Abstract
In a previous work we observed multilayered plate-like structures surrounding partially denatured HeLa chromosomes at metaphase ionic conditions. This unexpected finding has led us to carry out an extensive investigation of these structures. Our results show that plates can also be found in metaphase chromosomes from chicken lymphocytes. We have used atomic force microscopy (AFM) to image and investigate the mechanical properties of plates in aqueous solution. Plates are thin (~6.5 nm each layer) but compact and resistant to penetration by the AFM tip: their Young’s modulus is ~0.2 GPa and the stress required for surface penetration is ~0.03 GPa in the presence of Mg2+ (5–20 mM). Low-ionic strength conditions produce emanation of chromatin fibers from the edges of uncrosslinked plates. These observations and AFM results obtained applying high forces indicate that the chromatin filament is tightly tethered inside the plates. Images of metal-shadowed plates and cryo-electron microscopy images of frozen-hydrated plates suggest that nucleosomes are tilted with respect to the plate surface to allow an interdigitation between the successive layers and a thickness reduction compatible with the observed plate height. The similarities between denatured plates from chicken chromosomes and aggregates of purified chromatin from chicken erythrocytes suggest that chromatin has intrinsic structural properties leading to plate formation. Scanning electron micrographs and images obtained with the 200-kV transmission microscope show that plates are the dominant component of compact chromatids. We propose that metaphase chromosomes are formed by many stacked plates perpendicular to the chromatid axis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
It has been known for a long time that DNA molecules having an enormous length are packaged in metaphase chromosomes, but the detailed path of DNA within chromosomes is a major unresolved issue in molecular biology. In eukaryotic cells, DNA is associated with histone proteins forming nucleosomes (Wolffe 1998). Chromatin filaments are composed of nucleosome cores connected by a variable length of linker DNA that is associated with histone H1. Since, in the nucleosome core (Luger et al. 1997; Harp et al. 2000; White et al. 2001), the wrapping of 147 bp of DNA forming 1.7 turns around the histone octamer (H2A2H2B2H32H42) reduces only sevenfold the length of the DNA, it is obvious that the nucleosome filament must be folded to yield the high linear packing ratio [~10,000 (Sumner 2003)] and the high local DNA concentration [~0.17 g/ml (Daban 2003)] found in condensed chromosomes.
In principle, the dimensions of the nucleosome core (cylinder of 5.7 nm height and 11 nm diameter) are large enough to allow the visualization of these particles using transmission electron microscopy (TEM) and atomic force microscopy (AFM). Unfortunately, however, individual nucleosomes can only be seen in extended fibers prepared in low concentration buffers without divalent cations (Thoma et al. 1979; Leuba et al. 1994); in particular, nucleosomes are clearly visible in extended chromatin without linker histones (Yang et al. 1994). At cation concentrations approaching those found in vivo, TEM (Thoma et al. 1979; Bartolomé et al. 1994), cryo-electron microscopy (Cryo-EM; Robinson et al. 2006), and AFM (Zlatanova et al. 1994; Caño et al. 2006) results show that nucleosomes are not visible as separated units in the condensed fibers. More difficulties have been found for the visualization of the chromatin filament within the living cell. At present there are no clear imaging evidences demonstrating that the 30-nm chromatin fiber is present in vivo in the interphase nuclei (Horowitz-Scherer and Woodcock 2006; Tremethick 2007; van Holde and Zlatanova 2007).
These problems have led different laboratories to use diverse approaches to investigate the organization of nucleosomes in chromatin fibers. Two main kinds of structural models for the 30-nm chromatin fiber have been suggested from these studies. In crossed-linker models, the nucleosome filament forms a zigzag structure and linker DNA is extended in the fiber interior (Makarov et al. 1985; Subirana et al. 1985; Bordas et al. 1986; Williams et al. 1986; Leuba et al. 1994; Bednar et al. 1998; Mozziconacci and Victor 2003; Staynov and Proykova 2008). In the solenoid model (Widom and Klug 1985; Bradbury and Baldwin 1986), linker DNA is folded and the nucleosome filament forms a simple helix. Recently, Schalch et al. (2005) have obtained the X-ray crystal structure of a tetranucleosome with a short linker DNA and lacking histone H1; from this structure these authors have suggested a crossed-linker model in which two helices are formed by the stacking of nucleosome cores. The most compact models described so far for chromatin fibers are solenoids in which the nucleosomes of the successive helical turns are interdigitated (Bartolomé et al. 1994; Daban and Bermúdez 1998; Robinson et al. 2006; Wong et al. 2007; Kepper et al. 2008); whereas the typical solenoid contains ~6 nucleosomes per 11 nm fiber, the interdigitation considered in these models gives rise to densities up to 15 nucleosomes per 11 nm. In addition, it has to be taken into account that compact chromatin fibers may be discontinuous (Zentgraf and Franke 1984; Caravaca et al. 2005). It has also been proposed that less compact fibers with a zigzag folding are also capable of forming dense structures by lateral association involving interdigitation of nucleosomes (Grigoryev 2004; Engelhardt 2007). At high cation concentrations, model nucleosome arrays (Hansen 2002) and purified nucleosome core particles (Mangenot et al. 2003) give rise to diverse types of supramolecular aggregates.
Several structural models have been proposed for metaphase chromosomes. In the loop-sacaffold model (Paulson and Laemmli 1977; Marsden and Laemmli 1979; Saitoh and Laemmli 1994) and, generally, in other models including loops (Pienta and Coffey 1984; Manuelidis and Chen 1990; Filipski et al. 1990), the loops of 50–100 bp of DNA anchored to a protein scaffold form fibers of 30 nm which are further helically folded giving rise first to fibers of intermediate diameter (200–300 nm) and finally to the coiled metaphase chromatid (~600 nm). In the model of multiple helical coiling without loops of Taniguchi and Takayama (1986), the 30-nm chromatin fiber forms directly a superhelix of about 200 nm and then the coiled chromatid. In a more recent study about chromosome condensation during prophase, Kireeva et al. (2004) have suggested a model in which an intermediate 100–130 nm chromonema fiber folds into a 200–250 nm chromatin fiber; in metaphase this fiber forms the final chromatid, which is stabilized by axial condensin.
Nonhistone proteins may play different roles in the control of chromosomes structure during the cell cycle (Nasmyth and Haering 2005). Topoisomerase IIα and condensin I, which constitute the two main components of the scaffold in histone-depleted chromosomes (Maeshima and Laemmli 2003), condensin II which is also located in the chromosome axis (Ono et al. 2003), and cohesins (Ishiguro and Watanabe 2007) have been considered important elements for the assembly and segregation of metaphase chromosomes. Nevertheless, the exact structural role of these proteins is controversial (Belmont 2002; Gassmann et al. 2004). For instance, the results obtained by Poirier and Marko (2002a) in stretching experiments indicated that chromosomes from somatic cells have a mechanical integrity that is essentially dependent on DNA, and that the protein scaffold does not form a mechanically continuous backbone within chromatids. Furthermore, from the results obtained in studies performed with condensin-depleted cells it has been suggested that condensin is not essential for mitotic chromosome condensation (Hudson et al. 2003; Savvidou et al. 2005). Considering these apparently contradictory observations, in agreement with Hirano (2005) and Belmont (2006), it now seems clear that to elucidate the actual role of the different nonhistone proteins found in chromosomes it will be necessary to know the actual structure of chromatin within condensed chromatids.
As described above, all the structural models for metaphase chromosomes consider that chromatids are built with chromatin fibers. However, in a previous work of our laboratory (Caravaca et al. 2005), in which we investigated the bulk structure of HeLa metaphase chromosomes as a function of the ionic conditions, we found that chromatin fibers can only be detected by TEM when chromosomes are denatured by incubation with deionized water or with buffers containing extremely low cation concentrations. Similar low ionic strength conditions were used in the early experiments that led to the proposal of the initial fibrillar (DuPraw 1966) and loop-scaffold (Marsden and Laemmli 1979) models for chromosome structure. In contrast, we observed highly compact chromosomes in the presence of a wide range of cation concentrations. Moreover, after incubation at 37°C under these ionic conditions, we detected multilayered plate-like structures protruding from partially denatured HeLa metaphase chromosomes. Since the conditions in which we observed these planar structures include the cation concentrations (determined using ion microscopy by Strick et al. 2001) corresponding to metaphase chromosomes, we decided to carry out an extensive investigation of these structures, which had never been described before in the chromatin literature. The results obtained in this investigation open a completely new perspective on the structure of chromatin within metaphase chromosomes.
