Abstract
We report on the solvation properties and intermolecular interactions of a model protein (bovine serum albumine, BSA) in urea aqueous solutions, as obtained by combining small-angle neutron and X-ray scattering experiments. According to a global fit strategy, all the whole set of scattering curves are analysed by considering a unique model which includes the BSA structure, the protein-protein interactions and the thermodynamic exchange process of water/urea molecules at the protein solvent interface. As a main result, the equilibrium constant that accounts for the difference in composition between the bulk solvent and the protein solvation layer is derived. Results confirm that urea preferentially sticks to the protein surface, inducing a noticeable change in both the repulsive and the attractive interaction potentials.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The role of stabilizing or denaturing cosolvents in the determination of a protein stability and unfolding has been extensively studied (Bloom et al. 2006; Dill et al. 2007). In the presence of cosolvent, literature studies on protein solutions were mainly focused on two different mechanisms. The first one is the competition between water and cosolvent molecules in surrounding the protein surface, and it results in a preferential localization or exclusion of the cosolvent molecules from the solvation shell (Gekko and Timasheff 1981; Timasheff 1998; Zhang et al. 1996; Courtenay et al. 2000; Schellmann 2003). The second phenomenon is related to the modulation of the protein–protein interaction potential caused by changes in the solvent properties induced by cosolvent (Stradner et al. 2005; Pagan and Gunton 2005; Valente et al. 2005; Niebuhr and Koch 2005). Both preferential solvation (Karen et al. 2002; Baynes and Trout 2003; Shulgin and Ruckenstein 2006, 2007; Smith 2006) and protein–protein interaction (Ortore et al. 2005; Spinozzi et al. 2002; Narayanan and Liu 2003; Stradner et al. 2004; Farnum and Zukoski 1999; Pellicane et al. 2004; van Oss 2003) have been widely studied using experimental techniques, molecular dynamics simulations and theoretical models. However, despite a huge amount of literature devoted on these topics, there are only a few studies which try to correlate the modulation of protein–protein interactions with different protein solvation features (Javid et al. 2007; Sinibaldi et al. 2007).
Nonetheless, it is well known that protein-protein interactions are strongly correlated with the solvent properties and the role played in this phenomenon by cosolvents is fundamental for life (Yancey et al. 1982). The most used techniques to investigate protein-protein interactions are the determination of the second virial coefficient by means of dynamic light scattering (DLS) (Valente et al. 2005; Farnum and Zukoski1999) and the reconstruction of the structure factor by in-solution small-angle scattering (SAS) of X-ray (SAXS) (Spinozzi et al. 2002; Narayanan and Liu 2003; Niebuhr and Koch 2005) or Neutrons (SANS) (Hansen and McDonald 1986). On one side, DLS can be used to estimate if interactions are modified with respect to a reference condition; on the other side, SAS can be used to derive the particle-particle radial distribution function and to separate electrostatic contributions from other kinds of short range interactions (Spinozzi et al. 2002).
We recently showed that SANS technique could be successfully used to derive the composition of the solvation shell of proteins in a binary solvent (Sinibaldi et al. 2007) and to describe the protein-protein interactions in different solvent conditions. A global fit strategy is, however, necessary: SANS or SAXS experiments should be performed on a set of wisely chosen experimental conditions relative to different protein concentrations, cosolvent amounts and deuteration grade, and data should be analysed using the same protein structural model and a thermodynamic model for the solvation process. In particular, the use of the thermodynamic solvent exchange model introduced by Schellman (2003) appeared suitable to reduce the number of fitting parameters, and very useful to enable the simultaneous fit of all the SAS curves relative to all the different experimental conditions.
In this way, we were able to estimate the thermodynamic equilibrium constant (K) that describes the exchange between water and cosolvent molecules in the bulk and in contact with a protein surface. In particular, results showed that lysozyme in a water–glycerol mixture is preferentially hydrated, in fully agreement with previous literature indications obtained at infinite protein dilution (Gekko and Timasheff 1981), while in water-urea mixtures, urea appeared to be preferentially located at the lysozyme surface (Ortore et al. 2008), still in agreement with previous data (Timasheff and Xie 2003). Moreover, the spherical two body interaction theory was successfully used to fully characterize the modulation of lysozyme–lysozyme interactions in both water–glycerol and water–urea mixtures.
The urea case was particularly interesting. In fact, urea preferential interactions with protein surfaces were widely analysed, but usually the experimental techniques that have been considered (such as vapour pressure osmometry) concerned highly diluted protein conditions (Timasheff and Xie 2003). Our data were obtained at relatively high concentrations (Ortore et al. 2008) and the derived preferential interaction coefficients were in full agreement with those estimated by Timasheff and Xie (2003). The results on lysozyme interactions were quite interesting. Indeed, urea appeared to induce a strong change of the intensity of the coulombic interactions, that can be only explained assuming that the protein net charge is changing as a function of the increasing urea content at the protein surface. Moreover, our results also confirmed recent SAXS data showing that lysozyme dissolved in water–urea solutions experiences a lower short range attraction in respect to the protein in pure water solution (Niebuhr and Koch 2005).
