcpt8-rs4 (Fig. 5b) indicate functional proteins. Both sequences encode the three aa of the catalytic triad and all six conserved cysteines. Although Mcpt8-rs4 is evolving at a high evolutionary rate (Table 2b), the substrate-binding region for the deduced Mcpt8-rs4 protein is almost identical to rMCP-10 and rMCP-8, possibly indicating redundant substrate specificities. In contrast to both of these potentially functional Mcpt8-related sequences, Mcpt8-rs2 is most likely a pseudogene, as the open reading frame terminates after aa position 62. The predicted Mcpt8-rs3 protein seems to be a full-length protein, but contains a mutation of H57 to Q. Thus, this hypothetical protein most likely has no serine protease activity.
Discussion
The recently completed genome sequences of rat and dog have provided us with the possibility to compare the entire chymase locus in four different placental mammals: mouse rat, dog, and human.
The main conclusion from this analysis is that the mast cell chymase locus has diversified greatly, not only after the divergence of the last common ancestor of primates, carnivores, and rodents, but also during recent rodent evolution. Species-specific serial gene duplications have generated large differences in actual number of both functional and nonfunctional genes.
For example, multiple mast cell chymase genes in the rat and T cell granzyme genes in the mouse have evolved during the past 18 Myr, whereas no such development is observed in dog or human. The functional consequences of these new genes for the rodent immune system remain to be elucidated. However, the big numeral difference in chymase locus genes between rodents and primates, and the probable involvement of these genes in rodent-specific biology, imply that care should be taken when drawing parallels between rodent and human mast cell biology.
These results also exemplify the high plasticity of mammalian genomes and how big differences can appear in relatively short evolutionary time frames. For example, the chymase loci of human and dog are located on chromosomes with largely conserved synteny and display relatively few differences with almost the same numbers of active genes. In contrast, the region of conserved synteny between dog or human and rodents is limited to the chymase locus and its closer surroundings, showing that large-scale chromosomal rearrangements have occurred during speciation (Fig. 1a). The rodent chymase locus was also very actively evolving with large numbers of new genes, including gene subfamilies not found in primates, such as the β-chymases and the M/R8-family.
One of the major questions concerning the chymase locus was the origin of these new enzymes. Have they expanded in rodents from ancient ancestor genes present early in mammalian evolution, or did they originate in the rodent lineage after the separation from the other major lineages of placental mammals?
An important clue to this problem was the identification of a second chymase gene in dog (Caughey 2005), here termed Cma2. Phylogenetic analyses of exon nucleotide sequences group Cma2 with the β-chymases, making it likely that the common ancestor for α- and β-chymases was duplicated before the separation of carnivores from primates and rodents approximately 95 Myr ago (Springer et al. 2003). The lack of β-chymases in primates thereby appears to be due to the deletion of one of the copies in the primate lineage at a later time point (Caughey et al. 2000).
The dog Cma2 gene seems, however, to contain a deletion resulting in a premature stop codon. Therefore, this gene probably does not give rise to a functional protein. As the inactivation of the gene is due to a single deletion, it may have occurred very recently, maybe even after the domestication of the dog from wild gray wolves (C. lupus) at least 15,000 years ago (Savolainen et al. 2002; Vila et al. 1997). Hence, a functional Cma2 gene may be present in the gray wolf or even in other dog breeds, questions that are currently under investigation.
No member of the M/R8-family has so far been identified in a non-rodent species, which makes the origin of this family even more problematic. In addition, the number of family members and their tissue distribution varies remarkably. Only a single Mcpt8 gene is found in the mouse and there are seven members in the rat (Fig. 4). The mouse Mcpt8 gene is expressed specifically in basophils (Lützelschwab et al. 1998; Poorafshar et al. 2000), whereas the three previously characterized family members in the rat are all expressed in MMC (Lützelschwab et al. 1997). Two of the four novel R8-family sequences, Mcpt8-rs1 and -rs4, may also be functional, but no information is yet available concerning their expression pattern. As to protein function, neither the cleavage specificity nor any other function is yet known for any member of the M/R8-family. We show, however, that the R8-family genes are subject to a selective pressure in favor of their diversification. Such differences in the coding region between the family members indicate a functional significance for these new members.
