Abstract
Electrotransfer is a method by which molecules can be introduced into living cells via plasma membrane electropermeabilization. Here, we show that electropermeabilization affects the lateral mobility of Rae-1, a GPi anchored protein. Our results suggest that 10–20 % of the membrane surface is occupied by defects or pores and that these structures propagate rapidly (\(<\)1 min) over the cell surface. Electrotransfer of plasmid DNA (pDNA) also affects the lateral mobility of Rae-1. Furthermore, we clearly show that, once inserted into the plasma membrane, pDNA is completely immobile and excludes Rae-1; this indicates that the pDNA molecules are tightly packed together to form aggregates occupying at least the outer leaflet of the plasma membrane.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The permeability of a cell membrane can be transiently increased by applying external electric field pulses. This phenomenon, called electropermeabilization or electroporation, leads to the formation of “membrane defects” also defined as “electropores” in the cell membrane (Neumann et al. 1989; Chang et al. 1992; Weaver 1993). Since it is an elegant way to introduce exogenous molecules into the cytoplasm this method is routinely used in basic research (as well as for clinical applications) providing a very efficient way for the delivery of drugs, oligonucleotides and plasmids DNA (pDNA) into cells (Mir et al. 2006; Daud et al. 2008; Low et al. 2009; Escoffre and Rols 2012; Weiland et al. 2013). Electropermeabilization is a multi-step process which occurs in a series of different steps (Teissié et al. 2005):
-
1.
Induction step (ns). The electric field first causes the membrane potential difference to increase. When this reaches a critical value (about 200 mV) then local transient permeant structures appear.
-
2.
Expansion step (\(\upmu\)s). Defects within the membrane continue to expand for as long as the field intensity remains above the critical value.
-
3.
Stabilization step (ms). As soon as the field intensity is reduced below the critical value, a stabilization process takes place relatively quickly (within a few milliseconds) which results in the membrane existing as the “permeabilized state”.
-
4.
Resealing step (s, min). This state is then followed by slow resealing of the defects.
-
5.
Memory effect (h). Although some physical and morphological changes of the membrane persist on a longer time scale, the cell behavior essentially returns to normal.
If the kinetics of electropermeabilization seems to be well established, very little is known about the changes occurring at the cell and membrane molecular levels (Teissié et al. 2005). Nevertheless, although structural changes in the plasma membrane (i.e., formation of “membrane defects” or “electropores”) have never been directly visualized under the microscope, other techniques have been used to observe electropermeabilization. These include measurements of the conductivity of cell suspensions and pellets (Kinosita and Tsong 1979; Abidor et al. 1994; Pavlin et al. 2005, 2007), electro-optical relaxation experiments on lipid vesicles (Kakorin et al. 1996; Griese et al. 2002), charge pulse studies on lipid bilayers (Griese et al. 2002; Benz et al. 1979), measurements of membrane voltage on cells with potentiometric fluorescence dyes (Hibino et al. 1993), and monitoring the influx or efflux of molecules and fluorescent dyes (Gabriel and Teissié 1997, 1999; Pucihar et al. 2008; Rols and Teissié 1998; Mir et al. 1988; Tekle et al. 1994; Prausnitz et al. 1994, 1995). More recently, molecular simulation on pure lipid models revealed the possibility of pore formation during the pulse and pore evolution up to tenths of a nanosecond time scale (Levine and Vernier 2010; Tarek 2005; Tieleman 2004). Finally, numerical computation of the evolution of these pores has resulted in a more sophisticated, but still theoretical, description of the phenomenon (Krassowska and Filev 2007). On pure lipid models, Krassowka et al., predict that the mean size of “small” pores is approximately 1 nm for 97 % of cases whereas the size of “large” pores reach from 20 nm to as big as 400 nm (on 50 \(\upmu\)m vesicles pulsed with a 0.6 kV cm−1 electric field intensity for 1 ms). Although small molecules (i.e., \(<\)4 kDa) mostly cross the permeabilized cell membrane by a post-pulse diffusion process, pDNA first interacts with the electropermeabilized part of the membrane forming localized aggregates (Escoffre et al. 2011; Pucihar et al. 2008). If one takes into account the size of the pDNA (i.e., 3 MDa, 30 nm in diameter) and the net negative charge of pDNA, then dielectric exclusion must also be overcome. It is, thus, supposed that the permeabilization is due to conducting defects called “pores” and that these membrane structures must be large and stable (Parsegian 1969). Nevertheless, the cell membrane is organised in a much more complex manner than a model lipid bilayer, so one expects that the sites where pDNA electrotransfer occurs will be determined by the local electric field and also the local membrane composition and tension (Rosazza et al. 2011, 2012). In order to detect the effect of electropermeabilization on the plasma membrane, in the absence or presence of pDNA, we have monitored the lateral mobility of a Glycosyl Phosphatidylinositol (GPi) anchored protein Rae-1 (retinoic acid early-inducible protein 1) during the resealing step by means of fluorescence recovery after photobleaching (FRAP) experiments. Rae-1 (Major Histocompatibility Class-I homologous protein) is a GPi anchored protein which is located in the outer leaflet of the plasma membrane without any partitioning into lipid domains (Nomura et al. 1996; Zou et al. 1996). Such a GPi anchored protein can be considered as a good candidate to report changes in the plasma membrane lateral state and in its close interface with the outside of the cell. By measuring its mobility before and after application of permeabilizing electric field pulses (in absence of pDNA) we report a drastic and significant increase in the half-time of fluorescence recovery. This is in unison with a decrease of the mobile fraction, both at the anode and cathode facing pole of the cell. This experimental data suggests the formation of electro-induced structures in the plasma membrane (e.g., “pores” or “membrane defects”). We then demonstrate that when pDNA is inserted into the membrane after electropermeabilization, because Rae-1 is then unable to re-enter the membrane location occupied by the pDNA; this is consistent with the hypothesis that pDNA is accumulated in a tightly bound manner into clusters.