Materials and methods
Metaphase chromosomes and chromatin fragments
Chromosomes from HeLa cells were isolated and purified following previously described procedures (Caravaca et al. 2005). Cells were incubated at 37°C for 10 min with a hypotonic KCl (75 mM) solution, centrifuged at 275g for 5 min at room temperature, and then resuspended and Dounce-homogenized (15 times) in ice-cold TE buffer (15 mM Triethanolamine–HCl, pH 7.5, 2 mM EDTA, and 0.5 mM EGTA) containing 20 mM NaCl, 80 mM KCl, 0.2 mM spermine, 0.5 mM spermidine, and 1 mg/ml digitonin. The homogenate was centrifuged at 200g for 10 min at 4°C, and the pellet was homogenized and centrifuged using the same conditions. The two supernatants, containing the chromosomes, were pooled. As indicated in the text, in some experiments Triton X-100 (0.2% v/v) was used instead of digitonin; in other experiments detergents were omitted. The chromosome suspension was used directly in the electron microscopy experiments or was further purified using sucrose or glycerol step gradients in PE buffer (5 mM Pipes, pH 7.2, and 1 mM EGTA) containing 5 mM NaCl and 5 mM MgCl2. In some experiments, the purified chromosomes were dialyzed extensively against PE buffer containing 5 mM NaCl and 5 mM MgCl2, but without glycerol and sucrose, and mechanically disrupted by sonication, Polytron homogenization, or rapid passage through a 22-gauge syringe needle. Chicken chromosomes were obtained from peripheral blood lymphocyte cultures stimulated with phytohemagglutinin (Macgregor and Varley 1983). The preparation of chromosomes in TE buffer containing monovalent cations and polyamines was carried out following the procedure described for HeLa cells.
Chromatin fragments containing 20–30 nucleosomes were obtained from chicken erythrocytes as described elsewhere (Bartolomé et al. 1994; Caño et al. 2006). In some experiments, the chromatin samples were layered onto 5–20% sucrose gradients containing 10 mM Triethanolamine–HCl, pH 7.5, and 0.8 mM MgCl2 prepared in small tubes (4 ml) and centrifuged at 100,000g for 2 h at 4°C; the selected fractions corresponding to aggregates of relatively low molecular mass were crosslinked overnight at 4°C with 0.15% glutaraldehyde before the spreading and electron microscopy observation (Fig. 8b–d, h–j). In other experiments, the chromatin sample was dialyzed overnight at 4°C against 10 mM Triethanolamine–HCl, pH 7.5, and 0.8 mM MgCl2; the resulting samples were spread directly (without performing any previous centrifugation to remove aggregates) and crosslinked on the grid at room temperature with 2.5% glutaraldehyde for 2.5 h (samples corresponding to Fig. 8e–g, k).
Electron microscopy
The spreading of the chromosomes on carbon-coated grids was generally carried out by centrifugation. The grid was placed on top of a solidified layer of agarose (3%) or in the cap of an inverted tube, and the chromosome suspension above the grid was centrifuged at 4°C for 10 min at 1,500g. In some experiments mechanically disrupted chromosomes were spread by simple diffusion for 15 min to 48 h. In the indicated experiments, after spreading, the grids were placed on tubes containing 4 ml of the solutions described in the figure legends and incubated at 37°C for 30 min. Chromosomes were fixed directly on the electron microscopy grids with 2.5% glutaraldehyde for 10 min at room temperature. The spreading of chromatin samples was performed by direct deposition (Aragay et al. 1991). The grids with the glutaraldehyde-crosslinked extensions of chromosomes and chromatin were washed by immersion in a large volume of deionized water (0.5–4 ml) for 10–20 min, dehydrated in ethanol for 2–3 s, and allowed to dry on a filter paper. The results obtained with control samples prepared using the different conditions described below, but without chromosomes and chromatin, showed that the final washing with water eliminates all the salts (NaCl, KCl, and MgCl2) and other components of the buffers that after drying could produce crystalline precipitates. Generally, the grids were rotary-shadowed with platinum; in unidirectional shadowing experiments performed to measure the plate height, spherical latex particles (Bal-Tec) were added for the determination of the shadowing angle. Unless otherwise indicated carbon-coated copper grids were pretreated with Alcian blue. In some experiments the carbon film was activated by glow-discharge. Micrographs were obtained using the Hitachi transmission H7000 and scanning S570 electron microscopes and the Jeol 200-kV JEM-2011 and 120-kV JEM-1400 transmission microscopes. To facilitate the visualization of plate edges and other structures, images were prepared in negative contrast. Additional experimental details have been described elsewhere (Aragay et al. 1991; Bartolomé et al. 1994; Caravaca et al. 2005).
Cryo-EM
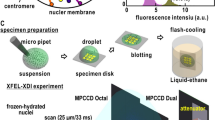
Frozen-hydrated preparations of mechanically disrupted chromosomes were obtained essentially following the general procedures described for aqueous suspensions of different biological materials (Dubochet et al. 1988; Bednar and Woodcock 1999). In these experiments we have used perforated Quantifoil R2/4 film (hole diameter ~2 μm) on a 400-mesh copper grid. After glow-discharge to make hydrophilic de holey film, a 5-μl drop of the sample was deposited onto the film, the grid was mounted on a plunger (Leica EM-CPC) and blotted with Whatman No. 4 filter paper. The aqueous suspension of chromatin plates within the holes was immediately vitrified by rapid immersion in liquid ethane. The specimen grid under liquid nitrogen was mounted on a Gatan 914 cryotransfer holder. Images were obtained with the Jeol JEM-1400 cryo-electron microscope operated at 100 kV, under low-dose conditions, and using different degrees of defocus (500–700 nm) to obtain an adequate phase contrast (Dubochet et al. 2007).
AFM
For the visualization of plates, chromosome preparations mechanically disrupted by passage through a syringe needle were adsorbed on freshly cleaved mica for 5 min and washed with the buffer corresponding to each experiment. Prior to use, the mica sheets (3 × 3 mm) were glued on top of a Teflon disc (9 mm diameter). For the visualization of whole chromosomes the freshly cleaved mica was placed in the cap of an inverted tube, and the chromosome suspension was spread by centrifugation as described above. Imaging was performed with a Nanoscope-IV Multimode microscope (Digital Instruments) equipped with a tapping-mode liquid cell and silicon nitride 100-μm long triangular cantilevers OMCL-TR400PSA (Olympus). All imaging and force spectroscopy experiments were performed in aqueous solution at room temperature. As expected, taking into account the high solubility of NaCl and MgCl2 in water, we have not observed crystalline precipitates in the control samples (prepared without chromatin and chromosomes) containing PE buffer and these salts at the concentrations used in our experiments (see legends of Fig. 5; Fig. S3 in the supplementary material).
The spring constants of the individual cantilevers used in our force spectroscopy studies were determined from thermal noise measurements performed with the MFP-3D microscope (Asylum Research); the values obtained were: 0.07 and 0.10 nN/nm for the tips used in experiments in the presence of 5 and 20 mM Mg2+, respectively (the nominal manufacture’s value is 0.08 nN/nm). The raw data plots (detector voltage vs. piezo displacement) were used to obtain the cantilever sensitivity (V/nm) and the corrected force curves showing the force versus tip displacement (Heinz and Hoh 1999). In these plots the force (F) values were calculated from the product of the cantilever spring constant and the deflection; the tip displacement was obtained by subtracting the cantilever deflection from the piezo displacement. The indentation (δ) of the tip into the surface of a soft sample can be modeled with the Hertz equation (Radmacher 2002): F = 4/3 E/(1 − ν2) R 1/2 δ3/2; where E is the elastic or Young’s modulus, ν the Poisson ratio (considered 0.5 in our calculations), and R the radius of the spherical tip apex used in our experiments (~15 nm, according to the manufacturer). We obtained the elastic modulus of chromatin plates under different ionic conditions by fitting this equation to the experimental values of F and δ in the region ranging from the point of the apparent beginning of the elastic indentation up to the point in which the abrupt plastic deformation is observed (see Fig. 6c). To obtain the stress at which the plate begins to exhibit this plastic behavior, we used the approximate contact area between the tip apex and the plate calculated according to Weisenhorn et al. (1993).
Results
SEM and 200-kV TEM images show that compact metaphase chromosomes contain multilayered plate-like structures
We have not used organic media [typically methanol–acetic acid (Sumner 1989)] for chromosome fixation. Furthermore, to avoid the relatively thick metal deposition on the surface of chromosomes produced by the current sputter coater instruments used in scanning electron microscopy (SEM), we coated the samples for these experiments with the thin platinum film produced by rotary shadowing; the same treatment was used to contrast TEM samples.
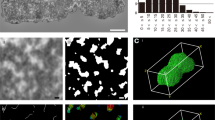
The overall morphology of chromosomes observed with SEM and TEM is similar, but the resolution obtained with the microscope operated at 200 kV (Fig. 1c, d, f) is higher than that obtained with the scanning microscope (Fig. 1a, b, e). Furthermore, the capability of the 200-kV microscope to image thick specimens has allowed us to obtain better images of condensed chromosomes than those obtained in our previous TEM study performed at low voltage (Caravaca et al. 2005). In agreement with this previous study, our results show that metaphase chromosomes in the presence of polyamines and Mg2+ do not have fibrillar morphology. Instead, our micrographs show many plate-like structures in the chromatids. Generally, the plates visible in the micrographs are apparently parallel to the chromatid axis. Presumably this is favored by the fact that chromosomes are soft structures (see below), even when they are condensed by the relatively high cation concentrations used in these experiments. The deposition of these soft structures on the electron microscopy grids causes the adsorption of the bottom layers to the carbon surface (see region d in Fig. 1f) and the orientation of the successive layers parallel to the carbon film. Nevertheless, we have observed plates with diverse orientations (see Fig. 1e, f) and in some regions (a, b, and c) it seems that plates are orthogonal to the chromatid axis.