Regarding proteolytic activity, it was suggested (Karlson 2003) that the substrate-binding cleft of rMCP-8, -9, and -10, similar to the situation in human α-tryptase (Huang et al. 1999; Marquardt et al. 2002), may be sterically blocked by the large arginine in position 216. It is interesting to note that one of the novel genes, Mcpt8-rs1, encodes the much smaller aa, cysteine in position 216, and thus might be proteolytically more active than the other family members. Mcpt8-rs3, on the other hand, encodes a mutation in one of the aa of the catalytic triad, H57Q. Mcpt8-rs3 is therefore probably not a functional protease. However, it may exert a different function, as was shown, e.g., for azurocidin, a member of the serine protease family without proteolytic activity, but with antibacterial and antifungal effect (Campanelli et al. 1990; Morgan et al. 1991).
The remaining novel sequences characterized here include both complete and truncated Mcpt2-like genes. These genes are all situated in the rat chymase locus interspersed with the R8-family genes (Fig. 4). We show that the two gene families have most likely coevolved by duplications of a unit containing a member of each family. Four of the novel Mcpt2-like sequences are complete in the sense that they contain all five exons. Two of the genes, Mcpt2-rs2a and -rs2c, are most likely functional. In these genes, two substitutions replace positively charged or neutral aa within the substrate-binding region with negatively charged ones (E192 and E220), which might confer a substrate-specificity distinctive from that of other Mcpt2-family members to the encoded proteins. It is also interesting to note that the predicted Mcpt2-rs2a and -2c display a low net charge of −0.6 and −0.9 at pH 7, respectively (Fig. 5). This is in contrast to most mast cell proteases (except tryptases), which at pH 7, have a net positive charge. The chymases expressed in mouse and rat connective tissue mast cells, rMCP-1 and mMCP-4, have an especially high positive net charge of 17.2 and 16.1, respectively, which facilitates their storage in mast cell granules in tight complex with the negatively charged heparin (Humphries et al. 1999; Pejler and Maccarana 1994). Due to their low net charge, Mcpt2-rs2a and -2c can probably not be efficiently involved in this type of charge-dependent storage. This indicates that these novel proteins may have an expression pattern differing from the mast cell proteases. It is interesting to note that rat vascular chymase, the only protein encoded in the chymase locus known to be expressed in a nonhematopoietic cell type, vascular smooth muscle cells, also has a low net charge of 1.4 at pH 7. We are currently investigating the expression pattern and possible function of these novel genes. However, no such data is available yet.
It is interesting to note that the second important group of mast cell protease genes, the tryptase genes, also resides in a region that has evolved by gene duplication and pseudogenization. In contrast to the chymase locus, the major diversification of the tryptase locus, however, seems to have occurred before the separation of rodents from primates, resulting in similar gene numbers in mouse, rat, and human (14, 16, and 14, respectively) (Wong et al. 2004). During approximately 95 Myr after the separation of rodents from primates (Springer et al. 2003), five genes in the tryptase locus that are functional in mouse and rat have become pseudogenes in human. Moreover, this locus has diversified extensively to encode transmembrane and soluble tryptic enzymes that are also expressed in various tissues and developmental stages (Wong et al. 2004). Possibly, the chymase locus in rodents, with many of the enzymes still having very similar structure, may be evolving toward a comparable degree of functional and structural diversification.
A comparison of the chymase locus in mouse and rat also illustrates how different the result of multiple rounds of gene duplications in relatively closely related species can be. In both species, duplications in the Mcpt and Gzm regions apparently occurred independently and in several rounds. In the Gzm region, these duplications resulted in seven functional granzyme genes in the mouse and only three in the rat. It is interesting to note that although this region contains less than half the number of functional genes in the rat, it is three times larger in the rat than in the mouse. This remarkably different outcome is explained by the fact that the expansion of the rat locus primarily involved duplications of partial genes, an aspect clearly demonstrating the random nature of gene duplication events.
In the Mcpt region, a duplication of a unit containing both a Mcpt2- and a Mcpt8-related sequence occurred in the rat, but not in the mouse, and created the basis for the major rat-specific expansion of these gene families. This relatively recent expansion massively increased the number of Mcpt2-related genes in the rat from three to six, and the number of Mcpt8-related genes from one to seven. Several of these novel genes also appear to be functional, and both gene families are apparently under selective pressure favoring their diversification.
Our findings illustrate the role of gene duplications as a very important factor in the evolution of genomes. Gene duplications may be of especially high importance in the process of speciation where duplications of genes with functions in immunity, smell, and taste may provide selective advantages in a novel environment. This concept is further supported by data from the natural killer cell gene complex (Baumgartner et al. 1996; Hao and Nei 2004; Nylenna et al. 2005), the vomeronasal pheromone receptor gene repertoire (Grus et al. 2005), and the bitter taste receptor genes (Shi et al. 2003) where numerous species-specific duplications and deletions were observed. The high evolutionary activity in the rodent chymase locus may reflect the encounter of new parasites and other pathogens when rodents established their ecological niches. In other words, species-specific duplication events and the function of the surviving genes may give us a clue about important bioecological changes in the course of speciation.