Material and methods
Expression of Rae-1 in CHO cell line
The eGFP-Rae-1 CHO cells have been generously made by Dr. B. Couderc (EA3035, Institut Claudius Regaud, France). CHO cells have been transfected by pDNA encoding eGFP-Rae-1 fusion protein (generous gift from Dr. A. Aucher and Dr. D. Hudrisier, IPBS-CNRS, UMR5089, France). The transfected cells are cultured under selective pressure with G418 (1 \(\upmu\)g/\(\upmu\)L) (InvivoGen, San Diego, CA, USA). The eGFP-Rae-1 expressing cells were sorted out by flow cytometry (FAScan; Beckman. Instruments, Inc., Fullerton, CA, USA). Cells were then grown as previously described (Phez et al. 2005).
pDNA labeling for electropermeabilization
A 4.7-kbp plasmid (pEGFP-C1, Clonetech, Palo Alto, CA, USA) carrying the green fluorescent protein gene controlled by the CMV promoter was prepared from Escherichia coli transformed bacteria by using the Maxiprep DNA purification system (Qiagen, Chatsworth, CA, USA). They were covalently labeled with the Cy-3 fluorophore using a Label-IT nucleic acid labeling kit (Mirus, Madison, WI, USA) according to the manufacturer's protocol. The fluorescent labeling did not affect the function of the expression cassette (not shown).
Electropermeabilization
Electropulsation was carried out with a CNRS cell electropulsator (Jouan, St Herblain, France), which delivers square-wave electric pulses. An oscilloscope (Enertec, St. Etienne, France) was used to monitor the pulse shape. The electropulsation chamber was built using two stainless-steel parallel rods (diameter 0.5 mm, length 10 mm, inter-electrode distance 5 mm) placed on a Lab-tek chamber (Mazères et al. 2009). The electrodes were connected to the voltage generator. A uniform electric field was generated. The chamber was placed on the stage of the confocal microscope (Zeiss, LSM 510, Germany). Electropermeabilization of cells was performed by application of millisecond electric pulses, conditions required to efficiently transfer macromolecules such as pDNA into cells (Rols and Teissié 1998). Ten pulses of 5 ms duration and 0.6 kV/cm amplitude were applied at a frequency of 1 Hz at room temperature. For FRAP experiments, the eGFP-Rae-1 CHO cells were seeded on a microscope glass coverslip chamber (Labtek II system, Nunc™, Denmark) at 0.5 × 106 cells per well 24 h before electropulsation. Cells were electropulsed in 200 \(\upmu\)L of pulsation buffer (10 mM K\(_2\)HPO\(_4\)/KH\(_2\)PO\(_4\), 1 mM MgCl\(_2\), 250 mM sucrose, pH 7.4). In the absence of pDNA, the cell electropermeabilization was monitored by adding propidium iodide at 100 \(\upmu\)M in the pulsation buffer. Eighty percent of cells located between the electrodes were permeabilized. The cell electropermeabilization in the presence of Cy3-pDNA was performed after adding of 2 \(\upmu\)g of Cy3-pDNA in 200 \(\upmu\)L of pulsation buffer. Cy3-pDNA molecules interacted with more than 60 % of cells located between the electrodes.
