Abstract
Chromosome shattering has been described as a special form of mitotic catastrophe, which occurs in cells with unrepaired DNA damage. The shattered chromosome phenotype was detected after application of a methanol/acetic acid (MAA) fixation protocol routinely used for the preparation of metaphase spreads. The corresponding phenotype in the living cell and the mechanism leading to this mitotic catastrophe have remained speculative so far. In the present study, we used V79 Chinese hamster cells, stably transfected with histone H2BmRFP for live-cell observations, and induced generalized chromosome shattering (GCS) by the synergistic effect of UV irradiation and caffeine posttreatment. We demonstrate that GCS can be derived from abnormal mitotic cells with a parachute-like chromatin configuration (PALCC) consisting of a bulky chromatin mass and extended chromatin fibers that tether centromeres at a remote, yet normally shaped spindle apparatus. This result hints at a chromosome condensation failure, yielding a “shattered” chromosome complement after MAA fixation. Live mitotic cells with PALCCs proceeded to interphase within a period similar to normal mitotic cells but did not divide. Instead they formed cells with highly abnormal nuclear configurations subject to apoptosis after several hours. We propose a factor depletion model where a limited pool of proteins is involved both in DNA repair and chromatin condensation. Chromosome condensation failure occurs when this pool becomes depleted.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lethal cellular damage can be caused by a variety of events, either immediately after the damage as in case of necrosis or it may be delayed and take place after an interval of several hours. Delayed cell death typically results from DNA damage either by environmental agents such as UV light, ionizing radiation, and genotoxic chemicals or by endogeneous causes, mostly reactive oxygen species resulting from cellular metabolism (for review, see Hoeijmakers 2001). In order to cope with potentially fatal DNA damage, organisms have developed control checkpoints and highly efficient DNA repair mechanisms (for review, see Sancar et al. 2004). If DNA damage cannot be repaired, cells are committed to delayed programmed cell death.
The best-studied type of such a kind of cell death is apoptosis, which can occur at any interphase stage (for reviews, see Chowdhury et al. 2006; Okada and Mak 2004). However, apoptosis is not the only way of delayed cell death. Other less well-defined types have been assigned to nonapoptotic endpoints by their phenotypic and biochemical traits (Blank and Shiloh 2007; Okada and Mak 2004). An example of this kind is mitotic cell death occurring after DNA damage during interphase. Recently, fragmentation of the entire chromosome complement was described as a cause for mitotic cell death observed after doxorubicin treatment of cells (Stevens et al. 2007). Apparently similar observations were already reported in UV-irradiated cells during the early 1940s (for an overview, see Chu 1965), and to our knowledge the term “chromosome shattering” was coined by Lovelace (1954) for mitotic cells in Tradescantia paludosa pollen with multiple chromosome fragmentation following UV irradiation.
Our present study was designed to better understand the process that brings about chromosome shattering. It is a continuation of experiments performed by Christoph Cremer and co-workers during the mid-1970s to the mid-1980s where chromosome shattering was demonstrated. In these experiments, cultured Chinese hamster cells were exposed either to whole-cell UV irradiaton (λ = 254 nm) or to partial irradiation of nuclei (about 5% of the total nuclear area) using a laser UV microbeam (λ = 257 nm). Irradiated cells were incubated with caffeine and allowed to proceed to mitosis. Chromosome preparations performed with standard protocols including hypotonic treatment, methanol/acetic acid fixation (MAA), and air-drying revealed major chromosomal damage, called “chromosome shattering.” Two types of chromosome shattering could be distinguished: generalized chromosome shattering (GCS) with shattering of the entire chromosome complement and partial chromosome shattering (PCS) where only a few mitotic chromosomes showed the shattered morphology, while the rest revealed an inconspicuous mitotic structure (Cremer et al. 1980a, 1981a; Zorn et al. 1976) (Fig. 1).
Metaphase spreads with damaged chromosomes obtained after laser UV microirradiation of nuclei in living Chinese hamster cells. Nuclei in living Chinese hamster cells were microirradiated (λ = 257 nm) at a single nuclear site comprising about 5% of the total nuclear area. Microirradiated cells were followed to the next mitosis (about 3–15 h) in medium with 1 mM caffeine. a, b Metaphase spread (a) and the corresponding karyogram (b) from a diploid, fibroblastoid Chinese hamster cell reveal a shattered chromosome 1 and a break in a chromosome 7 surrounded by intact chromosomes (Zorn et al. 1976). c Metaphase spread from a diploid fibroblastoid Chinese hamster cell shows an area of pulverized chromosomes (arrows) surrounded by chromosomes with chromatid gaps or breaks (Zorn et al. 1976). d Mitotic V79 cell showing fragmentation and pulverization of the whole chromosome complement (Cremer et al. 1981a). e Mitotic V79 cell with a pulverized chromosome complement (Cremer et al. 1981a). Scale bar 10 μm (reprinted with permission from Cremer and Cremer (2006)
Surprisingly, in these experiments GCS not only occurred after irradiation of the entire nucleus but also after partial irradiation when the same total UV incidence was applied. The percentage of cells showing the shattered chromosomal phenotype increased with the total incident dose delivered to the cell nucleus and the time of caffeine posttreatment, independent of the spatial distribution of this dose. These partial UV irradiation experiments were initially performed as an attempt to gain insight into the structural organization of chromosomes during interphase, which was largely unknown at that time. The analysis of metaphase chromosomes in the subsequent mitosis of locally irradiated cells obtained by appropriate UV doses and caffeine posttreatment showed sites of DNA damage restricted to a small subset of chromosomes, providing indirect, yet compelling evidence for a territorial organization of chromosomes during interphase (Cremer et al. 1982a, b; Zorn et al. 1979; Zorn et al. 1976; for review see Cremer and Cremer 2006; Meaburn and Misteli 2007). The discrepancy between localized UV damage and resulting general chromosome shattering in nuclei after local irradiation beyond a total UV incidence was puzzling and pointed to indirect effects of irradiated chromatin also affecting chromatin located remote from the microirradiation site (Cremer and Cremer 1986; Cremer et al. 1980a; for review see Cremer and Cremer 2006). As a hypothetical framework, a factor depletion model was proposed (Cremer and Cremer 1986; Cremer et al. 1980a, b) (see section “Discussion”).
Our present work sheds new light on the dynamic process leading to chromosome shattering. In order to pursue the behavior of chromatin during interphase and mitosis in living cells after UV irradiation/caffeine posttreatment, we stably transfected Chinese hamster cells with histone H2BmRFP. Our observations demonstrate that the phenotype of shattered chromosomes is neither visible directly in living cells nor in cells fixed with buffered paraformaldehyde (PFA), but only apparent after fixation with methanol/acetic acid (MAA) and air-drying, a protocol widely used to achieve metaphase spreads for cytogenetic analyses. We demonstrate that GCS results from abnormally shaped mitotic figures, consistently presenting a “parachute-like” configuration of bulk chromatin from which expanded chromatin fibers connect centromeres to a remote, yet normally shaped spindle apparatus. This indicates that the spindle apparatus was able to attach to centromeres, yet due to the lack of proper chromatin condensation, centromere movements led to the formation of extended chromatin fibers but failed to generate a metaphase plate. These abnormal mitotic cells proceeded to interphase within a normal time frame, but never showed anaphase chromatid segregation and typically missed cytokinesis. These interphase cells usually harbored one multilobulated nucleus and entered apoptosis after several hours.
Materials and methods
Cell lines and culture conditions
V79 cells (spontaneously immortalized lung fibroblasts from the Chinese hamster Cricetulus griseus) were stably transfected (Lipofectamin 2000, Biocompare, UK) with H2BmRFP (plasmid kindly provided by R. Brack-Werner, GSF, Munich, Germany). Cells were cultured on coverslips in DMEM/10% FCS/1% penicillin/streptomycin at 37°C/5% CO2.
UV irradiation, caffeine posttreatment
For UV irradiation, a mercury UV lamp (NU-4 KL; Benda, Germany) mainly emitting UV light at λ = 254 nm and a power of 5 W/m2 (=500 μW/cm2) was used. Cells grown on coverslips to a confluency of ~50% were briefly washed in PBS (to remove the phenol red contained in the medium) and quickly transferred to the irradiation field. Cells were irradiated for 2 s at a working distance of 15 cm (≈10 J/m2 = 0.01 nJ/μm2) corresponding to a total of ~0.5 nJ for an average nucleus with a square size of 50 μm2. Cells were then re-transferred into medium supplemented with 1 mM caffeine (Sigma-Aldrich, Germany) and further cultivated for 16 h.