Chromosomes from HeLa cells spread on carbon-coated grids were crosslinked with glutaraldehyde in 40 mM KCl, 0.1 mM spermine and 0.25 mM spermidine (a–e), and 5 mM NaCl and 10 mM MgCl2 (f). Chromosomes were visualized by SEM (a, b, e) and 200-kV TEM (c, d, f). The arrows in e and f point to plates with different orientations. Bars 500 nm
Multilayered plates can be seen protruding from HeLa chromosomes and chicken microchromosomes partially denatured using different procedures
The incubation at 37°C of metaphase chromosomes from chicken lymphocytes on the electron microscopy grid favors the extrusion of multilayered plates. Chicken chromosomes have a wide variety of sizes (International Chicken Genome Sequencing Consortium 2004). The TEM images presented in Fig. 2a and b correspond to partially denatured chicken microchromosomes. The plates extruded from these chromosomes show the same structural characteristics as those associated with HeLa chromosomes.
Examples of plates extruded from metaphase chromosomes of chicken lymphocytes and HeLa cells under different conditions. a, b Chicken microchromosomes associated with extruded plates; after spreading the samples were incubated for 30 min at 37°C on the grid with TE buffer containing 80 mM KCl, 20 mM NaCl and polyamines (0.2 mM spermine, 0.5 mM spermidine). HeLa chromosomes were purified on sucrose (c, f, g) and glycerol (d) gradients in PE buffer containing 5 mM NaCl and 5 mM MgCl2, dialyzed extensively against the same solution without sucrose and glycerol, and mechanically disrupted by passage through a syringe needle; in addition the sample corresponding to g was digested on the grid for 30 min at 37°C with 10 mg/mL of ribonuclease; the head arrows in c point to plates dissociated from a metaphase chromosome. e Plates from chicken metaphase chromosomes mechanically disrupted using the same procedure in TE buffer containing the same amount of monovalent cations and polyamines indicated above. The grid used in g was not activated with Alcian blue. TEM images were obtained with rotary-shadowed spreads, except d which was unidirectionally shadowed. Bars 200 nm (a, b, d–g); 600 nm (c)
Our results indicate that different mechanical treatments such as chromosome sonication, Polytron homogenization, and rapid passage through a syringe needle also favor the extrusion of plates. In particular, we observed that the mechanical stress produced by the latter procedure is able to split off part of the extruded plates from the chromatids (see Fig. 2c). Some examples of multilayered plates from metaphase chromosomes of HeLa cells and chicken lymphocytes mechanically disrupted, using a syringe needle, are presented in Fig. 2d–g. These plates have large areas with a very smooth surface, but we also observed large extrusions from mechanically disrupted chromosome containing many small plates (see Fig. S1A in the supplementary material). In most microscopic fields of our preparations of partially denatured chromosomes (from chicken lymphocytes and HeLa cells) we observed many multilayered plates of different sizes connected to chromatids and dissociated from them.
In order to exclude the possibility that the observed plates could be produced artefactually, we prepared our samples using diverse procedures. In our current preparation method we used digitonin and Dounce homogenization to break the cells, but we also observed plates in preparations performed using Triton X-100 instead of digitonin; we observed plates even in preparations without any detergent. Crude preparations of chromosomes without further purification on density gradients allowed us the visualization of plates (see examples in Fig. 2a, b, e). Plates were found in chromosome preparations containing high concentrations of sucrose or glycerol due to the purification on density gradients; we also observed plates when we used these chromosomes after an extensive dialysis to remove completely sucrose and glycerol (see examples in Fig. 2c, d, f, g; Fig. S1A). Plates remained unchanged when they were extensively digested with ribonuclease (Fig. 2g), indicating that RNA (which could be a chromosome component) is not involved in plate formation. Usually in our TEM and SEM experiments the spreading was by centrifugation, but in samples spread by simple diffusion we also observed plates (Fig. 3a). The carbon film on the electron microscopy grids was generally activated with Alcian blue. However, our results show that this treatment was not necessary for the observation of plates (Fig. 2g); we also observed plates using carbon films activated using the glow-discharge technique. Finally, although generally we carried out crosslinking of spread samples with glutaraldehyde, we detected plates even in uncrosslinked samples. In fact, in our AFM experiments (see below) plates were imaged without crosslinking. In addition, in the case of AFM the substrate was freshly cleaved mica instead of the carbon film used in our TEM and SEM experiments.
a TEM micrograph corresponding to plates spread on carbon-coated grids without centrifugation; the sample was adsorbed for 1 h. b, c Cryo-EM images of frozen-hydrated plates (unstained and unfixed). In b the Quantifoil film surrounding a hole with vitrified ice containing plates is indicated with an asterisk. Plates are shown at a higher magnification in c; large arrows point to the edges of some plates with several layers and small arrows point to the edges corresponding to monolayer plates. All the samples in this figure were prepared from HeLa chromosomes purified on sucrose gradients in PE buffer containing 5 mM NaCl and 5 mM MgCl2, dialyzed extensively against the same solution without sucrose, and mechanically disrupted by passage through a syringe needle. Bars 200 nm (a, b); 100 nm (c)
We have also observed plates in unfixed and unstained frozen-hydrated preparations. An example of Cryo-EM images of these samples is presented in Fig. 3. In these preparations there is no substrate and the plates in suspension in aqueous media are not oriented by any surface. Nevertheless, as can be seen in Fig. 3c, plates and in particular their edges are clearly distinguishable even in these preparations (see also Fig. 7). Table 1 summarizes all the assayed procedures and conditions leading to the observation of plates. This extensive study demonstrates that plates are native structures.
Extreme denaturing conditions cause the emanation of chromatin fibers from uncrosslinked plates
In order to characterize the plates we tried to find procedures capable of producing a significant denaturation, but without giving rise to a complete unfolding. In these studies we found that even uncrosslinked plates are highly stable. For instance, in the micrograph presented in Fig. S1B of the supplementary material, it can be seen that treatment of uncrosslinked plates on the electron microscopy grid with a relatively low concentration of the detergent SDS in the presence of Mg2+ produced a splitting into many small plates, but did not cause the complete disappearance of these planar structures.
We obtained a higher degree of denaturation by the incubation of the uncrosslinked plates with buffers of low ionic strength without divalent cations. Some micrographs obtained under these conditions are presented in Fig. 4. It can be seen that partially unfolded plates are associated with chromatin fibers of different compaction degrees emanating from them. Part of the surface of some plates (in particular see Fig. 4b) show unfolded layers with a grainy texture. The dimensions of the observed granules suggest that these unfolded layers could be formed by nucleosomes randomly placed on top of smooth layers. However, even under these denaturing conditions most of the visible layers have a compact texture.
Chromatin fibers emanating from plates partially denatured by treatment with solutions of low ionic strength. a–d HeLa metaphase chromosomes purified on sucrose gradients containing 5 mM NaCl and 5 mM MgCl2, were mechanically disrupted by passage through a syringe needle, spread on electron microscopy grids and incubated for 30 min at 37°C with 10 mM EDTA (pH 7.5) and 5 mM NaCl, crosslinked with glutaraldehyde and rotary shadowed. All TEM micrographs are at the same scale (bar 200 nm)
As can be seen in Fig. 4 and in Fig. S2 of the supplementary material, the fibers emanate from the edges of the plates, indicating that the chromatin filament is so tightly tethered inside the plates that it can only be liberated at the edges. We have also observed that chromatin fibers can interact laterally with the edges of the plates; in contrast, fibers can be on top of a plate without being absorbed by the chromatin in the top layer (see examples in Fig. S2). This suggests that the forces holding the chromatin filament within a layer are stronger than the interactions that exist between adjacent layers. As observed for different materials forming planar sheets (Callister and Rethwisch 2008), such affinity differences within and between layers are required for the formation of multilaminar structures. Furthermore, these observations are consistent with the sliding between layers that presumably occurs in the plates surrounding partially denatured chromatids (see “Discussion”). The differences in the length of relative sliding between layers probably facilitate the clear visualization of several stacked layers with well-defined edges in many plates (see for instance region d in Fig. 1f and Fig. 2e, g).
AFM experiments demonstrate that chromatin plates are stable in aqueous solution at room temperature
Even in the presence of divalent cations, metaphase chromosomes are very soft structures in aqueous solution. This, together with their large size, makes very difficult the imaging of chromosomes spread on mica by tapping-mode AFM; the resolution obtained using this technique with whole chromosomes is very poor (Fig. 5e). In contrast, plates adsorbed on mica can be imaged without any problem using tapping-mode AFM in the presence of 5 mM Mg2+ (Fig. 5a–d, g) and higher concentrations of this divalent cation (Fig. S3A–E, G in the supplementary material). In agreement with our TEM observations, the topographical images (Fig. 5a, c, g; Fig. S3A, C, E–G), the corresponding cross-section profiles (see examples in Fig. 5f), and in particular the amplitude images (Fig. 5b; Fig. S3B, D) indicate that the surface of plates is very smooth.