Fluorescence recovery after photobleaching
Fluorescence recovery after photobleaching experiments was conducted using a Zeiss LSM-510 confocal microscope. The image sequence was acquired at a 5 Hz frequency using the 488 nm line of an argon ion laser at a very low power to avoid photobleaching during recording. After 50 images, four regions of interests (ROI), of 1 \(\upmu\)m radius each, which correspond to 0.86 \(\upmu\)m waist (\(\omega\)) of a Gaussian beam, located in front of anode and cathode, respectively, and on left and right sides of the cell (see Table 1 for illustration), were rapidly photobleached (\(t < 300\) ms) at maximal laser power. Fluorescence recovery was monitored at 5 Hz by acquiring successive images during 40 s. The recovery curves were obtained by plotting the mean fluorescence intensity as a function of time in these four ROI, and were corrected for fluctuations in axial position by another ROI located into the cell and finally normalized to the mean value of each ROI before photobleaching. The curves were fitted using Eq. 1 which is a slightly modified 2D diffusion model for FRAP taking into account normalization and a mobile fraction \(M\) (Axelrod et al. 1976; Matthews and Favard 2007):
In Eq. 1 \(K\) measures the deepness of the bleach, \(t_{1/2}\) corresponds to the time where half the fluorescence has been recovered and is linked to the two dimensional diffusion coefficient \(D\) by
\(\beta\) being a discrete function of \(K\) tabulated in Yguerabide et al. (1982). \(F_0\) is the fluorescence intensity immediately after the end of the bleach, i.e., at \(t=0\). Equation 1 was used to its 20th order limited development for data fitting (\(n=20\)). \(M\), the mobile fraction, is defined as:
Fluorescence recoveries were acquired before electric pulses and at a time \(t\), 30 s \(< t <\) 90 s after electropermeabilization on the same sample using the same sequence. At this time t, more than 50 % of the cells were still permeabilized (Rols and Teissié 1998).
Results
Effects of electropermeabilization on the mobility of Rae-1 protein
The mobility of the Rae-1 protein was monitored by means of FRAP experiments before and after electropermeabilization. In Fig. 1a are depicted four different typical recovery curves obtained before (in green) and after (in red) electropermeabilization at the position facing the anode (square) and the cathode (triangle). Fitting of these recovery curves using Eq. 1 depicted as full lines in green (before electropermeabilization) or red (after electropermeabilization) lead to determination of two significant parameters (\(t_{1/2}\) and \(M\)).
a Typical recovery curves before and after electropermeabilization: FRAP curves are displayed for anode (square) and cathode (triangle) positions before (in green) and after (in red) electropermeabilization. b Distribution of \(t_{1/2}\) and \(M\) before and after electropermeabilization: \(M\) was plotted as a function of \(t_{1/2}\), before (in green) and after (in red) electropermeabilization for the cell pole facing the anode (upper) or the cathode (lower). Intensity scale correspond to the number of events in each class of (\(t_{1/2}\), \(M\)) pair. Isolevels are dispatched on the bottom of the graphs for easier readability
\(t_{1/2}\) is a function of the diffusion coefficient of the Rae-1 protein, the second one is a function of the number of mobile Rae-1 proteins during the time course of the experiment (see Sect. 2.4 for definition). In order to analyze the change in the mobility of the Rae-1 molecule, the mobile fraction was plotted as a function of the half-time of recovery at the anode (Fig. 1b, upper part) and the cathode (Fig. 1b, lower part) before (in green) and after (in red) electropermeabilization, for a set of 250 different cells.
As described in Fig. 1b, after electropermeabilization the mobility of the Rae-1 protein is reduced (\(M\) slightly decreases and \(t_{1/2}\) increases) on both sides of the cell facing the electrodes (i.e., anode and cathode). The mean values and standard deviations of \(t_{1/2}\) and \(M\), at the pole of the cell facing the anode and the cathode, before and after electropermeabilization, as well as the left and right part of the equatorial level of the cell (perpendicular to the electric field direction) are reported in the Table 1 using descriptive statistics. Student \(t\) tests were performed on paired values of cathode and anode showing that both parameters scored \(p < 0.001\) and can be considered as significantly different before and after electropermeabilization. Interestingly, an increase in the of \(t_{1/2}\) and a decrease of \(M\) has also been observed in the membrane regions not facing the electrodes.