Culture conditions and image acquisition for live-cell observation
For live-cell experiments, cells were grown on round coverslips (PeCon, Germany) to fit into the POC-R chamber (Visitron Systems, Germany) required for mounting on the microscopic stage. These coverslips were gridded with a diamond pencil to allow relocalization of individual cells after fixation and immunofluorescence staining. For long-term observations, medium was supplemented with 100 μM Trolox (6-hydroxy-2,5,7,8-tetramethylchroman-2-carboxylic acid) (Sigma-Aldrich, Germany) 24 h before starting a live-cell experiment. Immediately before onset of a live-cell observation, cells were incubated in phenol red-free live-cell medium: 1:1 vol/vol DMEM w/o phenol red (Gibco, Germany) and Ham’s F10 w/o phenol red (Biochrom, Germany) supplemented with 10% FCS, 1% penicillin/streptomycin, 25 mM HEPES final concentration (Calbiochem, CA, USA), 3.2 mM L-glutamin (Biochrom, Germany), and 100 μM Trolox.
For the identification of S phase, cells were pulse-labeled with BrdU (15 μM; Sigma-Aldrich, Germany) for 15 min starting 16 h after UV irradiation/caffeine posttreatment. After BrdU release, cells were further grown for 70 min before fixation.
Live-cell observations were performed with a Zeiss Axiovert 200 M fluorescence microscope, equipped with a 40×/1.3 plan-neofluar ph3 oil objective. A motorized z-stage and the software MetaMorph 6.2r6 (Universal Imaging Group) were used. To meet the requirements for long-term observations, the microscope was equipped with an incubation chamber and a heating system (Visitron Systems, Germany), a 12 V/100 W halogen lamp, and a 450-nm LP filter to minimize exposure to UV light. For the selected cells, one image every 10 or 15 min was recorded automatically with about 100 ms at 3.0 V in transmission mode and about 300–500 ms at 7.0 V for mRFP. A contrast-based autofocus was used in transmission mode.
Cell fixation
For in situ fixation using methanol/acetic acid (MAA), cells were briefly incubated in 1:2 vol/vol 0.56% KCl/1× PBS before being subject to hypotonic treatment in 0.56% KCl for 20 min. Then fixative (3:1 vol/vol methanol/acetic acid; −20°C) was added dropwise to the KCl solution. Several times, a portion of the solution was removed and replaced by fresh fixative until complete replacement of the fixative was accomplished. Slides were air-dried, subjected to an ethanol series (70/90/100%, 3 min each), and air-dried again.
For fixation in paraformaldehyde (PFA), cells were briefly washed in 1× PBS, fixed with 4% PFA for 10 min at RT, washed 2 × 5 min in 1× PBS, and stored in 1× PBS at 4°C for up to 1 week. In contrast to the MMA fixation, air-drying of PFA-fixed cells was avoided at all times to preserve the 3D nuclear structure.
Immunofluorescence and TUNEL test
For immunofluorescence, PFA-fixed cells were permeabilized with 1× PBS/0.5% Triton-X-100 for 10 min and blocked in 4% BSA/1× PBS/0.01% Tween20 for 10 min. For detection of BrdU, the cells were equilibrated in 50% formamide/2× SSC for 30 min before denaturation in the same solution for 2 min 30 s at 76°C. After cooling down to room temperature, samples were washed with 1× PBS, and immunofluorescence was performed.
The following primary antibodies were used: rabbit-anti-cleaved caspase 3 (Cell Signaling, MA, USA), mouse-anti-H3pS10 (kindly provided by Jan Postberg, University of Witten, Germany), rabbit-anti-H3pS10 (Abcam, UK), goat-anti-lamin B (Santa Cruz Biotechnology, CA, USA), mouse-anti-α-tubulin (clone DM-1A, Sigma-Aldrich, Germany), mouse-anti-BrdU (Roche, Germany), and human-anti-centromere (Antibodies Incorporated, CA, USA). This latter antibody stains different (not specifically indicated) kinetochore proteins, the protein complex assembled at each centromere. For simplicity, this antibody is referred to here as centromere-specific. For immunodetection, the following secondary antibodies were used: FITC-conjugated goat-anti-human, Cy3-conjugated goat-anti-rabbit, Cy3-conjugated donkey-anti-rabbit (Jackson Immuno Research, PA, USA), Alexa488-conjugated donkey-anti-goat (Molecular Probes, OR, USA), Alexa488-conjugated goat-anti-mouse (Molecular Probes, OR, USA), Cy3-conjugated sheep-anti-mouse, and Cy5-conjugated goat-anti-mouse (Dianova, Germany).
For detection of apoptosis on MAA-fixed cells, a TUNEL-assay (In Situ Cell Death Detection Kit, Fluorescein; Roche, Germany) was performed according to the protocol provided by the manufacturer. In this assay, free 3′-OH ends of fragmented DNA are enzymatically labeled with fluorescein-tagged deoxyuridine triphosphates using terminal deoxynucleotidyl transferase (TdT).
Cells were stained with DAPI (0.2 μg/ml in 1× PBS/0.01% Tween20) (Sigma-Aldrich, Germany) and mounted in Vectashield embedding medium (Vector Laboratories, CA, USA).
Fluorescence in situ hybridization (FISH)
Chromosome painting probes delineating Chinese hamster chromosomes 1, 2, and 5 were obtained by flow sorting from the Chinese hamster ovarian cancer (CHO)-derived cell line and amplified by DOP-PCR using the 6 MW primer (Telenius et al. 1992). Primary or secondary amplifications were PCR-labeled with digoxigenin (#1), biotin (#2), and dinitrophenol (DNP) (#5). Approximately 100 ng of each labeled probe and 500 ng of sheared unlabeled Chinese hamster genomic DNA as competitor DNA were dissolved per microliter of hybridization solution (50% formamide/2× SSC/10% dextran sulfate). Pretreatment of cells, 2D- or 3D-FISH, and washing steps were performed under standard conditions as described previously (Cremer et al. 2008). Biotin-labeled probes were detected by Avidin-Alexa488 (Molecular Probes, OR, USA) and FITC-conjugated goat-anti-avidin (Vector Laboratories, CA, USA), digoxigenin-labeled probes by Cy5-conjugated mouse-anti-DIG (Jackson Immuno Research, PA, USA) and Cy5-conjugated goat-anti-mouse (Dianova, Germany), and DNP-labeled probes by rabbit-anti-DNP (Sigma-Aldrich, Germany) and Cy3-conjugated goat-anti-rabbit (Jackson Immuno Research, PA, USA).
Image acquisition of fixed specimens
Images were collected either with an epifluorescence microscope (Axiophot 2, Zeiss, Germany) equipped with the software MetaVue 6.2r2 (Universal Imaging Group) or with a confocal laser scanning microscope (SP5 DMI 6000 CS, Leica, Germany) equipped with the software Leica Application Suite 1.7.0. Both microscopes were equipped with a 63×/1.4 plan-apochromat oil objective. Scanning was performed with a pixel size of 50 or 60 nm and an axial distance of 200 nm between optical sections. For each optical section, images were collected sequentially for all fluorochromes.
Image processing
Image processing was done in Adobe Photoshop 7.0 (Adobe Systems) or ImageJ 1.37c (Public Domain by Wayne Rasband). Confocal image stacks were shift corrected and deconvolved with Huygens Professional (Scientific Volume Imaging).