AFM study of plates from HeLa metaphase chromosomes in aqueous solution in the presence of 5 mM Mg2+. (a, c–e) Topographical images (tapping-mode); b Amplitude image of the same sample shown in a. g Three-dimensional image. The image in d was obtained after the application of a high force in the region marked with a square in c. f Height profiles generated along the lines ab, cd, and ef indicated in a, c, and d, respectively. The plates shown in a–d and g were obtained from chromosomes purified on sucrose gradients containing PE buffer, 5 mM NaCl and 5 mM MgCl2, dialyzed (at room temperature for 4 h) against the same solution without sucrose, mechanically disrupted by passage through a syringe needle, and then spread on mica and imaged without crosslinking. The chromosomes in e were spread by centrifugation on mica, crosslinked with glutaraldehyde, and washed with PE buffer without glucose containing 5 mM NaCl and 5 mM MgCl2. Bars: 200 nm (a–d); 2 μm (e)
Since all these images were obtained with uncrosslinked preparations, our results indicate that plates are stable by themselves in aqueous solution. In fact, plates have a mechanical stability high enough to allow contact-mode imaging (Fig. S3F). Furthermore, when higher forces were applied with the AFM tip to internal regions of different plates, there was no apparent change in their structure. In contrast, when high forces were applied to regions including the edges, the plates become severely damaged. One of such experiments is presented in Fig. 5c (image obtained before the high force was applied to the region marked with a square) and Fig. 5d (image obtained after the application of the force). These results are in agreement with our observations presented in the preceding section indicating that the chromatin filament is tightly bound in the internal regions of the plates and more prone to be unfolded in the periphery.
Chromatin plates are thin but resistant to penetration by the AFM tip
The apparent height of plates in aqueous solution containing 5 mM Mg2+ was obtained from cross-section profiles such as those shown in Fig. 5f. The histogram corresponding to these measurements is presented in Fig. 6d. The observed mean height of monolayer plates is 4.0 nm. This value is lower than that obtained by TEM in unidirectional shadowing experiments. Using this technique we have found that the mean height of monolayer plates is 7.4 nm (Fig. 6b) and the mean height of a step in multilayered plates is 5.3 nm (Fig. 6a). These results, which have been obtained from a large number of plates in 5 mM Mg2+, are similar to those found in our previous TEM measurements (Caravaca et al. 2005) performed with a lower number of plates under different conditions. The relatively low height found by AFM could be due to sample compression produced by the tip during imaging (Jiao and Schäffer 2004). However, the main cause of the apparent height reduction is probably related to the unorganized chromatin that is frequently observed around the plates (see Fig. 5; Fig. S3 in the supplementary material). In the presence of this chromatin background the tip cannot reach the mica surface in many measurements and consequently this may produce a reduction of the apparent height of the plates.
Height and mechanical properties of plates from HeLa metaphase chromosomes. Histograms of plate height obtained by TEM from unidirectional-shadowed preparations: a height of a step in multilayered plates (5.3 ± 1.8 nm, n = 26); b height of monolayer plates (7.4 ± 2.5 nm, n = 77). d Height histogram obtained by AFM measurements of monolayer plates in aqueous solutions (4.0 ± 0.9 nm, n = 100). All these height measurements were performed with plates prepared in PE buffer containing 5 mM NaCl and 5 mM MgCl2. c Examples of force versus tip displacement plots corresponding to chromatin plates in aqueous solutions containing PE buffer, 5 mM NaCl and the indicated Mg2+ concentrations. In 5 mM Mg2+, the first transition (c) corresponds to a penetration of 0.9 ± 0.4 nm that takes place at 2.0 ± 0.6 nN (n = 30); in the main transition (d) the penetration is 2.9 ± 0.6 nm and occurs at 4.4 ± 0.4 nN (n = 30). In 20 mM Mg2+ there is only one transition (c′) corresponding to a penetration of 2.5 ± 0.3 nm that occurs at 6.2 ± 0.7 nN (n = 32). The cantilever deflection is zero in regions a and a′; the regions b and b′ correspond to cantilever deflections produced by attractive and repulsive forces between the tip and the plate; in regions e and d′ the tip has reached the surface of the substrate and the deflection is very high
To obtain direct information about the mechanical properties of chromatin plates, we have performed detailed force measurements (see Fig. 6c). In 5 mM Mg2+, the corrected force curve indicates that, in addition to an apparent progressive deformation of the plate, there is a small transition at 2 nN (c, 0.9-nm jump), followed by a large abrupt transition (d, 2.9-nm jump) at 4.4 nN corresponding to the complete penetration of the tip through the plate. The force curve of plates in 20 mM Mg2+ is similar, but show a single abrupt transition (c′, 2.5-nm jump) at a higher force (6.2 nN). The stresses required for the initiation of the main transitions in 5 and 20 mM Mg2+ are 0.024 and 0.037 GPa, respectively. The attractive and repulsive forces between the tip and the plate are responsible for the cantilever deflections observed in the first part (regions b and b′) of the force curves (Heinz and Hoh 1999; Zlatanova et al. 2000). The region ranging from the point of the apparent beginning of the elastic indentation up to the point in which an abrupt plastic deformation is observed has allowed us to estimate the Young’s modulus of chromatin plates under different conditions using the Hertz equation (see “Materials and methods”). The values obtained for this elastic modulus are 0.16 ± 0.06 GPa (n = 24) and 0.23 ± 0.06 GPa (n = 29) in 5 and 20 mM Mg2+, respectively. In 5 mM Mg2+, the Young’s modulus corresponding to the region between the first abrupt transition (c) and the main transition (d) is 0.16 ± 0.06 GPa (n = 24).
Although it is not possible to know exactly which fraction of the cantilever deflection is due to the initial interactions in regions b and b′, the force curves can also be used to estimate the plate thickness. In 5 and 20 mM Mg2+, the displacements of the tip from the apparent beginning of the indentation up to the point of contact between the tip and the substrate are 6.8 ± 0.9 (n = 30) and 6.1 ± 1.3 (n = 32) nm, respectively. These estimates of plate thickness are compatible with the height measurements obtained by TEM. Thus, considering all our results, it can be concluded that the height of monolayer plates is approximately 6.5 nm.
Internal structure of chromatin plates
The plate thickness is similar to the height of a single nucleosome (~5.7 nm). This suggests that each step could be formed by a monolayer of nucleosomes with their flat faces oriented parallel to the plate surface. However, since such a regular organization should produce plates with many circular structures (having the nucleosome diameter: ~11 nm) clearly visible in the micrographs, but we do not see this repetitive structure in the surface of the plates, our results exclude this simple organization of nucleosomes in the plates.
In the edges of plates imaged by Cryo-EM and conventional TEM (Fig. 7), there are pairs of dense bars separated by about 3 nm. Since this distance corresponds approximately to the pitch of the helix formed by DNA in the nucleosome (Finch et al. 1977; Richmond et al. 1984; Uberbacher and Bunick 1985), we have interpreted that each pair of bars belongs to the two DNA turns of a nucleosome. These bar pairs can be seen roughly parallel or forming V-shaped figures. This is in agreement with the wedge shape of nucleosomes. The same structural features were observed previously in aggregates of purified nucleosome core particles imaged using conventional TEM (Finch et al. 1977) and Cryo-EM (Leforestier et al. 1999).
Cryo-EM images of frozen-hydrated plates (a, b) are shown in reverse contrast to facilitate the comparison with TEM images of plates contrasted by metal shadowing (c–e). The arrows indicate some regions in the edges of the plates in which the dense bars that presumably correspond to the turns of DNA in nucleosomes are visible (see “Results”). All the samples shown in this figure were prepared, as described in Fig. 3, from HeLa chromosomes in PE buffer containing 5 mM NaCl and 5 mM MgCl2. f Histogram corresponding to the distance between bars in the edges of rotary-shadowed plates from HeLa chromosomes (average distance 3.7 ± 0.5 nm; n = 247); the average distance obtained from Cryo-EM images of the same samples is 3.9 ± 0.4 nm (n = 72). Plates from chromosomes of chicken lymphocytes have an average distance between the bars in the edges of 3.5 ± 0.4 nm (n = 154). Bars 20 nm (a–c); 50 nm (d, e)
The bars are more easily seen in the periphery than in the internal regions of the plates (see arrows in Fig. 7a–e). At high magnification, the surface of the plates has an irregular texture, similar to that observed by other authors that have studied irregular aggregates of nucleosome core particles (Leforestier et al. 1999). We have performed distance measurements in many regions containing several consecutive dense bars in the edges of the plates. The average distance between bars in these regions is ~3.7 nm (see Fig. 7f and the legend of this figure). This distance is significantly larger than the distance (~2.8 nm) between the turns of DNA in the columns of stacked nucleosome core particles observed in TEM prepartions (Finch et al. 1977; Dubochet and Noll 1978), in core particle crystals (Uberbacher and Bunick 1985; White et al. 2001), and in ordered liquid crystalline phases formed by aggregates of these particles (Leforestier et al. 1999, 2001). Thus, our results indicate that nucleosomes in the plates have diverse orientations and follow a pattern more complex than the simple stacking observed in other laboratories with purified core particles.