Effects of pDNA electroinsertion on the mobility of the Rae-1 protein
Fluorescence microscopy was used to directly observe the pDNA electrotransfer process and has revealed that pDNA molecules are found as clusters of an apparent size close to the diffraction limit, i.e., 200 nm (Golzio et al. 2002) at the cell surface. We attempted to probe the size of these pDNA clusters more accurately by measuring changes in Rae-1 mobility in the presence of electrotransferred pDNA. Unfortunately, direct image analysis of eGFP-Rae-1 expressing cells showing an electroinsertion of Cy3-pDNA revealed that Rae-1 fluorescence was excluded from the Cy3-pDNA cluster avoiding any possibility to perform correct FRAP measurements (Fig. 2). Nevertheless, absence of measurable fluorescence does not directly mean absence of Rae-1 proteins. Indeed, since emission spectrum of eGFP widely overlaps absorption spectrum of Cy3, lack in fluorescence can also be due to a very efficient energy transfer of Perrin–Forster type from eGFP to Cy3. Therefore, as shown in Fig. 3, acceptor photobleaching FRET experiments were performed. The acceptor Cy3-pDNA was photobleached using the 546 nm line of an He–Ne laser on the confocal microscope and the fluorescence of the donor eGFP-Rae-1 protein (Fig. 3b, green line) was measured. If FRET is the process inducing lack of Rae-1 fluorescence in the Cy3-pDNA clusters, then the donor eGFP-Rae-1 fluorescence should increase while the acceptor Cy3-pDNA fluorescence is irreversibly decreasing. This was not the case, indicating that no FRET occurred between Cy3-pDNA and the eGFP-Rae-1 protein.
Images of GFP-Rae-1 and Cy3-pDNA after electroinsertion of Cy3-pDNA: Cy3-pDNA is located at the pole of the cells immediately after the end of electropulsation (a), covering a large area at the cell surface. Fluorescence of GFP-Rae1 is located at the plasma membrane of the CHO cells (b) but is clearly extinguished in the area occupied by Cy3-pDNA. This is confirmed by merging the two images (c) where no colocalization can be seen
More interestingly, during the time course of the experiment, no fluorescence recovery was observed either for Cy3-pDNA (Fig. 3b, red line) or for the eGFP-Rae-1 protein, confirming a total absence of mobility of Cy3-pDNA and indicating a total inaccessibility of eGFP-Rae-1 into these clusters, whereas eGFP-Rae-1 recoveries, when monitored outside the inserted pDNA region, were identical to the ones obtained in Sect. 3.1 (data not shown).
Photobleaching of Cy3-pDNA after electroinsertion. a Images of Cy3-pDNA (in red) and GFP-Rae-1 (in green) fluorescence acquired during the expected recovery process. The photobleached area is located inside the yellow circle. In b Fluorescence intensities integrated in the yellow circle of Cy3-pDNA (in red) and GFP-Rae-1 (in green) normalized to the value of Cy3-pDNA before the bleaching. These two curves clearly show that fluorescence recovery occurred neither for Cy3-pDNA, nor for eGFP-Rae-1, leading to the conclusion that the Cy3-pDNA is highly immobile and that Rae-1 is totally excluded from the Cy3-pDNA clusters located in the membrane
Discussion
Macroscopic effects of electropermeabilization
Role of the cell swelling induced by electropermeabilization on the estimation of the recovery time in FRAP experiments: a In order to illustrate extreme cases of cell swelling induced by electropermeabilization, images of one cell acquired during the FRAP experiments before (green) and after (red) electropermeabilization were superimposed are registered to an invariant position within the cell (yellow part of the image). b This scheme depicts the effect of cell swelling induced by electropermeabilization [from 1 (left) to 2 (right)]. The swelling results in a membrane, which is more under tension, as compared to the resting cell. Since the laser waist is constant and assuming that the axis of the laser is kept normal to the plane of the membrane, it can be seen from this scheme that the area analyzed during FRAP experiments is smaller after swelling than before. Therefore, without any change in diffusion constant of the observed molecule (\(D=Cte\)), the recovery time after electropermeabilization (\(\tau _2\)) will be lower than before (\(\tau _1\)).
The main visible effect on cell and membrane shape occurring immediately after electropermeabilization is cell swelling (Golzio et al. 1998; Romeo et al. 2013). Figure 4a depicts an image of the cell recorded at the end of the FRAP experiments before (in green) and after (in red) electropermeabilization, showing that the swelling was also observed during the experiments. Figure 4b illustrates how this osmotic swelling can locally change the tension of plasma membrane. The FRAP half-time recovery is linked to the time needed for the molecule to explore the mean square of a length defined by the waist of the laser (see Eq. 2). When applied to the measurement of the lateral diffusion in the cell plasma membrane, this definition assumes that the plasma membrane is perfectly flat and perpendicular to the laser path. Although swelling is expected not to affect the angle of intercept of the laser and the plasma membrane, it can dramatically change the ‘flatness’ of the plasma membrane (and is flatter than before swelling occurs). Therefore, if a molecule diffuses with the same diffusion coefficient (\(D\)) before and after electrically-mediated swelling, it will take less time for FRAP recovery in a membrane under tension (closer to the real waist) than not (further from the real waist). Our results here show exactly the opposite effect, emphasizing the existence of microscopic obstacles to free diffusion after electropermeabilization. Moreover, our study shows that these obstacles are also present at the equator of the cell (i.e., locations parallel to the electrodes) at most 90 s after electropermeabilization. This unexpected result is nevertheless confirmative, with a much faster time resolution, of the recent work of Chopinet et al. (2013) where it was shown using AFM imaging that propagation of membrane perturbation along the entire cell surface occurs.