Results
General chromosome shattering (GCS) is observed after MAA fixation but not after PFA fixation
V79 cells stably transfected with histone H2BmRFP were exposed to UV irradiation (254 nm/10 J/m2 = 0.01 nJ/μm2) and postincubated for 16 h in medium with 1 mM caffeine, a drug known to enhance DNA-damaging effects (Blasina et al. 1999; Johansson et al. 2006). Control cultures were grown in parallel. Cells were fixed either with methanol/acetic acid (MAA) and air-dried or with buffered paraformaldehyde (PFA) and kept in buffer (Fig. 2a, b). Patterns of DAPI-stained DNA were quantitatively evaluated in several hundred randomly chosen cells both in irradiated and control samples (Fig. 2c). In irradiated samples subject to either type of fixation, an obvious increase in micronuclei and multilobulated nuclei was observed (nos. 2 and 3 in Fig. 2a, b) and a decrease in normal mitoses compared to control cells. GCS (3.9%, no. 5 in Fig. 2a) and faintly stained shadow-like patterns (4.0%, no. 6 in Fig. 2a) were only observed in MAA-fixed but not PFA-fixed samples. The shadow-like patterns were hardly visible and easily missed without special scrutiny. They showed no structures identifiable by light-microscopic observation in contrast to the grainy pattern of “pulverized” chromatin typical for GCS (Fig. 2a). Unexpectedly, PFA-fixed irradiated samples showed cells with a parachute-like chromatin configuration (PALCC) (2.9%, no. 7 in Fig. 2b) composed of a bulky chromatin mass and extended chromatin fibers resembling suspension lines of a parachute. These PALCCs will be described in more detail below. In addition, PFA-fixed but not MAA-fixed irradiated samples showed apoptotic cells with one or several intensely stained nuclear blebs (4.2%, no. 8 in Fig. 2b). Incubation of nonirradiated cells in 1 mM caffeine showed the same result as untreated control cells (data not shown).
Nuclear phenotypes in V79-H2BmRFP cells fixed 16 h after UV irradiation and caffeine posttreatment according to a the methanol/acetic acid (MAA) protocol and b the paraformaldehyde (PFA) protocol. Note the larger average size of MAA-fixed nuclei. Numbers point to normal and abnormal cells (see c for explanation). Scale bar 15 μm, images were taken by widefield epifluorescence microscopy. For better overview both images are composites from several single images. c Circle diagrams with the relative percentages of different phenotypes in PFA-fixed control cells (left), in MAA-fixed irradiated cells (middle), and in PFA-fixed irradiated cells (right). Note that shattered chromosome complements (GCS) and shadow-like chromatin patterns were only found after MAA fixation, never after PFA fixation. In contrast, cells with parachute-like chromatin configurations (PALCCs) and apoptotic cell nuclei were only noted in PFA-fixed, never in MAA-fixed samples
Apoptotic cells are the source of shadow-like nuclei; abnormal cells with parachute-like chromatin configurations (PALCCs) are the source of GCS
To identify the source of GCS and shadow-like patterns observed only after MAA fixation, live-cell microscopy was performed (Fig. 3). Live-cell observation of V79 cells with stable expression of H2BmRFP was started 16 h after UV irradiation and caffeine posttreatment, and mRFP-mediated chromatin fluorescence was recorded of >50 abnormal cells in their live state (Fig. 3a) and during hypotonic treatment (Fig. 3b). After in situ fixation with MAA, mRFP fluorescence was lost. Identical cells were recorded a third time after DAPI staining of DNA (Fig. 3c) and again after staining with the TUNEL test (Fig. 3d). These experiments allowed the following conclusions: faintly stained shadow-like DNA patterns in MAA-fixed samples are the remains of apoptotic nuclei. The faint DAPI staining is likely due to a loss of a considerable fraction of excessively fragmented DNA in apoptotic nuclei during MAA fixation. Shadow-like patterns consistently showed positive TUNEL staining indicating extensive DNA fragmentation (Figs. 3d, 4a). GCS events on the other hand were derived from PALCCs. GCS located side by side with TUNEL-positive shadow-like patterns were consistently TUNEL-negative, arguing against a massive DNA fragmentation (Figs. 3d, 4b).
Sequential epifluorescence images taken from a V79-H2BmRFP culture 16 h after UV irradiation/caffeine posttreatment show the same cells in their live state (a), during hypotonic treatment (b), after MMA fixation (c), and after a TUNEL test (d). Fluorescence in a and b was recorded from H2BmRFP (red); in c and d mRFP fluorescence of chromatin was abrogated by acetic acid fixation and replaced by imaging of DAPI-stained DNA (red). a Two cells with PALCCs are framed by purple and green boxes; the blue box includes an apoptotic cell nucleus. Additional cells without boxes show inconspicuous interphase nuclei, micronuclei, and multilobulated nuclei. b, c The morphology of the two PALCCs and the apopotic cell nucleus is completely lost. PALCCs are replaced by GCS patterns with apparently pulverized chromatin after MAA fixation, whereas the chromatin of the apoptotic nucleus is hardly detectable as a shadow-like DNA pattern. The unframed interphase nuclei are somewhat larger but otherwise appear not much altered at this level of resolution. d The TUNEL test (green signal) is positive for the reminiscent shadow-like fluorescence of the apoptotic nucleus, but negative for GCS. Images were collected with a Zeiss Axiovert 200 M epifluorescence microscope. Scale bar 20 μm. Compare Fig. 4 for selected single cells
TUNEL assay on selected single cells. a Apoptotic cells in live-cell observations (left panel) and respective shadow-like nuclei after MAA fixation show positive staining in the TUNEL test. b PALCCs in live-cell observations (left panel) and respective shattered chromosome complements (GCS) after MAA fixation are not stained by the TUNEL assay. Red DAPI, green TUNEL. Images of the living cells were recorded with a Zeiss Axiovert 200 M epifluorescence microscope. Images of fixed cells represent z-projections of three consecutive confocal sections (ΣZ = 600 nm). Scale bar 5 μm
PALCCs represent abnormally shaped mitoses with failure of normal chromatin condensation
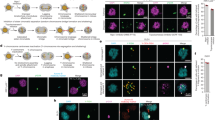
For a more detailed analysis of UV/caffeine-treated and PFA-fixed cells in comparison with untreated controls, we performed immunostaining against phosphorylated Serin-10 on histone H3 (H3pS10), a histone modification highly enriched during mitosis, against lamin B, an integral part of the nuclear envelope that disassembles during mitosis (Fig. 5a), against centromeres and α-tubulin, for the delineation of the mitotic spindle apparatus (Fig. 5b), and against cleaved caspase 3, a marker for apoptosis (Fig. 5c). Irrespective of the fact that PALCCs lacked distinct, condensed mitotic chromosomes (Fig. 5a, b, columns I, II), their mitotic status was supported by their conformance with normal mitoses (Fig. 5a, b, column III) in all other tested parameters. Like control mitotic cells, PALCCs showed strong positive staining for H3pS10 outlining the chromatin and disassembly of lamins typically observed in mitotic cells beyond the prophase stage (Fig. 5a, column I, II). α-Tubulin and centromeres revealed a normally shaped spindle apparatus, and most centromeres were found tethered to the spindle. However, instead of the typical arrangement of chromosomes and the spindle in normal mitotic cells, the spindle in PALCCs was often found at a distance up to several micrometers from the bulk chromatin with faintly stained chromatin fibers tapered towards the spindle-attached centromeres (Fig. 5b, column I, II). Centromeres in PALCC cells, as in normal mitotic cells, were attached to the spindle following the breakdown of the nuclear envelope. Presumably due to the failure of chromosome condensation, movements of centromeres in PALCCs pulled chromatin attached to the centromeres away from the bulk chromatin. In agreement with the results from the TUNEL test described above for shadow-nuclei and GCS in MAA-fixed samples, immunostaining of apoptotic nuclei after PFA fixation showed a strong staining of cleaved caspase 3, while PALCCs were consistently negative (Fig. 5c).