Abbreviations
- aa:
-
amino acid
- indels:
-
insertions and/or deletions
- Myr:
-
million years
- MC:
-
mast cell
- MMC:
-
mucosal mast cells
- CTMC:
-
connective tissue mast cells
- mMCP:
-
mouse mast cell protease (protein)
- rMCP:
-
rat mast cell protease (protein)
- Mcpt:
-
mast cell protease (protein)
- Mcpt :
-
mast cell protease (gene)
- M/R8-family:
-
mouse/rat Mcpt8-family
- R8-family:
-
rat Mcpt8-family
- M1/R2-family:
-
mouse Mcpt1-/rat Mcpt2-family
- R2-family:
-
rat Mcpt2-family
References
Baumgartner RA, Deramo VA, Beaven MA (1996) Constitutive and inducible mechanisms for synthesis and release of cytokines in immune cell lines. J Immunol 157:4087–4093
Benfey PN, Yin FH, Leder P (1987) Cloning of the mast cell protease, RMCP II. Evidence for cell-specific expression and a multi-gene family. J Biol Chem 262:5377–5384
Breathnach R, Chambon P (1981) Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem 50:349–383
Burset M, Seledtsov IA, Solovyev VV (2000) Analysis of canonical and non-canonical splice sites in mammalian genomes. Nucleic Acids Res 28:4364–4375
Campanelli D, Detmers PA, Nathan CF, Gabay JE (1990) Azurocidin and a homologous serine protease from neutrophils. Differential antimicrobial and proteolytic properties. J Clin Invest 85:904–915
Caughey GH (2005) Origins of functional diversity in mammalian mast cell peptidases. The Batsheva de Rothschild international workshop on mast cell signalling and function in health and disease. Eilat, Israel
Caughey GH, Schaumberg TH, Zerweck EH, Butterfield JH, Hanson RD, Silverman GA, Ley TJ (1993) The human mast cell chymase gene (CMA1): mapping to the cathepsin G/granzyme gene cluster and lineage-restricted expression. Genomics 15:614–620
Caughey GH, Raymond WW, Wolters PJ (2000) Angiotensin II generation by mast cell alpha- and beta-chymases. Biochim Biophys Acta 1480:245–257
Chandrasekharan UM, Sanker S, Glynias MJ, Karnik SS, Husain A (1996) Angiotensin II-forming activity in a reconstructed ancestral chymase. Science 271:502–505
Felsenstein J (1989) PHYLIP—phylogeny interference package (version 3.2). Cladistics 5:164–166
Gibbs RA, Weinstock GM, Metzker ML, Muzny DM, Sodergren EJ, Scherer S, Scott G, Steffen D, Worley KC, Burch PE, Okwuonu G, Hines S, Lewis L, DeRamo C, Delgado O, Dugan-Rocha S, Miner G, Morgan M,Hawes A, Gill R, Celera, Holt RA, Adams MD, Amanatides PG, Baden-Tillson H, Barnstead M, Chin S, Evans CA, Ferriera S, Fosler C, Glodek A, Gu Z, Jennings D, Kraft CL, Nguyen T, Pfannkoch CM, Sitter C, Sutton GG, Venter JC, Woodage T, Smith D, Lee HM, Gustafson E, Cahill P, Kana A, Doucette-Stamm L, Weinstock K, Fechtel K, Weiss RB, Dunn DM, Green ED, Blakesley RW, Bouffard GG, De Jong PJ, Osoegawa K, Zhu B, Marra M, Schein J, Bosdet I, Fjell C, Jones S, Krzywinski M, Mathewson C, Siddiqui A, Wye N, McPherson J, Zhao S, Fraser CM, Shetty J, Shatsman S, Geer K, Chen Y, Abramzon S, Nierman WC, Havlak PH, Chen R, Durbin KJ, Egan A, Ren Y, Song XZ, Li B, Liu Y, Qin X, Cawley S, Cooney AJ, D’Souza LM, Martin K, Wu JQ, Gonzalez-Garay ML, Jackson AR, Kalafus KJ, McLeod MP, Milosavljevic A, Virk D, Volkov A, Wheeler DA, Zhang Z, Bailey JA, Eichler EE, Tuzun E et al (2004) Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature 428:493–521
Grossman WJ, Revell PA, Lu ZH, Johnson H, Bredemeyer AJ, Ley TJ (2003) The orphan granzymes of humans and mice. Curr Opin Immunol 15:544–552
Grus WE, Shi P, Zhang YP, Zhang J (2005) Dramatic variation of the vomeronasal pheromone receptor gene repertoire among five orders of placental and marsupial mammals. Proc Natl Acad Sci USA 102:5767–5772
Guo C, Ju H, Leung D, Massaeli H, Shi M, Rabinovitch M (2001) A novel vascular smooth muscle chymase is upregulated in hypertensive rats. J Clin Invest 107:703–715