Change in the mobility of Rae-1: relation to the permeabilized area
Saxton (1982, 1987) has theoretically shown that the apparent diffusion coefficient of a tracer molecule decreases with the fractional area occupied by obstacles. The diffusion coefficient approaches zero if obstacles are immobile in a given area (i.e., the percolation threshold), or decrease to an asymptotic value if obstacles are very mobile (as compared to the tracer molecule). In this study, the apparent diffusion coefficient (i.e., half-time recoveries at constant given waists) of eGFP-Rae-1 protein is decreased as much as 10 % after electropermeabilization. This result is a confirmation of the existence of defects at least in the outer leaflet of the plasma membrane, since the tracer, eGFP-Rae-1 protein is located in this leaflet. This confirms previous results obtained by other approaches such as influx of fluorescent dyes into the cytosol (Gabriel and Teissié 1997, 1999; Pucihar et al. 2008; Rols and Teissié 1998; Mir et al. 1988; Tekle et al. 1994; Prausnitz et al. 1994, 1995).
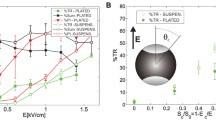
According to Saxton’s numerical simulations, a decrease of 10 % of the apparent diffusion coefficient means that 10 % (in the case of immobile pores or defects) to 20 % (in the case of very fast diffusing pores or defects) of the probed membrane area of the cell are occupied by these defects or pores. This latter case being certainly more adapted to our study since the defects or pores observed here are rapidly moving. In order to compare with experimental (Gabriel and Teissié 1997, 1999; Tekle et al. 2001; Portet et al. 2011) or numerical simulation (Krassowska and Filev 2007) data available in the literature, we have defined an aperture angle (\(0<\theta _{ap}<90^\circ\)) of permeabilization as the half angle of the solid angle in which the total permeabilized surface is included.
This aperture angle is directly linked to the surface permeabilized assuming the following:
-
the defects or pores are spontaneously created during each of the ten electric impulses and they exist for the time of the experiment.
-
they propagate rapidly over the cell surface (in our study defects are probed by the tracer at the equator of the cell 1 min after electropermeabilization)
Then the total permeabilized surface is consistent with the observation area in which the tracer over the membrane surface is monitored, i.e., between 10 % and 20 % of the total cell surface. Therefore, we show that if all the obstacles were fused into a unique one, the value of this aperture angle is found between 36° and 52°.
Portet et al., measured the average aperture angle of pores to be 6° by using pDNA translocation into giant unilamellar vesicles (GUVs) made of DOPC. Using GUVs made of DOPC with a radius close to the CHO cells (i.e., 15 μm) and electric fields twice our value (1.2 kV cm−1), Tekle et al. (2001) showed that up to 14 % of the total membrane surface can be lost when pulsing, leading to a value of 44°. Finally, Gabriel and Teissié (1999) measured the average aperture angle as the angle of the extent of the permeabilization immediately after the pulse by visualizing a small fluorophore entrance into the cell to be 56°. \(\theta _{\rm ap}\) values found from these different studies exhibit discrepancies. Many factors can account for the discrepancies in these studies, notably the duration and the strength of the external applied electric field, differences in the model used (i.e., cells Gabriel and Teissié 1999, 1997 or artificial lipid membranes Tekle et al. 2001; Portet et al. 2011), differences in the models radii or finally differences in external and internal buffers used for electropermeabilization. Nevertheless in Gabriel and Teissié (1999) the results have been obtained using exactly the same experimental conditions to ours (i.e., cell type, electropermeabilization buffer and electric field intensity range) and can, therefore, be directly compared. These results exhibit a value of \(\theta _{\rm ap}\) slightly superior to what is found here. Indeed, the lifetime distribution of the pores or defects in lipid membranes span a wide time range (between microseconds up to minutes), and this depends on several factors (e.g., field intensity, conductance, lipid membrane composition, artificial vesicles or cells) (Teissié et al. 2005). However, it is clear that the number and/or size of the pores or defects decreases with time after electropermeabilization. Since our experiments have been be conducted in average one minute after the end of pulse sequence, this effect could account for a smaller value of \(\theta _{\rm ap}\) as compared to the one found in Gabriel and Teissié (1999, 1997). Alternatively, Krassowska and Filev (2007) performed numerical simulation of the evolution in time and space of pores in a spherical pure lipid vesicle exposed to an electric field. They simulated the normalized average area occupied by these pores as a function of the external electric field applied to the cell. For an electric field used in this study of 0.2 kV cm−1 (that corresponds to our conditions regarding the 50 μm radius they use in their simulations) they found this area to be 0.07 % of the total area (=1°) which is far beyond our experimental results. Nevertheless, their values were obtained using only one pulse of 1 ms. If one applies this oversimplified hypothesis that the number, and, therefore, the area occupied by the pores is a function not only of the electric field intensity, but it is also of the total exposure time of cell to an electric field. Furthermore, if the created pores in each impulsion do not reseal during our total pulse sequence, the area occupied by the pores should be 50 times more important than in their study, resulting in an apparent angle of (=23°) which is closer to what we find here.