Correlation of PALCCs with mitotic cells (a, b). Columns I and II show two cells with typical PALCCs, column V a typical apoptotic cell nucleus recorded from a V79-H2BmRFP cell culture 16 h after UV irradiation/caffeine posttreatment. Columns III and IV present a normal metaphase and an interphase nucleus, respectively, from a control culture. From top to bottom, columns I–V in a show DAPI-stained DNA (1), immunostaining against H3pS10 (2), against lamin B (3), and merged images from 1–3 (4) (blue DAPI, red H3pS10, green lamin B). Note the correspondence of PALCCs with normal mitosis but not with normal interphase or apoptotic cells. Columns I–V in b show immunostaining against α-tubulin, a constituent of microtubules (1), and merged images with α-tubulin (red), centromeres (green), and DAPI-stained DNA (gray) (2). Columns I and II demonstrate the presence of mitotic spindles in cells with PALCCs. In contrast to a normal mitosis (column III) with its spindle arranged perpendicular to the plate, the bulky chromatin mass in PALCCs (columns I and II) locates at one side of the spindle apparatus, often several micrometers apart from it, and presents tethered centromeres. Column IV: Control interphase cell with centromeres visible in the nucleus and numerous bundles of microtubules expanding in the cytoplasm. Column V: Apoptotic cells lack a spindle and show only a few coarse degraded relicts of microtubules in the cytoplasm. Only a few centromeres are observed within the peripheral condensed chromatin. Images were taken with a confocal laser scanning microscope and represent single confocal sections (a) or z-projections of three consecutive confocal sections (ΣZ = 600 nm) (b). c 1–4 PALCC, 5–8 apoptotic cell. Importantly, immunodetection of cleaved caspase 3 (cl Casp 3) is negative in PALCCs (3), but strongly positive in apoptotic cells (7). In correspondence with images shown in a (columns I, II), H3pS10 outlines the PALCC (2); merged image (4) (green DAPI, blue H3pS10, red cleaved caspase 3). H3pS10 is negative in the apoptotic cell (compare a, column V) (6), but cleaved caspase 3 shows intensive staining (7); merged image (8) (green DAPI, blue H3pS10, red cleaved caspase 3). Images are taken by widefield epifluorescence microscopy. Scale bar 5 μm
We considered the possibility that PALCCs may not only represent abnormal mitotic events, but also be derived from abnormal premature chromosome condensation (PCC) of interphase cells, in particular from PCC of S-phase cells, which show a pulverized chromosome phenotype strikingly similar to GCS (Sperling and Rao 1974). Since PALCCs resulting from abnormal S-phase PCC should be able to incorporate BrdU when pulse-labeled with this thymidine analogue (Gotoh 2007 and our own unpublished data), 16 h after UV irradiation/caffeine posttreatment V79 cultures were treated with BrdU for 15 min and further grown for 70 min before MAA fixation (Fig. 6). About 50% of all nuclei with normal morphology had incorporated BrdU (data not shown) and could thus be assigned to S phase. However, BrdU incorporation was never observed in PALCCs (n = 30) (Figs. 6, 7), strongly indicating that PALCCs/GCS do not represent cells with PCC during S-phase.
Live-cell observation started 16 h after UV irradiation/caffeine posttreatment for identification of S-phase in PALCC/GCS after BrdU pulse labeling. Selected time points before, during, and after BrdU pulse labeling (1–6), after MAA fixation (7), and after immunodetection of BrdU (8) are displayed. PALCC formation is shown for cells emerging before (white and violet frames in 1), during (yellow frame in 2), or after BrdU pulse labeling (blue frame in 4). GCS after MAA fixation is shown for two cells (yellow and blue frame) (7). All cells with PALCC formation lack BrdU incorporation (8). Red DAPI, green BrdU. Scale bar 20 μm
Selected nuclei from the live-cell observation (compare Fig. 6) with PALCC formation during the observation period show consistent lack of BrdU incorporation. For comparison, an apoptotic cell, which is also negative for BrdU, is shown in the right panel. Time points refer to the BrdU pulse. Red DAPI, green BrdU. Scale bar 5 μm
Chromosome painting with chromosome-specific painting probes was performed in PFA- and MAA-fixed specimens to elucidate whether differentially painted chromosomes in PALCCs and GCS maintained distinct territories or became highly intermingled (Fig. 8). In control samples, painting probes clearly delineated distinct chromosomes in metaphase spreads and inconspicuous chromosome territories in interphase nuclei (Fig. 8a). This territorial organization was maintained in PALCCs (Fig. 8b). Occasionally, painting included dots along chromatin fibers extending from the bulk chromatin. Chromosome painting of GCS (Fig. 8c) yielded distinctly stained patterns of shattered chromosomes with some apparent disintegration or discontinuity. However, due to the limited sensitivity of chromosome painting probes, we were not able to distinguish between strong decondensation of segments, which therefore remained unstained, and a limited number of “true” fragments resulting from double-strand breaks. These results further support the conclusion that a general chromosome condensation failure underlies the formation of PALCCs and GCS. This failure becomes obvious in vivo when centromeres are pulled out from the bulk chromatin and in vitro after chromatin spreading.
Three-color painting of Chinese hamster chromosomes 1 (green), 2 (blue), and 5 (red) in V79-H2BmRFP cells. a Chromosome painting of MAA-fixed control metaphase spreads (top) reveals two homologues of each chromosome. The probe for #5 painted an additional fragment on an unidentified chromosome in all V79 cells, delineating a derivative chromosome in this cell line (arrow). Control interphase nuclei (bottom) show the typical territorial chromosome organization. b PALCCs in PFA-fixed cultures after UV/caffeine treatment show a predominantly territorial organization of painted chromosomes. Note some labeled dots along the chromatin fibers expanding from the bulk chromatin. c Painted GCS in MMA-fixed samples revealed areas predominantly stained in green, red, and blue. Accordingly, despite the shattered chromosome morphology, GCS still reflects the territorial chromosome organization of PALCCs. Images in panels a and c were recorded by widefield epifluorescence microscopy; images from panel b are z-projections from confocal images comprising ΣZ = 600 nm. Scale bar 5 μm
Chromosome painting in apoptotic nuclei of PFA-fixed samples suggested a complete destruction of the territorial chromosome organization in concordance with excessive chromatin and DNA fragmentation (Fig. 12_SONa in the “Electronic supplementary material”). In the shadow-like DNA patterns, which correspond to apoptotic nuclei in MAA-fixed samples, we observed an almost complete lack of hybridization signals likely due to a severe loss of DNA (Fig. 12_SONb). Taken together, our FISH data strongly emphasize the profound difference between chromatin structure in apoptotic nuclei and shadow-like DNA patterns on the one side and PALCCs and GCS on the other.
Fate of mitotic cells with parachute-like chromatin configurations
Finally, we monitored V79 cells expressing H2BmRFP in vivo in order to compare the fate of abnormal PALCC mitotic cells with mitotic cells from untreated cultures (Fig. 9). Long-term in vivo imaging of control cultures showed undisturbed cell growth with an average cell cycle duration of about 15 h, inconspicuous morphology of interphase nuclei (<1% of apoptotic cells), and mitoses arising over an observation period of about 48 h (Fig. 9a, c and SON movie_1 in the “Electronic supplementary material”). Figure 9b and SON movie_2 show an example of the typical process for PALCCs in irradiated cells. PALCCs became visible at the earliest 6 h after UV irradiation and caffeine posttreatment (data not shown) and always emerged directly from prophase cells of apparently normal appearance. Figure 9b (frame 2) shows an inconspicuous presumable G2 nucleus recorded 560 min after UV irradiation. Ten minutes later (frame 3), this cell had entered early prophase showing the typical pattern of prophase chromosomes starting to condense. An additional 10 min later (frame 4), the chromatin acquired a PALCC shape, which was maintained for about 30–45 min and then formed a single interphase cell with an apparently multilobulated nucleus (frames 6–7), demonstrating that abnormal mitotic cells with PALCC were able to proceed to G1 interphase within a time frame similar to normal mitotic cells. Yet these abnormal cells were in most cases subject to apoptosis after 4–12 h (Fig. 9b, frame 10 and Fig. 9d).
