Abstract
We have compiled the nucleotide sequences and their amino acid translations from a total of 89 Killer Immunoglobulin-like Receptor (KIR) alleles, derived from 17 different KIR genes. The alignments use the KIR3DL2*001 allele as a reference sequence. Each of the KIR sequences included in these alignments has been checked and where discrepancies have arisen between reported sequences, the original authors have been contacted where possible, and necessary amendments to published sequences have been incorporated into this alignment. Future sequencing may identify errors in this list and we would welcome any evidence that helps to maintain the accuracy of this compilation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
The sequences included in this compilation are taken from the publications listed in the Killer Immunoglobulin-like Receptor (KIR) Nomenclature Committee Report (Marsh et al. 2003). The KIR Nomenclature Committee has officially assigned the names to all the sequences included in these sequence alignments with the exception of two. Details of the officially named sequences including accession numbers and publication details can be found in the accompanying Nomenclature Report (Marsh et al. 2003). The two sequences listed under the names KIR2DL5(KIR2DLXa) (AF271607) and KIR2DL5(KIR2DLXb) (AF271608) have not yet been assigned official names as it is unclear whether they represent alleles of the KIR2DL5A or KIR2DL5B genes.
Each of the KIR sequences included in these alignments has been checked and where discrepancies have arisen between reported sequences, the original authors have been contacted where possible, and necessary amendments to published sequences have been incorporated into this alignment. Future sequencing may identify errors in this list and we would welcome any evidence that helps to maintain the accuracy of this compilation.
In the nucleotide (Fig. 1) and amino acid (Fig. 2) sequence alignments, a total of 89 sequences comprising 14 KIR genes and three pseudogenes have been aligned to the KIR3DL2*001 sequence. The KIR3DL2*001 sequence was used as a reference sequence as it provided a sufficiently long reference sequence which also possessed a high level of nucleotide identity and structural homology to the majority of the other KIR genes. The KIR sequences were retrieved from the EMBL Nucleotide Sequence Database or GenBank by means of the accession numbers given in the KIR Nomenclature Report (Marsh et al. 2003). Criteria used for the grouping of these sequences into genes is based on the number of immunoglobulin domains, the length of the cytoplasmic tail and sequence homology as proposed in previous publications (Long et al. 1996; Steffens et al. 1998; Vilches and Parham 2002). The sequence comparison was done by using a combination of Clustal (Thompson et al. 1994) and manual analysis after which sequence alignments were subjected to a reformatting tool available in house, which allowed us to show identity to a reference sequence and translate the nucleotide sequences into their corresponding protein sequences (Fig. 2).
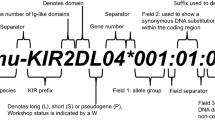
In the alignments, exon intron boundaries and protein domains have been marked with a pipe (|), asterisks (*) indicate positions where sequence is unavailable, periods (.) indicate an insertion or deletion. Identity to the reference sequence KIR3DL2*001 is shown by a hyphen (-). In the amino acid alignment, stop codons are indicated by an X. Minimum gaps in the sequence, indicated by a period (.), have been inserted to maintain the alignment between alleles of differing length, in such a way as to maintain the reading frame. The pseudo-exon 3 sequences for type I KIR2Ds have been included in the nucleotide alignment where available. The nucleotides are numbered starting at 1 for the first nucleotide of the codon for the initiation methionine. The numbering of the codons of the mature protein, after cleavage of the signal sequence, begins at +1, while the signal sequence is numbered backwards from −1. These alignments include KIR genes and alleles for which complete cDNA sequences or full genomic sequences are available, alternative splice variants escape the scope of this publication and partial cDNA sequences are not being included until further information for them is made available.
These sequences are also available from the IPD/KIR Database, http://www.ebi.ac.uk/ipd/kir. The database provides an online repository for the KIR sequences officially named by the KIR Nomenclature Committee. The IPD/KIR Database provides online versions of the sequence alignments and the nomenclature reports. In time it is envisaged that the website will also provide tools for the submission of new and confirmatory KIR sequences to the KIR Nomenclature Committee. The IPD/KIR Database is part of the Immuno Polymorphism Database (IPD), which provides a suite of tools, and databases for the study of polymorphisms in the immune system.
References
Long EO, Colonna M, Lanier LL (1996) Inhibitory MHC class I receptors on NK and T cells: a standard nomenclature. Immunol Today 17:100
Marsh SGE, Parham P, Dupont B, Geraghty DE, Trowsdale J, Middleton D, Vilches C, Carrington M, Witt C, Guethlein LA, Shilling H, Garcia CA, Hsu KC, Wain H (2003) Killer Immunoglobulin-like Receptor (KIR) Nomenclature Report. Immunogenetics DOI 10.1007/s00251-003-0571-z
Steffens U, Vyas Y, Dupont B, Selvakumar A (1998) Nucleotide and amino acid sequence alignment for human killer cell inhibitory receptors (KIR), 1998. Tissue Antigens 51:398–413
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Vilches C, Parham P (2002) KIR: diverse, rapidly evolving receptors of innate and adaptive immunity. Annu Rev Immunol 20:217–251
Acknowledgements
We would like to thank Peter Stoehr and the staff of the European Bioinformatics Institute for their support of the IPD/KIR Database.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Garcia, C.A., Robinson, J., Guethlein, L.A. et al. Human KIR sequences 2003. Immunogenetics 55, 227–239 (2003). https://doi.org/10.1007/s00251-003-0572-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-003-0572-y