Effect of pDNA electroinsertion on the lateral mobility of Rae-1
The apparent size of the pDNA clusters inserted into the membrane after electropermeabilization (larger than 200 nm in diameter, Golzio et al. 2002; Phez et al. 2005) should lead to a drastic increase in the half-time recovery (\(t_{1/2}\)) and a drastic decrease in the mobile fraction (\(M\)) due to eGFP-Rae-1 proteins trapped in these pDNA clusters. Unfortunately, the very low level to total absence of the fluorescence of Rae-1 in the pDNA clusters at the membrane did not facilitate the FRAP measurements.
This absence of fluorescence could be of two different origins:
-
Loss in fluorescence of the eGFP-Rae-1 protein due to very efficient energy transfer of the Perrin–Förster type (FRET) between eGFP fused Rae-1 protein and Cy-3 labeling the pDNA.
-
Exclusion of Rae-1 molecules from the pDNA clusters.
Photobleaching FRET experiments have been performed in order to discriminate between these two possibilities. The absence of recovery in the fluorescence of Rae-1 proteins when the Cy-3 molecules are bleached clearly shows that eGFP-Rae-1 proteins are totally excluded from pDNA clusters induced by electropermeabilization. On the other hand, no recovery occurs for pDNA fluorescence, confirming that the pDNA clusters are highly immobile, as previously described (Escoffre et al. 2011). These new results here favor a pDNA aggregate that causes exclusion of some of the membrane components, if not all, to its periphery.
Conclusion
In order to sense at the molecular scale the effect of electropermeabilization and pDNA electroinsertion on the plasma membrane lateral organization, the mobility of a tracer (Rae-1 protein) has been assessed in living cells. This study show that the membrane reporters provide a means to sense the defects induced by electropermeabilization and estimate the surface permeabilization during exposure to electric fields. More interestingly, this study demonstrates that these defects propagate all over the surface of the plasma membrane in less than 2 min. Finally, the nature of pDNA clusters immediately after electroinsertion have been shown to exist in highly dense clusters, since they prevent Rae-1 protein penetration.
References
Abidor IG, Li LH, Hui SW (1994) Studies of cell pellets: II. Osmotic properties, electroporation, and related phenomena: membrane interactions. Biophys J 67(1):427–435
Axelrod D, Koppel D, Schlessinger J, Elson E, Webb W (1976) Mobility measurement by analysis of fluorescence photobleaching recovery kinetics. Biophys J 16(9):1055–1069
Benz R, Beckers F, Zimmermann U (1979) Reversible electrical breakdown of lipid bilayer membranes: a charge-pulse relaxation study. J Membr Biol 48(2):181–204
Chang D, Chassy B, Saunders J, Sowers A (1992) Guide to electroporation and electrofusion. San Diego
Chopinet L, Roduit C, Rols MP, Dague E (2013) Destabilization induced by electropermeabilization analyzed by atomic force microscopy. Biochim Biophys Acta 1828(9):2223–2229
Daud AI, DeConti RC, Andrews S, Urbas P, Riker AI, Sondak VK, Munster PN, Sullivan DM, Ugen KE, Messina JL, Heller R (2008) Phase i trial of interleukin-12 plasmid electroporation in patients with metastatic melanoma. J Clin Oncol 26(36):5896–5903
Escoffre JM, Rols MP (2012) Electrochemotherapy: progress and prospects. Curr Pharm Des 18(23):3406–3415
Escoffre JM, Portet T, Favard C, Teissié J, Dean DS, Rols MP (2011) Electromediated formation of DNA complexes with cell membranes and its consequences for gene delivery. Biochim Biophys Acta-Biomembr 1808(6):1538–1543
Gabriel B, Teissié J (1997) Direct observation in the millisecond time range of fluorescent molecule asymmetrical interaction with the electropermeabilized cell membrane. Biophys J 73(5):2630–2637
Gabriel B, Teissié J (1999) Time courses of mammalian cell electropermeabilization observed by millisecond imaging of membrane property changes during the pulse. Biophys J 76(4):2158–2165