Live-cell observations of mitotic events in normal and UV/caffeine-treated V79-H2BmRFP cells. a Long-term observation (30.5 h) of untreated control cells (for complete image sequence over 48 h, see movie_1 in the “Electronic supplementary material”). Selected frames 1–10 demonstrate inconspicuous nuclear morphology and mitotic events completed within about 1 h. For example, white arrows indicate a mother nucleus (2), mitosis (3), and two daughter nuclei (4). One daughter cell undergoes a subsequent mitosis about 11 h later (5, 6; green arrows). The other daughter cell follows 45 min later (7, 8, yellow arrows). b Long-term observation (~16 h) of an irradiated cell (for complete image sequence, see movie_2). Frames 1–10 show a cell with a nucleus of inconspicuous morphology (1, 2), which enters prophase 9.5 h after UV irradiation and caffeine posttreatment (3), forms a typical PALCC during the following 10 min (4), and after another 40 min a cell with a single multilobulated nucleus (6). This cell remains for another ~5 h (7, 8) before entering apoptosis (9, 10). Images were recorded with a Zeiss Axiovert 200 M epifluorescence microscope. Scale bar 10 μm. c Time intervals between two consecutive mitoses measured for V79-H2BmRFP control cells (n = 9). d Time intervals between the formation of PALCCs and the onset of apoptosis (n = 23). Comparison of c and d indicates that apoptosis happens during interphase
Multilobulated nuclei showed a reconstituted lamina, lack of H3pS10 (Fig. 10a), and microtubular filaments re-organized as a filamentous structure in the cytoplasm (Fig. 10b), corresponding to typical features of interphase nuclei. Remarkable however was the restriction of centromeres to one or two lobuli of the reconstituted multilobulated nucleus (Fig. 10b). This conformation with centromere clustering is likely a consequence of failed segregation. Chromosome painting of multilobulated nuclei revealed maintenance of the territorial structure of chromosomes (Fig. 10c). Taken together, these observations demonstrate that mitotic cells with PALCCs, in spite of their failure of chromosome segregation and cell division, were still able to generate a single highly abnormal interphase cell within the time range necessary to complete a normal mitotic event. This finding further supports our conclusion that PALCCs preceding the formation of multilobulated nuclei contain insufficiently condensed but otherwise still intact chromosomes.
Multilobulated nuclei derived from abnormal mitotic cells with PALCCs. a Multilobulated nucleus with DAPI-stained DNA (1) stains positive for lamin B (2) and negative for H3pS10 (3); merged image (4) (blue DAPI, green lamin B, red H3pS10). DAPI staining shows a typical interphase pattern with more intensely stained chromatin at the nuclear envelope and around nucleoli. b Two multilobulated nuclei demonstrate cytoplasmic microtubules typical for interphase (1, 3) and clusters of centromeres (green) in one or two DAPI-stained nuclear lobes (gray), while centromeres are virtually absent in other lobes (2, 4); microtubules stained by α-tubulin shown here in red (for comparison see Fig. 5b, columns I, II, frame 2). c Two DAPI-stained multilobulated nuclei (1, 3) with painted chromosomes 1, 2, and 5 (2, 4) clearly demonstrate chromosome territories. Images were taken with a confocal laser scanning microscope and represent single confocal sections (a), z-projections of three consecutive confocal sections (ΣZ = 600 nm) (b), or z-projections of six confocal sections (ΣZ = 1.2 μm) (c). Scale bar 5 μm
Discussion
Mitotic cells with PALCCs/GCS: a special case of mitotic catastrophe followed by postmitotic apoptotic cell death
To our knowledge, this is the first study addressing the dynamic process of chromosome shattering detected in cultured Chinese hamster cells after UV irradiation and caffeine posttreatment. Based on live-cell observations in combination with immunohistochemistry and FISH, we demonstrate that abnormal mitotic cells with parachute-like chromatin configurations (PALCCs) were the in vivo structures from which generalized chromosome shattering (GCS) was obtained after methanol/acetic acid fixation (MAA) and air-drying by artificial spreading of this frail chromatin structure. We conclude that PALCCs are built up from chromatin with a defective condensation or tensegrity. PALCCs are enriched in H3pS10, a hallmark for mitosis (see below for further discussion excluding an interphase origin of cells with PALCCs), and surrounded by disassembled lamin, but do not contain typically condensed mitotic chromosomes. Chromatin fibers extending from the bulk chromatin of PALCCs contain functional centromeres tethered at a remote, but normally shaped spindle apparatus. Traction forces produced by the spindle apparently removed attached centromeres and extracted the chromatin fibers from the bulk chromatin. The inconspicuous spindle formation confirms earlier observations from in vitro experiments that lamin disassembly and spindle formation can obviously be achieved autonomously from chromatin condensation (Newport and Spann 1987). The temporal dissociation between chromosome condensation and succeeding breakdown of the nuclear envelope has been described by Beaudouin et al. (2002). Although PALCCs demonstrate a form of “mitotic catastrophe” that prevents proper chromosome segregation and cell division, the abnormal mitotic cells are still able to proceed to an abnormal interphase cell with a multilobulated nucleus before they suffer a postmitotic cell death by classical apoptosis. The dynamics of this process is schematically summarized in Fig. 11.
The development of “mitotic catastrophes” into multilobulated nuclei has been previously observed by a number of studies (for review, see Blank and Shiloh 2007; Roninson et al. 2001). Our findings are, however, inconsistent with some previously suggested chronologies of chromosome fragmentation and mitotic cell death (for comprehensive reviews, see Blank and Shiloh 2007; Roninson et al. 2001), but it must be emphasized that processes observed in different cell types and species summarized under the heading “mitotic catastrophe” may indeed differ substantially from each other. Blank et al. (2006) and Ianzini and Mackey (1997) have argued for a major role of premature chromosome condensation (see below), while Nitta et al. (2004) pointed out a delayed arrest in mitosis before cell death occurs. Stevens et al. (2007) proposed a model that distinguishes between chromosome fragmentation and mitotic catastrophe. In early mouse ATR−/− blastocysts carrying a double knock-out of the kinase ATR, Brown and Baltimore (2000) observed mitotic activity until day 2, followed by a caspase-independent chromosome fragmentation and finally caspase-dependent apoptosis after day 3. The phenotypic similarity of these observations with our findings is striking, but presently we cannot say whether the mechanisms are also similar or even identical.
The impact of fixation methods on chromatin structure
Our finding that MAA but not PFA fixation of mitotic cells with PALCCs results in GCS represents an extreme example for the different outcomes of observable chromatin structure depending on the fixation method. PFA fixation generates a matrix of insolubilized proteins cross-linked with trapped nucleic acids (Kiernan 2000) and was shown to preserve to a large extent the 3D morphology and large-scale chromatin structures of living cells when air-drying is carefully avoided at all times (Hepperger et al. 2007; Solovei et al. 2002). Large-scale preservation is in agreement with our observation of highly similar nuclear phenotypes between live cells and PFA-fixed cells in contrast to the MMA fixation protocol. The latter is routinely applied for the preparation of metaphase chromosome spreads and includes incubation in a hypotonic solution before fixation and air-drying thereafter. Methanol precipitates nucleic acids and proteins by dehydration, and the concomitant shrinking is compensated by hypotonic pretreatment. Finally, by adding acetic acid, proteinaceous structures are further destroyed (Bignold 2002). MAA fixation combined with air-drying leads to severe flattening of nuclei and to the well-known spreading of chromosomes in mitotic cells (Hepperger et al. 2007). GCS can thus be considered as an MAA-induced artificial spreading of chromatin, but reflects at the same time the high sensitivity of unmatured chromatin present in PALCCs.
To our knowledge, previous reports demonstrating chromosome shattering were all based on observations of MAA-fixed cells (Bezrookove et al. 2003; Brown and Baltimore 2000; Cremer and Cremer 1986; Cremer et al. 1980a; Hittelman and Pollard 1984; Lovelace 1954; Nghiem et al. 2001; Stevens et al. 2007), although the dependence of this phenotype on the fixation protocol was never addressed. We emphasize that fixation methods should be taken into account with regard to the observable chromatin structure and the possibility that different fixation protocols can lead to discrepant results should be considered.
PALCCs do not originate from premature chromosome condensation
Premature chromosome condensation (PCC) can be induced at any stage of the cell cycle (Schlegel and Pardee 1986). G1-PCC single chromatids and G2-PCC chromosomes are typically longer than their mitotic counterparts. In S-phase cells, however, PCC yields chromatin configurations that strongly resemble GCS (Sperling and Rao 1974). PCC can be triggered by fusion of an interphase cell with a mitotic cell (Sperling and Rao 1974), by chemical agents (Gotoh 2009), in cell systems with a double knock-out of the kinase ATR (ATR−/−) (Brown and Baltimore 2000), or after ATR inhibition (Nghiem et al. 2001). Since in our experiments we posttreated UV-irradiated cells with caffeine, a drug known to enhance DNA damage effects possibly by the inhibition of ATR (Blasina et al. 1999; Johansson et al. 2006), we considered the possibility that cells with PALCCs/GCS might emerge from interphase cells with abnormal PCC induction. Enrichment of H3pS10, a hallmark for mitosis (Hendzel et al. 1997), was observed in cells with PALCCs but cannot be used as a stringent criterion to distinguish between PALCCs arising from PCC or from abnormal mitosis since H3pS10 is also enriched in cells with PCC (Huang et al. 2006 and our own unpublished data).