Therefore, such urea effects merit to be further investigated, considering a different protein, but also using a broader experimental approach. In particular, the combination of SANS and SAXS techniques is a very suitable tool for this purpose: contrast variation SANS data offer the possibility to easily derive direct information about the solvation layer, while the higher quality of SAXS curves, especially in the low Q range, allows a better investigation of the structure factor, hence of the protein–protein interactions (Svergun et al. 1998; Spinozzi et al. 2000). In this paper, we then investigate by SAXS and SANS experiments a model protein, the bovine serum albumin (BSA) at concentration up to 125 g L−1, in the presence of a molar content of urea well below those causing protein unfolding. The two independent sets of scattering experiments have been performed on samples prepared according to an identical protocol to avoid uncertainties due to different deuteration grades. The direct evaluation of the thermodynamic exchange constant from SAS curves has been performed using the original global fitting procedure already adopted in different cases (Ortore et al. 2005; Sinibaldi et al. 2007; Spinozzi et al. 2006).
As a result, in the present study we obtained a quantitative description of protein solvation layer in a water urea mixture, obtained according to a precise solvation model. The fitting results show an increase of the presence of urea in the local domain, as well as the changes induced by denaturant in the protein–protein interaction potential. In analogy to our previous work (Sinibaldi et al. 2007), the preferential interaction coefficient Γ for the different investigated samples has been also calculated. The behaviour of Γ as a function of the urea concentration agrees well with experimental trend previously estimated by vapour pressure osmometry (Zhang et al. 1996).
Materials and methods
Samples preparation
Protein solutions for SANS measurements were prepared at weight concentrations c of 10, 30 and 100 g L−1 (i.e. 15, 45 and 150 mM) by dissolving the requested amount of BSA powder (Bovine Serum Albumin, 99% purity, Sigma Aldrich) in 50 mM phosphate buffer (pH 7.0) prepared using deuterated water and containing different amounts of hydrogenated urea. Since our aim is to study BSA before urea denaturation (Ahmad and Qasim 1995), three different urea compositions were considered, namely 0, 0.5 and 1.5 M. For SAXS experiments protein powders were dissolved in the same buffer prepared in deuterated water to avoid uncertainities related to deuteration effects. In this case, four values of protein concentrations (25, 50, 100 and 125 g L−1, corresponding to protein molar concentration from 38 mM up to 188 mM) and two different solvent compositions (0 and 1.5 M urea) were considered. In all cases, the resulting deuteration grade, x D = n D/(n H + n D), where n D and n H are the number densities (number of atoms per total volume) of hydrogen and deuterium atoms, is slightly different for each sample, as reported in Figs. 2 and 3. The number of investigated experimental conditions was 9 for SANS and 8 for SAXS experiments (see Table 1).
SAS experiments
SANS measurements were carried out at the Laboratoire Leon Brillouin (LLB Saclay, France), using the PAXE instrument. The neutron wavelength λ n was 6 Å, the sample-detector distance 2.3 m and the resulting scattering vector Q ranged on 0.035 and 0.25 Å−1. The sample holder was a 1 mm quartz cell and the small angle scattering signal was revealed by a two-dimensional BF3-detector with 64 × 64 cells. Scattered intensities were radially averaged and corrected for background, buffer contribution, detector inhomogeneities and sample transmission by means of software available at LLB. Scattering cross sections were converted to absolute units by a measurement of the absolute intensity of the incident beam (Teixeira 1992).
SAXS measurements were performed at the DESY synchrotron radiation source (Hamburg, Germany) at the A2 beamline. The X-ray wavelength λ X was 1.5 Å, the sample-detector distance 1.15 m, and the corresponding Q range was between 0.02 and 0.3 Å−1. Samples were measured in cells 1 mm thickness with thin mica windows. The small angle scattering signal was recorded using a two dimensional CCD camera with 1024 × 1024 pixels. Images were radially averaged and corrected for the dark signal, sample transmission and buffer contribution.
Theoretical analysis
SANS and SAXS curves were analysed according to the same theoretical principles, only considering for each case the scattering length density or the electron density of the sample, respectively. Hereafter for simplicity the definition scattering density will refer alike to the scattering length density and to the electron density.
The main equation for monodisperse and randomly oriented particles in solution concerns the macroscopical differential scattering cross section \(\frac{\hbox{d}\Sigma}{\hbox{d}\Omega}(Q):\)
where n p is the protein number density, S(Q) the effective structure factor, P(Q) the protein averaged squared form factor and B a flat background.