On the other hand, taking into account that our measurements indicate that the plate height is lower than the nucleosome diameter, it can be suggested that nucleosomes are tilted to allow the interdigitation of nucleosomes between adjacent layers. The apparent height of each interdigitated layer can be lower than the nucleosome diameter and consequently may produce planar structures compatible with the observed plate thickness. There are other experimental results that favor this compact structure: (1) We know at present that the successive helical turns of nucleosomes forming the chromatin fiber can be interdigitated, and that the apparent height of each interdigitated turn with tilted nucleosomes is only a fraction of the nucleosome diameter (Daban and Bermúdez 1998; Robinson et al. 2006; Wong et al. 2007; Kepper et al. 2008; see next section); (2) Tilted nucleosomes in each layer could interact through their faces with the nucleosomes of the adjacent layers, and at present there are many experimental evidences indicating that this lateral interaction between nucleosomes is the driving force for the formation of condensed chromatin structures (see “Discussion”).
Purified chromatin fragments in the presence of Mg2+ can associate to form small plate-like structures
We observed using TEM (Bartolomé et al. 1994; Bermúdez et al. 1998), nondenaturing gel electrophoresis in the presence of Mg2+ (Bartolomé et al. 1995), and AFM in aqueous solution (Caño et al. 2006) that small chromatin fragments from chicken erythrocytes in solutions containing different concentrations of mono- and divalent cations form compact circular structures of about 35 nm in diameter. Some of these compact bodies are indicated with white arrows in Fig. 8b. We modeled these structures as interdigitated compact solenoids (Daban and Bermúdez 1998). Rhodes and coworkers (Robinson et al. 2006) have recently performed a very detailed study, using TEM and Cryo-EM, of the structure of reconstituted chromatin fragments containing DNA of defined sequence. The results obtained by these authors have led them to propose an interdigitated compact model for chromatin fibers similar to that suggested in our early studies. More recently, other laboratories have studied the conformational and dynamic properties of interdigitated solenoids (Wong et al. 2007; Kepper et al. 2008).
Unfolded chromatin plates from chicken metaphase chromosomes (a) contain structural elements similar to those seen in aggregates of small chromatin fragments from chicken erythrocytes (b–k). a After spreading, chromosomes of chicken lymphocytes were incubated in the presence of polyamines for 30 min at 37°C (the same conditions as in Fig. 2a, b). b–k Different aggregation degrees of small chromatin fragments from chicken erythrocytes in 10 mM Triethanolamine–HCl, pH 7.5, and 0.8 mM MgCl2. The white arrows in b point to the highly folded chromatin fragments described previously in our laboratory (similar structures are observed in a and some of them are also indicated with arrows); the association of these compact fragments gives rise to dimers (some of them are indicated with arrowheads in b) and higher aggregates (b–k). All TEM micrographs shown in this figure are at the same scale (bar 200 nm)
We have observed that metaphase chromosomes with a relatively high degree of denaturation may contain, in addition to many small plates, compact circular structures similar to those observed in experiments performed with purified small chromatin fragments. This can be seen particularly clear in Fig. 8a, in which some of these circular structures are indicated with white arrows. Note that circular structures are on top of small chromatin plates or laterally associated with them. In addition, note that many small plates show a grainy texture similar that observed in the unfolded layers considered above (Fig. 4b); the dimensions of the observed granules are compatible with the size of nucleosomes placed with diverse relative orientations.
On the other hand, we found previously that intermediate Mg2+ concentrations (2–20 mM) cause the association of small chromatin fragments producing very large aggregates (Caravaca et al. 2005; Caño et al. 2006). In this work, in order to obtain small aggregates, we have used lower Mg2+ concentrations. We have observed in different experiments that, in the presence of 0.8 mM Mg2+, the increase of the chromatin concentration and the incubation time favor the aggregation of the purified small chromatin fragments. As shown in the selected micrographs presented in Fig. 8b–k, chromatin fragments can associate laterally giving rise to dimers (indicated with white arrowheads) and to relatively small aggregates. Note that in these aggregates the 35-nm circular bodies change significantly their shape and form small plate-like structures (see micrographs 8c, d, g–i); in some cases it seems that the aggregated structures contain several layers of small plates (see micrographs 8e, f, j, k). Since these structures produced by purified chromatin are similar to those seen in the denatured plates presented in Fig. 8a, it can be suggested that chromatin has intrinsic structural properties that lead to the formation of plates.
Discussion
Plate structure
Plates associated with metaphase chromosomes were reported for the first time in a previous work of our laboratory (Caravaca et al. 2005). This initial TEM study was performed exclusively with human chromosomes (HeLa cells). Here we have observed plates in chromosomes of chicken, a species with a large evolutionary distance from humans. The very different buffers, ionic conditions, preparation procedures, spreading techniques, and substrates assayed in our work allow us to conclude that plates are not produced artefactually (see Table 1). In particular, our AFM and Cryo-EM results showing that plates exist in aqueous solution and are stable without crosslinking indicate that plates are native structures. The chromatin fibers that emanate from plates incubated at low ionic strength (this work) and with high NaCl concentrations (Caravaca et al. 2005), together with the high similarities observed between partially denatured plates and the aggregates produced by the spontaneous association of purified chromatin fragments indicate that the chromatin filament is able to fold giving rise to planar structures. The denaturation experiments and AFM studies performed applying high forces show that the chromatin filament is tightly tethered inside the plates, but can be more easily unfolded in the peripheral regions.
Although almost all the structural studies in the chromatin field have been devoted to fibers, in a previous study of a simplified system (nucleosome cores lacking histone H1 and linker DNA), Leforestier et al. (2001) using Cryo-EM techniques demonstrated that isolated nucleosome core particles at a high concentration associate giving rise to plate-like structures with many layers formed by columns of core particles; the electrostatic interactions involved in this lamellar structure has been studied in a theoretical work by Cherstvy and Everaers (2006). More recently, Engelhardt (2007) has proposed that in extended chromatin fibers the lateral association produced by interdigitation of nucleosomes could form dense structures composed of several sheets, and Hancock (2008) has found experimentally that, in the presence of high concentrations of crowding agents (25% polyethylene glycol), purified polynucleosomes can self-associate forming large sheets of chromatin.
Our TEM and Cryo-EM results exclude the possibility that chromatin plates are formed by monolayers of nucleosomes with their flat faces oriented parallel to the plate surface. Plates could also be formed by columns of nucleosomes having their axes parallel to the plate surface. This structure was observed in lamellar aggregates obtained with isolated core particles (Leforestier et al. 2001; see above). However, this simple pattern is also excluded because the plate thickness found in our TEM and AFM measurements (~6.5 nm) is lower than the column diameter (equal to the nucleosome diameter: ~11 nm). Moreover, columns of nucleosomes in the plates should be clearly visible in the electron micrographs, but our images and distance measurements show that nucleosomes in the plates follow a complex orientation pattern. Our results strongly suggest that nucleosomes are tilted with respect to the plate surface to allow the interdigitation between layers and a reduction of thickness compatible with the observed plate height. This thickness reduction is consistent with the remarkable reduction of fiber length observed in interdigitated solenoids (Daban and Bermúdez 1998; Robinson et al. 2006; Wong et al. 2007). Note, in addition, that tilted nucleosomes in each layer may interact through their faces with the nucleosomes of the adjacent layers. Results from different laboratories (McDowall et al. 1986; Mangenot et al. 2003; Daban 2000, 2003), suggesting that face-to-face interactions between nucleosomes is the driving force for the formation of chromatin structures with a high local concentration of DNA, also favor our interpretation. Furthermore, Kepper et al. (2008) have demonstrated using Monte Carlo dynamic simulations that interdigitated solenoids having nucleosomes with a relatively high tilt angle, favoring face-to-face interactions, are more stable than other possible conformations. Lateral association of nucleosome cores has also been found in a tetranucleosome crystal (see “Introduction”; Schalch et al. 2005) and high-resolution X-ray diffraction analysis (Luger et al. 1997; Harp et al. 2000; White et al. 2001) indicate that this association involves and acidic surface formed by histones H2A and H2B and basic residues of the N-termimal tail of histone H4. Finally, more recently, it has been demonstrated (Zhou et al. 2007; Chodaparambil et al. 2007) that this acidic region is responsible for the regulation of both the folding of well-defined nucleosome arrays and their self-association leading to condensed three-dimensional structures (Caterino and Hayes 2007).
Plates in chromosomes
Our SEM micrographs and particularly the images obtained with the 200-kV transmission microscope show that plate-like structures are the dominant component of compact chromatids. Other authors (Adolph and Kreisman 1983; Sone et al. 2002) obtained similar SEM images of metaphase chromosomes prepared in aqueous solutions containing divalent cations and polyamines, but they did not suggest any role for the planar structures observed in their micrographs. Considering our results indicating that both, the plates that occupy the bulk of the chromatids and the plates extruded from them, have a multilayered appearance, we propose that metaphase chromosomes are formed by stacked plates. We do not know how the single DNA molecule which constitutes each chromatid connects the successive chromatin layers. Nevertheless, the close correlation between the cytogenetic map of chromosomes and the genome sequence (International Human Genome Sequencing Consortium 2001; The BAC Resource Consortium 2001) implies that the chromatin filament must fill each chromatid progressively from one end to the other. This constraint precludes the possibility of chromosome formation with large plates extended parallel to the chromatid axis. Therefore, it is reasonable to consider that in native chromosomes plates are orthogonal to the chromatid axis. In fact, in spite of their thinness and soft texture, in some micrographs we have observed plates that are apparently perpendicular to the chromatid axis. The plates observed in our TEM and AFM images have different sizes, but many of them (see for instance Figs. 2f, 4b, 5a, c) have a surface area large enough to span entirely the cross-section of a chromatid. A scheme of the thin-plate model suggested from our results is presented in Fig. 9.