Three further observations, however, strongly support that PALCCs represent a mitotic event: (1) Cells with PALCCs harbor a normal mitotic spindle, whereas failure of mitotic spindle formation was observed for chemically induced PCC (Ghosh et al. 1992 and our own unpublished data). (2) PALCCs/GCS lack BrdU incorporation after BrdU pulse labeling prior to fixation. While a significant fraction of PCCs shows BrdU staining (Gotoh 2007 and our own unpublished data), cells yielding GCS always remained unlabeled, although a large fraction of nuclei with BrdU patterns typical for different stages of S-phase was observed in the same culture. This excludes the contribution of PCC during S-phase to the formation of PALCCs and GCS. (3) We can further exclude the possibility that MAA fixation of apopotic cells contributed to the GCS phenotype. MAA fixation of apoptotic cell nuclei resulted in faintly stained chromatin shadows positive for TUNEL staining, while GCS showed a more grainy, pulverized chromatin structure and consistent negative TUNEL staining.
Our conclusion that GCS represents a true mitotic event is consistent with the results of early experiments from the Cremer group described above, who based this conclusion on the G2 DNA content in cells with GCS (Cremer and Gray 1982). In addition, GCS was obtained with peak frequencies after 9–16 h, i.e., at times when cells irradiated during G1/early S-phase were expected to go into mitosis (Cremer and Cremer 1986; Cremer et al. 1980a, 1981a, b).
The factor depletion model: an experimentally testable, theoretical framework to explain chromosome condensation failure and delayed cell death
The induction of GCS, not only by whole-cell UV irradiation but also after UV microirradiation restricted to a small nuclear area, emphasized the possibility of clastogenic effects triggered not only directly in irradiated chromatin but also in unirradiated territories remote from the site of nuclear microirradiation. In order to explain chromosome shattering within a theoretical framework, the Cremer group has proposed a factor depletion model (Cremer and Cremer 1986; Cremer et al. 1980a, b; for review see Cremer and Cremer 2006). This model predicts that a limited pool of proteins is directly involved in both postreplicative DNA repair of UV damage and an unknown process of postreplicative chromatin maturation necessary to allow proper chromosome condensation during prophase. GCS occurs when the factor pool becomes depleted because of an excess of competing DNA repair sites to such an extent that factors are no longer available at sites of postreplicative chromatin maturation. According to the factor depletion model, it is simply the number of these sites irrespective of their nuclear distribution. This assumption made it possible to explain GCS obtained after partial UV irradiation and caffeine posttreatment.
When the model was first proposed in the 1980s, caffeine was considered as an inhibitor of postreplication repair (Van Den Berg and Roberts 1976). Accordingly, it was suggested that unknown proteins involved in both postreplication repair and postreplicative chromatin maturation might be candidates for factors predicted by this model. Yet the necessary technology to test this prediction was not available 20 years ago. More recently, caffeine was described as an inhibitor of the kinases ATM and ATR (Blasina et al. 1999; Sarkaria et al. 1999). These kinases play an important role in the G1/S and G2/M checkpoints, which allow only correctly replicated mature interphase chromosomes to proceed into mitosis (Lin and Dutta 2007). DNA damage results in activation of ATM and/or ATR, which then leads to cell-cycle arrest, either via p53, p21, and cdk2/cyclinE (G1/S-checkpoint) or p53, p21, and cdk1/cyclinA or via chk1 and chk2, cdc25c and cdk1/cyclinB (G2/S-checkpoint) (Blasina et al. 1999; Hurley and Bunz 2007; Pollard and Earnshaw 2004; Sarkaria et al. 1999). Although the assumption that caffeine inhibits ATM and ATR was challenged by Cortez (2003), we favor the idea that PALCCs/GCS occur in cells, which proceed into S-phase and mitosis despite severe DNA damage as a result of the inhibitory effect of caffeine on the above-mentioned signaling pathways. The effect of caffeine on G2/M progression was already observed by Lau and Pardee (1982) and Musk et al. (1988). Furthermore caffeine was shown to inhibit the rescue of stalled replication forks by translesion DNA synthesis, thereby causing a switch to bypass via homologous recombination (Johansson et al. 2006), a mechanism that involves numerous DNA repair complexes (for review, see Hoeijmakers 2001). This observation seems also to be correlated with the inhibition of ATM and ATR, as it was shown that chk1 is necessary for the degradation of PCNA, which is crucial for translesion DNA synthesis (Johansson et al. 2006).
The lack of proper chromosome condensation starting during prophase could reflect either a failure of postreplicative chromatin maturation during G2 or a lack of proteins that induce chromatin condensation after entry into mitosis. Both hypotheses are consistent with our present findings and the factor depletion model and provide starting points for a search for candidate proteins.
We now know that both DNA repair and chromatin condensation during mitosis involve alterations of chromatin structure (for review, see Karagiannis and El-Osta 2007). Chromatin decondensation seems a general response to DNA damage, possibly due to the relocation of damaged DNA to the perichromatin region, to allow DNA repair in this functional nuclear compartment residing at the surface of compact chromatin domains and lining the interchromatin space (Solimando et al. 2009). In recent years genes have been identified that code for proteins involved both in DNA repair as well as in mitosis-related chromatin condensation. Such proteins include condensins and cohesins, protein complexes with a core of SMC proteins, that are essential in chromosome condensation at mitosis but also in checkpoint-control pathways (Chen et al. 2004; Legagneux et al. 2004; Lehmann 2005).
An implicit assumption of the factor depletion model is that cell death evolved as an adaptive trait. For example, the survival of cells with a high mutational load due to unrepaired DNA damage may yield an unacceptable risk of tumor development. It therefore makes sense that in addition to apoptosis other mechanisms evolved to eliminate potentially dangerous cells. We assume that a given pool size reflects an optimal adaptive balance between selective disadvantages brought about by the loss of too many cells—in the case of a too small pool size—and the survival of too many potentially dangerous cells. Accordingly, the apparently simple solution to increase the pool size of critical factors beyond an optimal limit was prevented by selection.
The formation of mitotic cells with PALCCs/GCS by the synergistic effect of UV irradiation and caffeine posttreatment—we observed a similar effect of GCS when cells with BrdU-labeled DNA were posttreated with caffeine (unpublished data)—depends on an artificial experimental situation that does not exist in natural evolution. Our experiments should thus be understood as model cases for other situations that triggered the evolution of chromosome condensation failure as an adaptive mechanism. The factor depletion concept provides a general theoretical framework applicable to different types of DNA repair, where competition for limited factor pools could result in different types of delayed cell death. Recently, Greubel et al. (2008) built an ion microbeam for studies of the nuclear topography and kinetics of double-strand break (DSB) repair at the single-cell level. They showed that a first and a second delayed single ion exposure at different nuclear sites led to comparable accumulations of phospho-ATM, γ-H2AX, and Mdc1 at both earlier (e) and later (l) microirradiated sites. In contrast, accumulations of 53BP1 and the recombination protein Rad51 were strongly reduced at l-sites. This apparent competition effect was accompanied by a reduced amount of 53BP1 in undamaged areas of the irradiated nuclei. These results could also be interpreted by a factor depletion model where a critically limited pool size for certain factors involved in DSB repair combined with strong binding at irradiated sites leads to the exhaustion of unbound factors and thus to an undersupply of factors at l-sites. γ-H2AX, a histone modification well known for its essential function in DSB repair, has recently been suggested to have additional functions related to higher-order chromatin organization by the recruitment of chromatin remodeling complexes (Ismail and Hendzel 2008). This may explain the requirement of H2AX phosphorylation during mitosis, which depends on ATM (McManus and Hendzel 2005). Inhibition of ATM by caffeine could thus critically reduce the allocation of γ-H2AX for proper chromosome condensation since it is already depleted by preceding DNA damage events.