Gurish MF, Nadeau JH, Johnson KR, McNeil HP, Grattan KM, Austen KF, Stevens RL (1993) A closely linked complex of mouse mast cell-specific chymase genes on chromosome 14. J Biol Chem 268:11372–11379
Hao L, Nei M (2004) Genomic organization and evolutionary analysis of Ly49 genes encoding the rodent natural killer cell receptors: rapid evolution by repeated gene duplication. Immunogenetics 56:343–354
Hoit BD, Shao Y, Kinoshita A, Gabel M, Husain A, Walsh RA (1995) Effects of angiotensin II generated by an angiotensin converting enzyme-independent pathway on left ventricular performance in the conscious baboon. J Clin Invest 95:1519–1527
Huang R, Hellman L (1994) Genes for mast-cell serine protease and their molecular evolution. Immunogenetics 40:397–414
Huang RY, Blom T, Hellman L (1991) Cloning and structural analysis of MMCP-1, MMCP-4 and MMCP-5, three mouse mast cell-specific serine proteases. Eur J Immunol 21:1611–1621
Huang C, Li L, Krilis SA, Chanasyk K, Tang Y, Li Z, Hunt JE, Stevens RL (1999) Human tryptases alpha and beta/II are functionally distinct due, in part, to a single amino acid difference in one of the surface loops that forms the substrate-binding cleft. J Biol Chem 274:19670–19676
Humphries DE, Wong GW, Friend DS, Gurish MF, Qiu WT, Huang C, Sharpe AH, Stevens RL (1999) Heparin is essential for the storage of specific granule proteases in mast cells. Nature 400:769–772
Hunt JE, Friend DS, Gurish MF, Feyfant E, Sali A, Huang C, Ghildyal N, Stechschulte S, Austen KF, Stevens RL (1997) Mouse mast cell protease 9, a novel member of the chromosome 14 family of serine proteases that is selectively expressed in uterine mast cells. J Biol Chem 272:29158–29166
Ide H, Itoh H, Tomita M, Murakumo Y, Kobayashi T, Maruyama H, Osada Y, Nawa Y (1995) Cloning of the cDNA encoding a novel rat mast-cell proteinase, rMCP-3, and its expression in comparison with other rat mast-cell proteinases. Biochem J 311(Pt 2):675–680
Jurka J, Klonowski P, Dagman V, Pelton P (1996) CENSOR—a program for identification and elimination of repetitive elements from DNA sequences. Comput Chem 20:119–121
Karlson U (2003) Cutting edge-cleavage specificity and biochemical characterization of mast cell serine proteases. PhD thesis, Department of Cell and Molecular Biology, Uppsala University. ISBN 91-554-5699-5
Karlson U, Pejler G, Tomasini-Johansson B, Hellman L (2003) Extended substrate specificity of rat mast cell protease 5, a rodent alpha-chymase with elastase-like primary specificity. J Biol Chem 278:39625–39631
Kido H, Izumi K, Otsuka H, Fukusen N, Kato Y, Katunuma NA (1986) Chymotrypsin-type serine protease in rat basophilic leukemia cells: evidence for its immunologic identity with atypical mast cell protease. J Immunol 136:1061–1065
Le Trong H, Parmelee DC, Walsh KA, Neurath H, Woodbury RG (1987) Amino acid sequence of rat mast cell protease I (chymase). Biochemistry 26:6988–6994
Lützelschwab C, Pejler G, Aveskogh M, Hellman L (1997) Secretory granule proteases in rat mast cells. Cloning of 10 different serine proteases and a carboxypeptidase A from various rat mast cell populations. J Exp Med 185:13–29
Lützelschwab C, Huang MR, Kullberg MC, Aveskogh M, Hellman L (1998) Characterization of mouse mast cell protease-8, the first member of a novel subfamily of mouse mast cell serine proteases, distinct from both the classical chymases and tryptases. Eur J Immunol 28:1022–1033
Marquardt U, Zettl F, Huber R, Bode W, Sommerhoff C (2002) The crystal structure of human alpha1-tryptase reveals a blocked substrate-binding region. J Mol Biol 321:491–502
Morgan JG, Sukiennicki T, Pereira HA, Spitznagel JK, Guerra ME, Larrick JW (1991) Cloning of the cDNA for the serine protease homolog CAP37/azurocidin, a microbicidal and chemotactic protein from human granulocytes. J Immunol 147:3210–3214
Neurath H (1986) The versatility of proteolytic enzymes. J Cell Biochem 32:35–49