Golzio M, Mora MP, Raynaud C, Delteil C, Teissié J, Rols MP (1998) Control by osmotic pressure of voltage-induced permeabilization and gene transfer in mammalian cells. Biophys J 74(6):3015–3022
Golzio M, Teissié J, Rols MP (2002) Direct visualization at the single-cell level of electrically mediated gene delivery. Proc Natl Acad Sci USA 99(3):1292–1297
Griese T, Kakorin S, Neumann E (2002) Conductometric and electrooptic relaxation spectrometry of lipid vesicle electroporation at high fields. Phys Chem Chem Phys 4(7):1217–1227
Hibino M, Itoh H, Kinosita K (1993) Time courses of cell electroporation as revealed by submicrosecond imaging of transmembrane potential. Biophys J 64(6):1789–1800
Kakorin S, Stoylov S, Neumann E (1996) Electro-optics of membrane electroporation in diphenylhexatriene-doped lipid bilayer vesicles. Biophys Chem 58(1–2):109–116
Kinosita K, Tsong TY (1979) Voltage-induced conductance in human erythrocyte membranes. Biochim Biophys Acta 554(2):479–497
Krassowska W, Filev PD (2007) Modeling electroporation in a single cell. Biophys J 92(2):404–417
Levine ZA, Vernier PT (2010) Life cycle of an electropore: field-dependent and field-independent steps in pore creation and annihilation. J Membr Biol 236(1):27–36
Low L, Mander A, McCann K, Dearnaley D, Tjelle T, Mathiesen I, Stevenson F, Ottensmeier CH (2009) DNA Vaccination with Electroporation Induces Increased Antibody Responses in Patients with Prostate Cancer. Hum Gene Ther 20(11):1269–1278
Matthews C, Favard C (2007) Theory, principles and applications of fluorescent technologies in cellular biology and cancer research. Bull Cancer (French) 94(1):115–125
Mazères S, Sel D, Golzio M, Pucihar G, Tamzali Y, Miklavcic D, Teissié J (2009) Non invasive contact electrodes for in vivo localized cutaneous electropulsation and associated drug and nucleic acid delivery. J Control Release 134(2):125–131
Mir L, Banoun H, Paoletti C (1988) Introduction of definite amounts of nonpermeant molecules into living cells after electropermeabilization—direct access to the cytosol. Exp Cell Res 175(1):15–25
Mir LM, Gehl J, Sersa G, Collins CG, Garbay JR, Billard V, Geertsen PF, Rudolf Z, O’Sullivan GC, Marty M (2006) Standard operating procedures of the electrochemotherapy: instructions for the use of bleomycin or cisplatin administered either systemically or locally and electric pulses delivered by the Cliniporator (TM) by means of invasive or non-invasive electrodes. EJC Suppl 4(11):14–25
Neumann E, Sowers A, Jordan C (1989) Electroporation and electrofusion in cell biology, Plenum, New York
Nomura M, Zou Z, Joh T, Takihara Y, Matsuda Y, Shimada K (1996) Genomic structures and characterization of rae1 family members encoding gpi-anchored cell surface proteins and expressed predominantly in embryonic mouse brain. J Biochem 120(5):987–995
Parsegian V (1969) Energy of an ion crossing of a low dielectric membrane: Solutions to four relevant electrostatic problems. Nature 221:844–846
Pavlin M, Kanduser M, Rebersek M, Pucihar G, Hart FX, Magjarevic R, Miklavcic D (2005) Effect of cell electroporation on the conductivity of a cell suspension. Biophys J 88(6):4378–4390
Pavlin M, Leben V, Miklavcic D (2007) Electroporation in dense cell suspension-theoretical and experimental analysis of ion diffusion and cell permeabilization. Biochim Biophys Acta 1770(1):12–23
Phez E, Faurie C, Golzio M, Teissié J, Rols M (2005) New insights in the visualization of membrane permeabilization and DNA/membrane interaction of cells submitted to electric pulses. Biochim Biophys Acta Gen Subj 1724(3):248–254
Portet T, Favard C, Teissié J, Dean DS, Rols MP (2011) Insights into the mechanisms of electromediated gene delivery and application to the loading of giant vesicles with negatively charged macromolecules. Soft Matter 7(8):3872–3881
Prausnitz MR, Milano CD, Gimm JA, Langer R, Weaver JC (1994) Quantitative study of molecular transport due to electroporation: uptake of bovine serum albumin by erythrocyte ghosts. Biophys J 66(5):1522–1530