Abbreviations
- FISH:
-
Fluorescence in situ hybridization
- GCS:
-
Generalized chromosome shattering
- H2BmRFP:
-
Histone H2B conjugated to red fluorescent protein
- H3pS10:
-
Histone H3 with phosphorylated serine at position 10
- MAA:
-
Methanol/acetic acid
- PALCC:
-
Parachute-like chromatin configuration
- PCC:
-
Premature chromosome condensation
- PCS:
-
Partial chromosome shattering
- PFA:
-
Paraformaldehyde
- SON:
-
Supplemental online
References
Beaudouin J, Gerlich D, Daigle N, Eils R, Ellenberg J (2002) Nuclear envelope breakdown proceeds by microtubule-induced tearing of the lamina. Cell 108:83–96. doi:10.1016/S0092-8674(01)00627-4
Bezrookove V, Smits R, Moeslein G, Fodde R, Tanke HJ, Raap AK, Darroudi F (2003) Premature chromosome condensation revisited: a novel chemical approach permits efficient cytogenetic analysis of cancers. Genes Chromosomes Cancer 38:177–186. doi:10.1002/gcc.10268
Bignold LP (2002) Hypothesis for the influence of fixatives on the chromatin patterns of interphase nuclei, based on shrinkage and retraction of nuclear and perinuclear structures. Br J Biomed Sci 59:105–113
Blank M, Shiloh Y (2007) Programs for cell death: apoptosis is only one way to go. Cell Cycle 6:686–695
Blank M, Lerenthal Y, Mittelman L, Shiloh Y (2006) Condensin I recruitment and uneven chromatin condensation precede mitotic cell death in response to DNA damage. J Cell Biol 174:195–206. doi:10.1083/jcb.200604022
Blasina A, Price BD, Turenne GA, McGowan CH (1999) Caffeine inhibits the checkpoint kinase ATM. Curr Biol 9:1135–1138. doi:10.1016/S0960-9822(99)80486-2
Brown EJ, Baltimore D (2000) ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev 14:397–402
Chen ES, Sutani T, Yanagida M (2004) Cti1/C1D interacts with condensin SMC hinge and supports the DNA repair function of condensin. Proc Natl Acad Sci USA 101:8078–8083. doi:10.1073/pnas.0307976101
Chowdhury I, Tharakan B, Bhat GK (2006) Current concepts in apoptosis: the physiological suicide program revisited. Cell Mol Biol Lett 11:506–525. doi:10.2478/s11658-006-0041-3
Chu EH (1965) Effects of ultraviolet radiation on mammalian cells. I. Induction of chromosome aberrations. Mutat Res 2:75–94. doi:10.1016/0027-5107(65)90010-2
Cortez D (2003) Caffeine inhibits checkpoint responses without inhibiting the ataxia-telangiectasia-mutated (ATM) and ATM- and Rad3-related (ATR) protein kinases. J Biol Chem 278:37139–37145. doi:10.1074/jbc.M307088200
Cremer C, Cremer T (1986) Induction of chromosome shattering by ultraviolet light and caffeine: the influence of different distributions of photolesions. Mutat Res 163:33–40. doi:10.1016/0027-5107(86)90055-2
Cremer T, Cremer C (2006) Rise, fall and resurrection of chromosome territories: a historical perspective. Part II. Fall and resurrection of chromosome territories during the 1950s to 1980s. Part III. Chromosome territories and the functional nuclear architecture: experiments and models from the 1990s to the present. Eur J Histochem 50:223–272
Cremer C, Gray JW (1982) DNA content of cells with generalized chromosome shattering induced by ultraviolet light plus caffeine. Mutat Res 94:133–142. doi:10.1016/0027-5107(82)90175-0
Cremer C, Cremer T, Simickova M (1980a) Induction of chromosome shattering and micronuclei by ultraviolet light and caffeine. I. Temporal relationship and antagonistic effects of the four deoxyribonucleosides. Environ Mutagen 2:339–351. doi:10.1002/em.2860020304
Cremer T, Cremer C, Zimmer J, Zorn C (1980b) UV-micro-irradiation of Chinese hanster cells and posttreatment with caffeine: indication for clastogenic effects remote from the irradiation site. In: Altmann H, Riklis E, Slor H (eds) DNA-repair and late effects. NRCN Publication, Israel, pp 53–62
Cremer C, Cremer T, Jabbur G (1981a) Laser-UV-microirradiation of Chinese hamster cells: the influence of the distribution of photolesions on unscheduled DNA synthesis. Photochem Photobiol 33:925–928. doi:10.1111/j.1751-1097.1981.tb05514.x
Cremer C, Cremer T, Zorn C, Zimmer J (1981b) Induction of chromosome shattering by ultraviolet irradiation and caffeine: comparison of whole-cell and partial-cell irradiation. Mutat Res 84:331–348. doi:10.1016/0027-5107(81)90202-5
Cremer T, Cremer C, Baumann H, Luedtke EK, Sperling K, Teuber V, Zorn C (1982a) Rabl’s model of the interphase chromosome arrangement tested in Chinese hamster cells by premature chromosome condensation and laser-UV-microbeam experiments. Hum Genet 60:46–56. doi:10.1007/BF00281263
Cremer T, Cremer C, Schneider T, Baumann H, Hens L, Kirsch-Volders M (1982b) Analysis of chromosome positions in the interphase nucleus of Chinese hamster cells by laser-UV-microirradiation experiments. Hum Genet 62:201–209. doi:10.1007/BF00333519
Cremer M, Grasser F, Lanctot C, Muller S, Neusser M, Zinner R, Solovei I, Cremer T (2008) Multicolor 3D fluorescence in situ hybridization for imaging interphase chromosomes. Methods Mol Biol 463:205–239. doi:10.1007/978-1-59745-406-3_15
Ghosh S, Paweletz N, Schroeter D (1992) Failure of kinetochore development and mitotic spindle formation in okadaic acid-induced premature mitosis in HeLa cells. Exp Cell Res 201:535–540. doi:10.1016/0014-4827(92)90307-T
Gotoh E (2007) Visualizing the dynamics of chromosome structure formation coupled with DNA replication. Chromosoma 116:453–462. doi:10.1007/s00412-007-0109-5
Gotoh E (2009) Drug-induced premature chromosome condensation (PCC) protocols: cytogenetic approaches in mitotic chromosome and interphase chromatin. Methods Mol Biol 523:83–92
Greubel C, Hable V, Drexler GA, Hauptner A, Dietzel S, Strickfaden H, Baur I, Krucken R, Cremer T, Dollinger G, Friedl AA (2008) Competition effect in DNA damage response. Radiat Environ Biophys 47:423–429. doi:10.1007/s00411-008-0182-z
Hendzel MJ, Wei Y, Mancini MA, Van Hooser A, Ranalli T, Brinkley BR, Bazett-Jones DP, Allis CD (1997) Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma 106:348–360. doi:10.1007/s004120050256
Hepperger C, Otten S, von Hase J, Dietzel S (2007) Preservation of large-scale chromatin structure in FISH experiments. Chromosoma 116:117–133. doi:10.1007/s00412-006-0084-2
Hittelman WN, Pollard M (1984) Visualization of chromatin events associated with repair of ultraviolet light-induced damage by premature chromosome condensation. Carcinogenesis 5:1277–1285. doi:10.1093/carcin/5.10.1277
Hoeijmakers JH (2001) Genome maintenance mechanisms for preventing cancer. Nature 411:366–374. doi:10.1038/35077232
Huang X, Kurose A, Tanaka T, Traganos F, Dai W, Darzynkiewicz Z (2006) Sequential phosphorylation of Ser-10 on histone H3 and ser-139 on histone H2AX and ATM activation during premature chromosome condensation: relationship to cell-cycle phase and apoptosis. Cytometry A 69:222–229. doi:10.1002/cyto.a.20257
Hurley PJ, Bunz F (2007) ATM and ATR: components of an integrated circuit. Cell Cycle 6:414–417
Ianzini F, Mackey MA (1997) Spontaneous premature chromosome condensation and mitotic catastrophe following irradiation of HeLa S3 cells. Int J Radiat Biol 72:409–421. doi:10.1080/095530097143185
Ismail IH, Hendzel MJ (2008) The gamma-H2A.X: is it just a surrogate marker of double-strand breaks or much more? Environ Mol Mutagen 49:73–82. doi:10.1002/em.20358
Johansson F, Lagerqvist A, Filippi S, Palitti F, Erixon K, Helleday T, Jenssen D (2006) Caffeine delays replication fork progression and enhances UV-induced homologous recombination in Chinese hamster cell lines. DNA Repair (Amst) 5:1449–1458. doi:10.1016/j.dnarep.2006.07.005