Thin-plate model for chromatin condensation in metaphase chromosomes. a Our results suggest that chromatids are composed of the chromatin plates studied in this work. Presumably, in native chromosomes the thin plates are stacked regularly along the chromatid axis producing chromosomes with a smooth surface; to simplify the drawing we schematize this structure only in region a. b Chromosome isolation and spreading, and the diverse treatments described in the text cause the sliding and distortion of the thin chromatin plates; this is schematized in a′ in the side view. The plates surrounding the chromatids are oriented by the carbon film (red) and become clearly visible; these plates are indicated with light color in the top view (left part of a′). The distorted plates in the chromosomes are only partially visible in the periphery of the chromatids; this is also schematized in the top view (right part of a′)
As expected the Young’s modulus found for chromatin plates (~0.2 GPa) is much lower than that observed for typical solid materials and ionic crystals (103–10 GPa) (Fraxedas et al. 2002; Callister and Rethwisch 2008). The elastic stiffness of plates is similar to that observed for DNA and globular proteins in force spectroscopy studies (Young’s modulus ~0.5 GPa; Zlatanova et al. 2000), but it is much higher than that observed for chromosomes in stretching experiments (Young’s modulus ~103 Pa; Poirier and Marko 2002b). Taking into account all these values and according to these authors, mitotic chromosomes can be considered as soft structures having an elastic stiffness much lower than that of the materials from which they are composed. In this context, it can be suggested that our observations indicating that the interactions that exist between adjacent layers in plates are weaker than those holding the chromatin filament within a layer could be related to the elastic properties observed for whole chromosomes in stretching experiments.
Possible relationship between plates and chromosome bands
There are classical cytogenetic observations that could be directly related to the existence of stacked chromatin plates oriented perpendicular to the chromatid axis. The identification of individual chromosomes is based on diverse staining procedures that give rise to characteristic banding patterns (Sumner 1989; Castleman 2000), in which the visible bands are roughly orthogonal to the chromatid axis. Even the thinnest bands show this orientation. The transverse orientation of all the bands is schematized in the well-established ideograms corresponding to the banding patterns of human chromosomes (ISCN 2005). Typical banding procedures use chromosomes fixed with methanol-acetic acid. This could alter the native structure of chromosomes. Nevertheless, equivalent patterns having bands roughly orthogonal to the chromatid axis have been obtained with native chromosomes (not treated with organic solvents) stained with different fluorescent dyes (Saitoh and Laemmli 1994). The cytogenetic bands are much thicker than the thin chromatin plates studied here, but it has been reported that chromosome stretching causes the splitting of the typical bands into much thinner subbands which are also oriented perpendicular to the chromatid axis (Hliscs et al. 1997). Taken together, these observations suggest that chromosome bands could be produced by selective staining of chromatin plate clusters having the DNA base composition adequate for the interaction with the dyes used in the different banding protocols. The relative weakness of the interactions between chromatin layers (see above) could be related to the easy band splitting observed in stretching experiments. The thinness, soft texture, and high tendency of chromatin plates to associate forming multilayered structures make impossible to determine experimentally the number of plates in a chromatid. However, taking into account our plate height measurements and assuming a close contact between consecutive plates in highly condensed chromosomes, we can estimate that a cytogenetic band having a thickness of 300 nm should correspond to a cluster of about 45 chromatin plates.
Functional implications
The folding of the chromatin filament forming plates within chromosomes at metaphase ionic conditions is not compatible with the consideration that the 30-nm chromatin fiber is the fundamental structural element of metaphase chromosomes. Nevertheless, our model for chromatin condensation does not exclude the possible existence of additional organization levels of the chromatin filament. In particular, the attachment of specific DNA sequences to nonhistone proteins may give rise to the loops that become visible in the presence of the divalent cation chelator EDTA (Marsden and Laemmli 1979) and after the removal of histones (Paulson and Laemmli 1977). Our denaturation experiments suggest that chromatin fibers and plates are interconvertible; presumably, in the future, it will be possible to integrate both structural forms into a dynamic model for chromatin function during the cell cycle.
The dense aspect of chromatids in the presence of divalent cations and polyamines, together with the large relative amount of histones (Uchiyama et al. 2005) and DNA (Daban 2000) in metaphase chromosomes, suggest that chromatin plates occupy most of the volume and consequently may be responsible for the basic physical properties of the metaphase chromosome. This is in agreement with the fact that the observed condensation/decondensation behavior of metaphase chromosomes in aqueous solution induced by different cations (Poirier et al. 2002) can be correlated with the compaction/unfolding behavior observed for purified chromatin samples in different laboratories (Zlatanova et al. 1994; Bartolomé et al. 1995; Hansen 2002; Mangenot et al. 2003). Thus, according to these considerations, nonhistone proteins in condensed metaphase chromosomes have to interact with a dense chromatin structure instead of the extended loops generally considered in the current chromatin literature. Nevertheless, the observed condensation/decondensation properties of metaphase chromosomes also indicate that a significant part of the chromosome volume is occupied by water, which could facilitate the diffusion of proteins with different functions. This possibility is consistent with in vivo photobleaching experiments showing that there is a rapid exchange of histone H1 (Lever et al. 2000; Misteli et al. 2000) and topoisomerase II (Tavormina et al. 2002; Christensen et al. 2002) in mitotic chromosomes. Therefore, although chromatin is highly condensed in chromosomes, the resulting structure is dynamic from a functional point of view.
At present it is not possible to describe the structural relationships between chromatin plates and characteristic chromosome elements such as centromeres and telomeres. Nor is it known how chromatin plates are related to other large-scale properties of chromosome structure such as the observed diameter of chromatids and the possible existence of a hole along the chromatid axis for the location of nonhistone proteins. Many questions remain open, but we think that the study of chromatin plates will inspire completely new experimental approaches that will be very useful for the understanding of the structure and functional properties of eukaryotic chromosomes.
Abbreviations
- AFM:
-
Atomic force microscopy
- Cryo-EM:
-
Cryo-electron microscopy
- SEM:
-
Scanning electron microscopy
- TEM:
-
Transmission electron microscopy
References
Adolph KW, Kreisman LR (1983) Surface structure of isolated metaphase chromosomes. Exp Cell Res 147:155–166. doi:10.1016/0014-4827(83)90280-X
Aragay AM, Fernandez-Busquets X, Daban JR (1991) Different mechanisms for in vitro formation of nucleosome core particles. Biochemistry 30:5022–5032. doi:10.1021/bi00234a026
Bartolomé S, Bermúdez A, Daban JR (1994) Internal structure of the 30 nm chromatin fiber. J Cell Sci 107:2983–2992
Bartolomé S, Bermúdez A, Daban JR (1995) Electrophoresis of chromatin on nondenaturing agarose gels containing Mg2+. Self-assembly of small chromatin fragments and folding of the 30-nm fiber. J Biol Chem 270:22514–22521. doi:10.1074/jbc.270.38.22514
Bednar J, Woodcock CL (1999) Cryoelectron microscopic analysis of nucleosomes and chromatin. Methods Enzymol 304:191–213. doi:10.1016/S0076-6879(99)04012-4
Bednar J, Horowitz RA, Grigoryev SA, Carruthers LM, Hansen JC, Koster AJ, Woodcock CL (1998) Nucleosomes, linker DNA, and linker histone form a unique structural motif that directs the higher-order folding and compaction of chromatin. Proc Natl Acad Sci USA 95:14173–14178. doi:10.1073/pnas.95.24.14173