Newlands GF, Gibson S, Knox DP, Grencis R, Wakelin D, Miller HR (1987) Characterization and mast cell origin of a chymotrypsin-like proteinase isolated from intestines of mice infected with Trichinella spiralis. Immunology 62:629–634
Nylenna O, Naper C, Vaage JT, Woon PY, Gauguier D, Dissen E, Ryan JC, Fossum S (2005) The genes and gene organization of the Ly49 region of the rat natural killer cell gene complex. Eur J Immunol 35:261–272
Pejler G, Maccarana M (1994) Interaction of heparin with rat mast cell protease 1. J Biol Chem 269:14451–14456
Perona JJ, Craik CS (1995) Structural basis of substrate specificity in the serine proteases. Protein Sci 4:337–360
Poorafshar M, Helmby H, Troye-Blomberg M, Hellman L (2000) MMCP-8, the first lineage-specific differentiation marker for mouse basophils. Elevated numbers of potent IL-4-producing and MMCP-8-positive cells in spleens of malaria-infected mice. Eur J Immunol 30:2660–2668
Puente X (2005) A genomic view of the complexity of mammalian proteolytic systems. Biochemical Society Focused Meetings
Puente XS, Lopez-Otin C (2004) A genomic analysis of rat proteases and protease inhibitors. Genome Res 14:609–622
Savolainen P, Zhang YP, Luo J, Lundeberg J, Leitner T (2002) Genetic evidence for an East Asian origin of domestic dogs. Science 298:1610–1613
Serafin WE, Reynolds DS, Rogelj S, Lane WS, Conder GA, Johnson SS, Austen KF, Stevens RL (1990) Identification and molecular cloning of a novel mouse mucosal mast cell serine protease. J Biol Chem 265:423–429
Shi P, Zhang J, Yang H, Zhang YP (2003) Adaptive diversification of bitter taste receptor genes in Mammalian evolution. Mol Biol Evol 20:805–814
Solivan S, Selwood T, Wang ZM, Schechter NM (2002) Evidence for diversity of substrate specificity among members of the chymase family of serine proteases. FEBS Lett 512:133–138
Springer MS, Murphy WJ, Eizirik E, O’Brien SJ (2003) Placental mammal diversification and the Cretaceous-Tertiary boundary. Proc Natl Acad Sci USA 100:1056–1061
Takai S, Shiota Nm, Kobayashi S, Matsumura E, Miyazaki M (1997) Induction of chymase that forms angiotensin II in the monkey atherosclerotic aorta. FEBS Lett 412:86–90
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Trong HL, Newlands GF, Miller HR, Charbonneau H, Neurath H, Woodbury RG (1989) Amino acid sequence of a mouse mucosal mast cell protease. Biochemistry 28:391–395
Vernersson M, Aveskogh M, Hellman L (2004) Cloning of IgE from the echidna (Tachyglossus aculeatus) and a comparative analysis of epsilon chains from all three extant mammalian lineages. Dev Comp Immunol 28:61–75
Vila C, Savolainen P, Maldonado JE, Amorim IR, Rice JE, Honeycutt RL, Crandall KA, Lundeberg J, Wayne RK (1997) Multiple and ancient origins of the domestic dog. Science 276:1687–1689
Wintroub BU, Schechter NB, Lazarus GS, Kaempfer CE, Schwartz LB (1984) Angiotensin I conversion by human and rat chymotryptic proteinases. J Invest Dermatol 83:336–339
Wong GW, Yasuda S, Morokawa N, Li L, Stevens RL (2004) Mouse chromosome 17A3.3 contains 13 genes that encode functional tryptic-like serine proteases with distinct tissue and cell expression patterns. J Biol Chem 279:2438–2452
Yang Z (1997) PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci 13:555–556
Yang Z, Nielsen R, Goldman N, Pedersen AM (2000) Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155:431–449
Acknowledgements
We thank Dr. Robert Fredriksson for excellent advice on bioinformatic tools. We thank professor Lars Pilström and Jenny Reimer for critical reading of the manuscript. This work was funded by the Swedish Research Council.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Gallwitz, M., Hellman, L. Rapid lineage-specific diversification of the mast cell chymase locus during mammalian evolution. Immunogenetics 58, 641–654 (2006). https://doi.org/10.1007/s00251-006-0123-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-006-0123-4