Karagiannis TC, El-Osta A (2007) Chromatin modifications and DNA double-strand breaks: the current state of play. Leukemia 21:195–200. doi:10.1038/sj.leu.2404478
Kiernan J (2000) Formaldehyde, formalin, paraformaldehyde and glutaraldehyde: what they are and what they do. Microsc Today 8:8–12
Lau CC, Pardee AB (1982) Mechanism by which caffeine potentiates lethality of nitrogen mustard. Proc Natl Acad Sci USA 79:2942–2946. doi:10.1073/pnas.79.9.2942
Legagneux V, Cubizolles F, Watrin E (2004) Multiple roles of condensins: a complex story. Biol Cell 96:201–213. doi:10.1016/j.biolcel.2004.01.003
Lehmann AR (2005) The role of SMC proteins in the responses to DNA damage. DNA Repair (Amst) 4:309–314. doi:10.1016/j.dnarep.2004.07.009
Lin JJ, Dutta A (2007) ATR pathway is the primary pathway for activating G2/M checkpoint induction after re-replication. J Biol Chem 282:30357–30362. doi:10.1074/jbc.M705178200
Lovelace R (1954) Chromosome shattering by ultraviolet radiation (2650A). Pro Natl Acad Sci USA 40:1129–1135
McManus KJ, Hendzel MJ (2005) ATM-dependent DNA damage-independent mitotic phosphorylation of H2AX in normally growing mammalian cells. Mol Biol Cell 16:5013–5025. doi:10.1091/mbc.E05-01-0065
Meaburn KJ, Misteli T (2007) Cell biology: chromosome territories. Nature 445:379–781. doi:10.1038/445379a
Musk SR, Downes CS, Johnson RT (1988) Caffeine induces uncoordinated expression of cell cycle functions after ultraviolet irradiation. Accelerated cycle transit, sister chromatid exchanges and premature chromosome condensation in a transformed Indian muntjac cell line. J Cell Sci 90(Pt 4):591–599
Newport J, Spann T (1987) Disassembly of the nucleus in mitotic extracts: membrane vesicularization, lamin disassembly, and chromosome condensation are independent processes. Cell 48:219–230. doi:10.1016/0092-8674(87)90425-9
Nghiem P, Park PK, Kim Y, Vaziri C, Schreiber SL (2001) ATR inhibition selectively sensitizes G1 checkpoint-deficient cells to lethal premature chromatin condensation. Proc Natl Acad Sci USA 98:9092–9097. doi:10.1073/pnas.161281798
Nitta M, Kobayashi O, Honda S, Hirota T, Kuninaka S, Marumoto T, Ushio Y, Saya H (2004) Spindle checkpoint function is required for mitotic catastrophe induced by DNA-damaging agents. Oncogene 23:6548–6558. doi:10.1038/sj.onc.1207873
Okada H, Mak TW (2004) Pathways of apoptotic and non-apoptotic death in tumour cells. Nat Rev Cancer 4:592–603. doi:10.1038/nrc1412
Pollard TD, Earnshaw W (2004) Cell biology. Elsevier, Philadelphia
Roninson IB, Broude EV, Chang BD (2001) If not apoptosis, then what? Treatment-induced senescence and mitotic catastrophe in tumor cells. Drug Resist Updat 4:303–313. doi:10.1054/drup.2001.0213
Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S (2004) Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem 73:39–85. doi:10.1146/annurev.biochem.73.011303.073723
Sarkaria JN, Busby EC, Tibbetts RS, Roos P, Taya Y, Karnitz LM, Abraham RT (1999) Inhibition of ATM and ATR kinase activities by the radiosensitizing agent, caffeine. Cancer Res 59:4375–4382
Schlegel R, Pardee AB (1986) Caffeine-induced uncoupling of mitosis from the completion of DNA replication in mammalian cells. Science 232:1264–1266. doi:10.1126/science.2422760
Solimando L, Luijsterburg MS, Vecchio L, Vermeulen W, van Driel R, Fakan S (2009) Spatial organization of nucleotide excision repair proteins after UV-induced DNA damage in the human cell nucleus. J Cell Sci 122:83–91. doi:10.1242/jcs.031062
Solovei I, Cavallo A, Schermelleh L, Jaunin F, Scasselati C, Cmarko D, Cremer C, Fakan S, Cremer T (2002) Spatial preservation of nuclear chromatin architecture during three-dimensional fluorescence in situ hybridization (3D-FISH). Exp Cell Res 276:10–23. doi:10.1006/excr.2002.5513
Sperling K, Rao PN (1974) The phenomenon of premature chromosome condensation: its relevance to basic and applied research. Humangenetik 23:235–258. doi:10.1007/BF00272508
Stevens JB, Liu G, Bremer SW, Ye KJ, Xu W, Xu J, Sun Y, Wu GS, Savasan S, Krawetz SA, Ye CJ, Heng HH (2007) Mitotic cell death by chromosome fragmentation. Cancer Res 67:7686–7694. doi:10.1158/0008-5472.CAN-07-0472
Telenius H, Pelmear AH, Tunnacliffe A, Carter NP, Behmel A, Ferguson-Smith MA, Nordenskjold M, Pfragner R, Ponder BA (1992) Cytogenetic analysis by chromosome painting using DOP-PCR amplified flow-sorted chromosomes. Genes Chromosomes Cancer 4:257–263. doi:10.1002/gcc.2870040311
Van Den Berg HW, Roberts JJ (1976) Inhibition by caffeine of post-replication repair in Chinese hamster cells treated with cis platinum (II) diamminedichloride: the extent of platinum binding to template DNA in relation to the size of low molecular weight nascent DNA. Chem Biol Interact 12:375–390. doi:10.1016/0009-2797(76)90052-1
Zorn C, Cremer T, Cremer C, Zimmer J (1976) Laser UV microirradiation of interphase nuclei and post-treatment with caffeine. A new approach to establish the arrangement of interphase chromosomes. Hum Genet 35:83–89. doi:10.1007/BF00295622
Zorn C, Cremer C, Cremer T, Zimmer J (1979) Unscheduled DNA synthesis after partial UV irradiation of the cell nucleus. Distribution in interphase and metaphase. Exp Cell Res 124:111–119. doi:10.1016/0014-4827(79)90261-1
Acknowledgments
H. Strickfaden and M. Cremer were supported by CIPSM. The authors are grateful to Fritzi Beck for providing slides with immunostaining of lamins and H3pS10.
Author information
Authors and Affiliations
Corresponding authors
Additional information
This article has been submitted as a contribution to the festschrift entitled “Uncovering cellular sub-structures by light microscopy” in honour of Professor Cremer’s 65th birthday.
Electronic supplementary material
Below is the link to the electronic supplementary material.
249_2009_496_MOESM1_ESM.tif
Fig. 12_SONa, b Three-color painting of Chinese hamster chromosomes 1 (green), 2 (blue), and 5 (red) in V79-H2BmRFP cells. a Apoptotic cells in PFA-fixed cultures after UV/caffeine treatment show a diffuse hybridization signal with strong intermingling of fluorescence signals reflecting a true fragmentation of chromosomes with a concomitant loss of the territorial structure. b Shadow-like chromatin patterns in MAA-fixed cultures lack any detectable hybridization signals. Scale bar = 5 μm Supplementary material 1 (tif 3043 kb)
249_2009_496_MOESM2_ESM.mpg
movie_1 Complete image sequence of the live-cell observation of V79-H2BmRFP control cells showing undisturbed cell proliferation up to 48 h. Images are taken each 15 min. The blurring after time point 1,860 min is due to a slight shifting of the z-stage after changing the medium. Picture sharpness in the late time points decreases due to the increasing cell density that hampers the autofocus function. The appearance of some apoptotic cells beyond time point 41 h is most probably due to the increased cell density. Scale bar = 10 µm Supplementary material 1 (MPG 57526 kb)
249_2009_496_MOESM3_ESM.mpg
movie_2 Complete image sequence of the live-cell observation of V79-H2BmRFP after UV irradiation/caffeine posttreatment. Scale bar = 10 µm Supplementary material 2 (MPG 13601 kb)
Rights and permissions
About this article
Cite this article
Hübner, B., Strickfaden, H., Müller, S. et al. Chromosome shattering: a mitotic catastrophe due to chromosome condensation failure. Eur Biophys J 38, 729–747 (2009). https://doi.org/10.1007/s00249-009-0496-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00249-009-0496-z