Belmont AS (2002) Mitotic chromosome scaffold structure: new approaches to an old controversy. Proc Natl Acad Sci USA 99:15855–15857. doi:10.1073/pnas.262672799
Belmont AS (2006) Mitotic chromosome structure and condensation. Curr Opin Cell Biol 18:632–638. doi:10.1016/j.ceb.2006.09.007
Bermúdez A, Bartolomé S, Daban JR (1998) Partial denaturation of small chromatin fragments: direct evidence for the radial distribution of nucleosomes in folded chromatin fibers. J Cell Sci 111:1707–1715
Bordas J, Perez-Grau L, Koch MHJ, Vega MC, Nave C (1986) The superstructure of chromatin and its condensation mechanism. II Theoretical analysis of the X-ray scattering patterns and model calculations. Eur Biophys J 13:175–185. doi:10.1007/BF00542561
Bradbury EM, Baldwin JP (1986) Neutron scatter and diffraction techniques applied to nucleosome and chromatin structure. Cell Biophys 9:35–66
Callister WD, Rethwisch G (2008) Fundamentals of materials science and engineering. Wiley, New York
Caño S, Caravaca JM, Martín M, Daban JR (2006) Highly compact folding of chromatin induced by cellular cation concentrations. Evidence from atomic force microscopy studies in aqueous solution. Eur Biophys J 35:495–501. doi:10.1007/s00249-006-0057-7
Caravaca JM, Caño S, Gállego I, Daban JR (2005) Structural elements of bulk chromatin within metaphase chromosomes. Chromosome Res 13:725–743. doi:10.1007/s10577-005-1008-3
Castleman KR (2000) Image analysis: quantitative interpretation of chromosome images. In: Balbock R, Graham J (eds) Image processing and analysis. A practical approach. Oxford University Press, Oxford, pp 69–109
Caterino TL, Hayes JJ (2007) Chromatin structure depends on what’s in the nucleosome’s pocket. Nat Struct Mol Biol 14:1056–1058. doi:10.1038/nsmb1107-1056
Cherstvy AG, Everaers R (2006) Layering, bundling, and azimuthal orientations in dense phases of nucleosome core particles. J Phys Condens Matter 18:11429–11442. doi:10.1088/0953-8984/18/50/003
Chodaparambil JV, Barbera AJ, Lu X, Kaye KM, Hansen JC, Luger K (2007) A charged and contoured surface on the nucleosome regulates chromatin compaction. Nat Struct Mol Biol 14:1105–1107. doi:10.1038/nsmb1334
Christensen MO, Larsen MK, Barthelmes HU, Hock R, Andersen CL, Kjeldsen E, Knudsen BR, Westergaard O, Boege F, Mielke C (2002) Dynamics of human DNA topoisomerases IIα and IIβ in living cells. J Cell Biol 157:31–44. doi:10.1083/jcb.200112023
Daban JR (2000) Physical constraints in the condensation of eukaryotic chromosomes. Local concentration of DNA versus linear packing ratio in higher order chromatin structures. Biochemistry 39:3861–3866. doi:10.1021/bi992628w
Daban JR (2003) High concentration of DNA in condensed chromatin. Biochem Cell Biol 81:91–99. doi:10.1139/o03-037
Daban JR, Bermúdez A (1998) Interdigitated solenoid model for compact chromatin fibers. Biochemistry 37:4299–4304. doi:10.1021/bi973117h
Dubochet J, Noll M (1978) Nucleosome arcs and helices. Science 202:280–286. doi:10.1126/science.694532
Dubochet J, Adrian M, Chang JJ, Homo JC, Lepault J, McDowall AW, Schultz P (1988) Cryo-electron microscopy of vitrified specimens. Q Rev Biophys 21:129–228
Dubochet J, Zuber B, Eltsov M, Bouchet-Marquis C, Al-Amoudi A, Livolant F (2007) How to “read” a vitreous section. Methods Cell Biol 79:385–406. doi:10.1016/S0091-679X(06)79015-X
DuPraw EJ (1966) Evidence for a ‘folded-fibre’ organization in human chromosomes. Nature 209:577–581. doi:10.1038/209577a0
Engelhardt M (2007) Choreography for nucleosomes: the conformational freedom of the nucleosomal filament and its limitations. Nucleic Acids Res 35:e106. doi:10.1093/nar/gkm560
Filipski J, Leblanc J, Youdale T, Sikorska M, Walker PR (1990) Periodicity of DNA folding in higher order chromatin structures. EMBO J 9:1319–1327
Finch JT, Lutter LC, Rhodes D, Brown RS, Rushton B, Levitt M, Klug A (1977) Structure of nucleosome core particles of chromatin. Nature 269:29–36. doi:10.1038/269029a0
Fraxedas J, Garcia-Manyes S, Gorostiza P, Sanz F (2002) Nanoindentation: toward the sensing of atomic interactions. Proc Natl Acad Sci USA 99:5228–5232. doi:10.1073/pnas.042106699
Gassmann R, Vagnarelli P, Hudson D, Earnshaw WC (2004) Mitotic chromosome formation and the condensin paradox. Exp Cell Res 296:35–42. doi:10.1016/j.yexcr.2004.03.006
Grigoryev SA (2004) Keeping fingers crossed: heterochromatin spreading through interdigitation of nucleosome arrays. FEBS Lett 564:4–8. doi:10.1016/S0014-5793(04)00258-3
Hancock R (2008) Self-association of polynucleosome chains by macromolecular crowding. Eur Biophys J 37:1059–1064. doi:10.1007/s00249-008-0276-1
Hansen JC (2002) Conformational dynamics of the chromatin fiber in solution: determinants, mechanisms, and functions. Annu Rev Biophys Biomol Struct 31:361–392. doi:10.1146/annurev.biophys.31.101101.140858
Harp JM, Hanson BL, Timm DE, Bunick GJ (2000) Asymmetries in the nucleosome core particle at 2.5 Å resolution. Acta Crystllogr D 56:1513–1534. doi:10.1107/S0907444900011847
Heinz WF, Hoh JH (1999) Spatially resolved force spectroscopy of biological surfaces using the atomic force microscope. Trends Biotechnol 17:143–150. doi:10.1016/S0167-7799(99)01304-9
Hirano T (2005) Condensins: organizing and segregating the genome. Curr Biol 15:R265–R275. doi:10.1016/j.cub.2005.03.037
Hliscs R, Mühlig P, Claussen U (1997) The nature of G-bands analyzed by chromosome stretching. Cytogenet Cell Genet 79:162–166. doi:10.1159/000134710
Horowitz-Scherer RA, Woodcock CL (2006) Organization of interphase chromatin. Chromosoma 115:1–14. doi:10.1007/s00412-005-0035-3
Hudson DF, Vagnarelli P, Gassmann R, Earnshaw WC (2003) Condensin is required for nonhistone protein assembly and structural integrity of vertebrate mitotic chromosomes. Dev Cell 5:323–336. doi:10.1016/S1534-5807(03)00199-0
International Chicken Genome Sequencing Consortium (2004) Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432:695–716. doi:10.1038/nature03154
International Human Genome Sequencing Consortium (2001) Initial sequencing and analysis of the human genome. Nature 409:860–921. doi:10.1038/35057062
ISCN (2005) An international system for human cytogenetic nomenclature. Shaffer LG, Tommerup N (eds) S Karger AG, Basel
Ishiguro K, Watanabe Y (2007) Chromosome cohesion in mitosis and meiosis. J Cell Sci 120:367–369. doi:10.1242/jcs.03324
Jiao Y, Schäffer TE (2004) Accurate height and volume measurements on soft samples with the atomic force microscope. Langmuir 20:10038–10045. doi:10.1021/la048650u
Kepper N, Foethke D, Stehr R, Wedemann G, Rippe K (2008) Nucleosome geometry and internucleosomal interactions control the chromatin fiber conformation. Biophys J 95:3692–3705. doi:10.1529/biophysj.107.121079
Kireeva N, Lakonishok M, Kireev I, Hirano T, Belmont AS (2004) Visualization of early chromosome condensation: a hierarchical folding, axial glue model of chromosome structure. J Cell Biol 166:775–785. doi:10.1083/jcb.200406049
Leforestier A, Fudaley S, Livolant F (1999) Spermidine-induced aggregation of nucleosome core particles: evidence for multiple liquid crystalline phases. J Mol Biol 290:481–494. doi:10.1006/jmbi.1999.2895
Leforestier A, Dubochet J, Livolant F (2001) Bilayers of nucleosome core particles. Biophys J 81:2414–2421
Leuba SH, Yang G, Robert C, Samori B, van Holde K, Zlatanova J, Bustamante C (1994) Three-dimensional structure of extended chromatin fibers as revealed by tapping-mode scanning force microscopy. Proc Natl Acad Sci USA 91:11621–11625. doi:10.1073/pnas.91.24.11621
Lever MA, Th’ng JPH, Sun X, Hendzel MJ (2000) Rapid exchange of histone H1.1 on chromatin in living human cells. Nature 408:873–876. doi:10.1038/35048603
Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ (1997) Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389:251–260. doi:10.1038/38444
Macgregor HC, Varley JM (1983) Working with animal chromosomes. Wiley, New York
Maeshima K, Laemmli UK (2003) A two-step scaffolding model for mitotic chromosome assembly. Dev Cell 4:467–480. doi:10.1016/S1534-5807(03)00092-3
Makarov V, Dimitrov S, Smirnov V, Pashev I (1985) A triple helix model for the structure of chromatin fiber. FEBS Lett 181:357–361. doi:10.1016/0014-5793(85)80292-1
Mangenot S, Leforestier A, Durand D, Livolant F (2003) Phase diagram of nucleosome core particles. J Mol Biol 333:907–916. doi:10.1016/j.jmb.2003.09.015
Manuelidis L, Chen TL (1990) A unified model of eukaryotic chromosomes. Cytometry 11:8–25. doi:10.1002/cyto.990110104
Marsden MPF, Laemmli UK (1979) Metaphase chromosome structure: evidence for a radial loop model. Cell 17:849–858. doi:10.1016/0092-8674(79)90325-8