Prausnitz MR, Corbett JD, Gimm JA, Golan DE, Langer R, Weaver JC (1995) Millisecond measurement of transport during and after an electroporation pulse. Biophys J 68(5):1864–1870
Pucihar G, Kotnik T, Miklavich D, Teissié J (2008) Kinetics of transmembrane transport of small molecules into electropermeabilized cells. Biophys J 95(6):2837–2848
Rols MP, Teissié J (1998) Electropermeabilization of mammalian cells to macromolecules: Control by pulse duration. Biophys J 75(3):1415–1423
Romeo S, Wu YH, Levine ZA, Gundersen MA (1828) Vernier PT (2013) Water influx and cell swelling after nanosecond electropermeabilization. Biochim Biophys Acta 8:1715–1722
Rosazza C, Escoffre JM, Zumbusch A, Rols MP (2011) The actin cytoskeleton has an active role in the electrotransfer of plasmid dna in mammalian cells. Mol Ther 19(5):913–921
Rosazza C, Phez E, Escoffre JM, Cézanne L, Zumbusch A, Rols MP (2012) Cholesterol implications in plasmid dna electrotransfer: evidence for the involvement of endocytotic pathways. Int J Pharm 423(1):134–143
Saxton M (1982) Lateral diffusion in an archipelago—effects of impermeable patches on diffusion in a cell-membrane. Biophys J 39(2):165–173
Saxton M (1987) Lateral diffusion in an archipelago—the effect of mobile obstacles. Biophys J 52(6):989–997
Tarek M (2005) Membrane electroporation: a molecular dynamics simulation. Biophys J 88(6):4045–4053
Teissié J, Golzio M, Rols M (2005) Mechanisms of cell membrane electropermeabilization: a minireview of our present (lack of ?) knowledge. Biochim Biophys Acta Gen Subj 1724(3):270–280
Tekle E, Astumian RD, Chock PB (1994) Selective and asymmetric molecular transport across electroporated cell membranes. Proc Natl Acad Sci USA 91(24):11512–11516
Tekle E, Astumian R, Friauf W, Chock P (2001) Asymmetric pore distribution and loss of membrane lipid in electroporated DOPC vesicles. Biophys J 81(2):960–968
Tieleman DP (2004) The molecular basis of electroporation. BMC Biochem 5:10
Weaver J (1993) Electroporation: a general phenomenon for manipulating cells and tissues. J Cell Biochem 51:426–435
Weiland O, AhlTn G, Diepolder H, Jung MC, Levander S, Fons M, Mathiesen I, Sardesai NY, Vahlne A, Frelin L, SSllberg M (2013) Therapeutic dna vaccination using in vivo electroporation followed by standard of care therapy in patients with genotype 1 chronic hepatitis C. Mol Ther 21(9):1796–1805
Yguerabide J, Schmidt JA, Yguerabide EE (1982) Lateral mobility in membranes as detected by fluorescence recovery after photobleaching. Biophys J 40(1):69–75
Zou Z, Nomura M, Takihara Y, Yasunaga T, Shimada K (1996) Isolation and characterization of retinoic acid-inducible cDNA clones in F9 cells: a novel cDNA family encodes cell surface proteins sharing partial homology with MHC class I molecules. J Biochem 119(2):319–328
Acknowledgments
This work has been performed in collaboration with the “Toulouse Réseau Imagerie” core IPBS facility (Genotoul, Toulouse, France), which is supported by the Association Recherche Cancer, Region Midi Pyrenees, the European union (FEDER) and Grand Toulouse cluster. This research project was conducted in the scope of EBAM European Associated Laboratory and of the COST Action TD1104. JM Escoffre was the recipient of an allocation de recherche du Ministère de l’Enseignement Supérieur et de la Recherche. The authors are grateful to Dr. M. Golzio and Dr. E. Bellard for critical reading of the manuscript and Dr. T. Melvin for English rewriting. C. Favard is a membership of CNRS consortii GDR2588 “MIV” and GDR3070 “CellTiss”.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Escoffre, J.M., Hubert, M., Teissié, J. et al. Evidence for electro-induced membrane defects assessed by lateral mobility measurement of a GPi anchored protein. Eur Biophys J 43, 277–286 (2014). https://doi.org/10.1007/s00249-014-0961-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00249-014-0961-1