McDowall AW, Smith JM, Dubochet J (1986) Cryo-electron microscopy of vitrified chromosomes in situ. EMBO J 5:1395–1402
Misteli T, Gunjan A, Hock R, Bustin M, Brown DT (2000) Dynamic binding of histone H1 to chromatin in living cells. Nature 408:877–881. doi:10.1038/35048610
Mozziconacci J, Victor JM (2003) Nucleosome gaping supports a functional structure for the 30 nm chromatin fiber. J Struct Biol 143:72–76. doi:10.1016/S1047-8477(03)00102-3
Nasmyth K, Haering CH (2005) The structure and function of SMC and kleisin complexes. Annu Rev Biochem 74:595–648. doi:10.1146/annurev.biochem.74.082803.133219
Ono T, Losada A, Hirano M, Myers MP, Neuwald AF, Hirano T (2003) Differential contributions of condensin I and condensin II to mitotic chromosome architecture in vertebrate cells. Cell 115:109–121. doi:10.1016/S0092-8674(03)00724-4
Paulson JR, Laemmli UK (1977) The structure of histone-depleted metaphase chromosomes. Cell 12:817–828. doi:10.1016/0092-8674(77)90280-X
Pienta KJ, Coffey DS (1984) A structural analysis of the role of the nuclear matrix and DNA loops in the organization of the nucleus and chromosome. J Cell Sci Suppl 1:123–135
Poirier MG, Marko JF (2002a) Mitotic chromosomes are chromatin networks without a mechanically contiguous protein scaffold. Proc Natl Acad Sci USA 99:15393–15397. doi:10.1073/pnas.232442599
Poirier MG, Marko JF (2002b) Micromechanical studies of mitotic chromosomes. J Muscle Res Cell Motil 23:409–431. doi:10.1023/A:1023402321367
Poirier MG, Monhait T, Marko JF (2002) Reversible hypercondensation and decondensation of mitotic chromosomes studied using combined chemical-micromechanical techniques. J Cell Biochem 85:422–434. doi:10.1002/jcb.10132
Radmacher M (2002) Measuring the elastic properties of living cells by the atomic force microscope. Methods Cell Biol 68:67–90. doi:10.1016/S0091-679X(02)68005-7
Richmond TJ, Finch JT, Rushton B, Rhodes D, Klug A (1984) Structure of the nucleosome core particle at 7 Å resolution. Nature 311:532–537. doi:10.1038/311532a0
Robinson PJJ, Fairall L, Huynh VA, Rhodes D (2006) EM measurements define the dimensions of the “30-nm” chromatin fiber: evidence for a compact, interdigitated structure. Proc Natl Acad Sci USA 103:6506–6511. doi:10.1073/pnas.0601212103
Saitoh Y, Laemmli UK (1994) Metaphase chromosome structure: bands arise from a differential folding path of the highly AT-rich scaffold. Cell 76:609–622. doi:10.1016/0092-8674(94)90502-9
Savvidou E, Cobbe N, Steffensen S, Cotterill S, Heck MMS (2005) Drosophila CAP-D2 is required for condensin complex stability and resolution of sister chromatids. J Cell Sci 118:2529–2543. doi:10.1242/jcs.02392
Schalch T, Duda S, Sargent DF, Richmond TJ (2005) X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature 436:138–141. doi:10.1038/nature03686
Sone T, Iwano M, Kobayashi S, Ishihara T, Hori N, Takata H, Ushiki T, Uchiyama S, Fukui K (2002) Changes in chromosomal surface structure by different isolation conditions. Arch Histol Cytol 65:445–455. doi:10.1679/aohc.65.445
Staynov DZ, Proykova YG (2008) Topological constraints on the possible structures of the 30 nm chromatin fibre. Chromosoma 117:67–76. doi:10.1007/s00412-007-0127-3
Strick R, Strissel PL, Gavrilov K, Levi-Setti R (2001) Cation-chromatin binding as shown by ion microscopy is essential for the structural integrity of chromosomes. J Cell Biol 155:899–910. doi:10.1083/jcb.200105026
Subirana JA, Muñoz-Guerra S, Aymamí J, Radermacher M, Frank J (1985) The layered organization of nucleosomes in 30 nm chromatin fibers. Chromosoma 91:377–390. doi:10.1007/BF00291012
Sumner AT (1989) Chromosome banding. In: Lacey AJ (ed) Light microscopy in biology. A practical approach. IRL Press, Oxford, pp 279–314
Sumner AT (2003) Chromosomes: organization and function. Blackwell, Oxford
Taniguchi T, Takayama S (1986) High-order structure of metaphase chromosomes: evidence for a multiple coiling model. Chromosoma 93:511–514. doi:10.1007/BF00386792
Tavormina PA, Côme MG, Hudson JR, Mo YY, Beck WT, Gorbsky GJ (2002) Rapid exchange of mammalian topoisomerase IIα at kinetochores and chromosome arms in mitosis. J Cell Biol 158:23–29. doi:10.1083/jcb.200202053
The BAC Resource Consortium (2001) Integration of cytogenetic landmarks into the draft sequence of the human genome. Nature 409:953–958. doi:10.1038/35057192
Thoma F, Koller T, Klug A (1979) Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J Cell Biol 83:403–427. doi:10.1083/jcb.83.2.403
Tremethick DJ (2007) Higer-order structure of chromatin: the elusive 30 nm fiber. Cell 128:651–654. doi:10.1016/j.cell.2007.02.008
Uberbacher EC, Bunick GJ (1985) X-ray structure of the nucleosome core particle. J Biomol Struct Dyn 2:1033–1055
Uchiyama S, Kobayashi S, Takata H, Ishihara T, Hori N, Higashi T, Hayashihara K, Sone T, Higo D, Nirasawa T, Takao T, Matsunaga S, Fukui K (2005) Proteome analysis of human metaphase chromosomes. J Biol Chem 280:16994–17004. doi:10.1074/jbc.M412774200
Van Holde KE, Zlatanova J (2007) Chromatin fiber structure: where is the problem now? Semin Cell Dev Biol 18:651–658. doi:10.1016/j.semcdb.2007.08.005
Weisenhorn AL, Khorsandi M, Kasas S, Gotzos V, Butt HJ (1993) Deformation and height anomaly of soft surfaces studied with an AFM. Nanotechnology 4:106–113. doi:10.1088/0957-4484/4/2/006
White CL, Suto RK, Luger K (2001) Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. EMBO J 20:5207–5218. doi:10.1093/emboj/20.18.5207
Widom J, Klug A (1985) Structure of the 300 Å chromatin filament X-ray diffraction from oriented samples. Cell 43:207–213. doi:10.1016/0092-8674(85)90025-X
Williams SP, Athey BD, Muglia LJ, Schappe RS, Gough AH, Langmore JP (1986) Chromatin fibers are left-handed double helices with diameter and mass per unit length that depend on linker length. Biophys J 49:233–248
Wolffe AP (1998) Chromatin structure and function, 3rd edn. Academic Press, London
Wong H, Victor JM, Mozziconacci J (2007) An all-atom model of the chromatin fiber containing linker histones reveals a versatile structure tuned by the nucleosome repeat length. PLoS One 2:e877. doi:10.1371/journal.pone.0000877
Yang G, Leuba SH, Bustamante C, Zlatanova J, van Holde K (1994) Role of linker histones in extended chromatin fibre structure. Nat Struct Biol 1:761–763. doi:10.1038/nsb1194-761
Zentgraf H, Franke WW (1984) Differences of supranucleosomal organization in different kinds of chromatin: cell type-specific globular subunit containing different numbers of nucleosomes. J Cell Biol 99:272–286. doi:10.1083/jcb.99.1.272
Zhou J, Fan JY, Rangasamy D, Tremethick DJ (2007) The nucleosome surface regulates chromatin compaction and couples it with transcriptional repression. Nat Struct Mol Biol 14:1070–1076. doi:10.1038/nsmb1323
Zlatanova J, Leuba SH, Yang G, Bustamante C, van Holde K (1994) Linker DNA accessibility in chromatin fibers of different conformations: a reevaluation. Proc Natl Acad Sci USA 91:5277–5280. doi:10.1073/pnas.91.12.5277
Zlatanova J, Lindsay SM, Leuba SH (2000) Single molecule force spectroscopy in biology using the atomic force microscope. Prog Biophys Mol Biol 74:37–61. doi:10.1016/S0079-6107(00)00014-6
Acknowledgments
We thank Francesca Vidal and Gerard Oncins for advice about preparation of chicken chromosomes and force curve analysis, respectively, and José M. Valpuesta and Montserrat Samsó for advice on Cryo-EM techniques. We also acknowledge the help of Ismael Díez and Jordi Díaz (Serveis Científico-Tècnics, UB) in the AFM experiments. TEM and SEM images were obtained in the Servei de Microscòpia (UAB). Chicken blood was obtained in the Servei de Granges i Camps Experimentals (UAB) and in Gimave S. A. (Ripollet, Barcelona). Cell culture was performed in the Servei de Cultius Cel·lulars (IBB). I.G. and P.C.-H. were supported by predoctoral fellowships from the Generalitat de Catalunya, and S.C. by a predoctoral fellowship from the Ministerio de Educación y Ciencia (MEC). This work was supported in part by grant BFU2005-3883 (MEC).
Author information
Authors and Affiliations
Corresponding author
Additional information
A preliminary report of this work was presented at the 23rd Reunión Bienal de la Sociedad de Microscopía de España, Bilbao, July 2007.
J. M. Caravaca and S. Caño contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Gállego, I., Castro-Hartmann, P., Caravaca, J.M. et al. Dense chromatin plates in metaphase chromosomes. Eur Biophys J 38, 503–522 (2009). https://doi.org/10.1007/s00249-008-0401-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00249-008-0401-1