The protein form factor describes the scattering arising from the protein in solution together with its solvation shell. BSA was then considered as a particle of homogeneous scattering density (core domain, p) covered by a shell of different scattering density (local domain, l) and thickness δ l , dissolved in an homogeneous solvent (bulk, b) (Feigin and Svergun 1987; Spinozzi et al. 2000, (2002; Sinibaldi et al. 2007). The corresponding form factor is:
where ρp, ρ l and ρb are the scattering densities of the different domains, V p and V l are the protein and the local domain scattering volumes and P ij (Q) is the partial form factor (Spinozzi et al. 2000, 2002). Scattering densities of the different domains were calculated according to the amino acid composition for the protein (ρp) (Jacrot 1976) and to the amounts of water and urea for the local domain (ρ l ) and the bulk (ρb), respectively (Sinibaldi et al. 2007). Urea partial molecular volume was considered to be equal in the bulk and in the local domain (ν u,b = ν u,l); by contrast, the volume of water molecule in contact with protein surface (νw,s) was considered as a fitting parameter, as it has been established that it could be different from the value observed in the bulk (ν w,b), (Svergun et al. 1998; Sinibaldi et al. 2007). The estimation of each P ij (Q) factor was achieved by the Monte Carlo method previously described (Spinozzi et al. 2000). Because the BSA crystalline structure is not yet available, the crystallographic structure of human serum albumin [HSA, PDB entry 1 bke (Curry et al. 1998)], which shares 90% sequence homology with the BSA (Svergun et al. 2001), was used.
In each investigated condition, the bulk and local domain solvent compositions have been derived considering an equilibrium exchange of water and urea molecules between the two domains as described by Schellman (2003). In particular, each protein is considered to present at its surface m solvation sites that can be occupied either by water or cosolvent molecules and no one could result unoccupied. The number of the m accessible sites can be calculated by dividing the volume of an envelope of 3 Å thickness around the protein surface with the volume of a water molecule (Sinibaldi et al. 2007). According to Schellman model (2003), each cosolvent molecule can occupy just one site, so that cosolvent molecules extend over the first layer eventually formed by water molecules, as schematically shown in Fig. 1.
Schematic representation of a solvated BSA molecule based on PDB structure 1 bke. Bulk composition was 1.5 M, while the thermodynamic constant was K = 0.5. The light- and dark-blue circles represent water molecules in the bulk and in the solvation layer, respectively; the three connected circles represent urea molecules in the bulk (pink) and in contact with the protein (purple)
The solvation layer (s) is different from the local domain (l), which is a layer of larger thickness δ l around protein surface. Local domain thickness is obtained from the fit procedure, according to whom it is not possible to distinguish between the geometry of a solvation layer and the one of a local domain, since SAS signal is studied via an isotropic average (Karen et al. 2002; Sinibaldi et al. 2007). The solvation layer composition is determined by a thermodynamic equilibrium constant K responsible for the molecular exchange process with bulk, and the space between the solvation layer and the local domain is filled with a mixture whose composition is equal to the one relative to bulk.
The process of solvent exchange happens on each site i of the protein surface:
where u b, u s, and w b, w s represent urea and water molecules in the bulk phase and in contact with protein surface, that is in the solvation layer, respectively. Hence, the thermodynamic exchange equilibrium constant is
where x w,s and x w,b are the molar fraction of water in the region close to the protein surface and in the bulk, respectively.
The effective structure factor S(Q) describes protein–protein interactions and in our case could provide information about the modifications induced by the cosolvent in the two body interactions. S(Q) was calculated under the random phase approximation (Narayanan and Liu 2003), adopting an interaction potential which includes an hard sphere potential, a coulombic screened potential and an attractive Yukawian term (Sinibaldi et al. 2007; Javid et al. 2007). As a few parameters, concerning protein-protein interactions, can be affected by urea, we need to detail at least two terms. The first is the coulombic screened potential, which is written as:
where Z is the protein charge, R is the protein radius, ε is the bulk dielectric constant [which depends on the amount of urea in solution (Lide 1996)], and κD is the Debye constant, which depends on the ionic strength I S of the urea–water mixture. Note that to ensure the electroneutrality condition of the solution, counterion concentration calculated according to the protein charge has been also taken into account (Spinozzi et al. 2002). The second is the attractive potential, which is written as
where J is the depth of the potential at contact (r = 2R) and d is the decay length.
As reported above, we resorted to a global fit strategy to apply the thermodynamic exchange solvation model (Sinibaldi et al. 2007; Ortore et al. 2008). The main common parameters are the thermodynamic constant K and the local domain thickness δ l [the entire set of equations needed to calculate the three phase form factor P(Q) as a function of K are reported in a previous paper (Sinibaldi et al. 2007)]. For what concerns the interaction parameters, we considered the protein charge Z to be independent on the protein concentration, but free to vary with urea content (Bhattacharya et al. 2004). On the other side, the depth and the decay length of the attractive potential (J and d pairs) were considered as free parameters for each experimental condition.
Preferential binding coefficient
The preferential binding coefficient is defined as: \(\Upgamma_{{\rm p} j}=\lim_{n_{\rm p}\rightarrow 0}\left(\frac{\partial n_j}{\partial n_p}\right)_{{\rm P,T},\mu_j} (j=w,u),\) where n j refers to the number density of the j-th species and the indices P, T and μ refer to pressure, temperature and chemical potential, respectively. The preferential binding coefficient can be expressed as Γpj = n j (G pj −G wu), where G ij are KB integrals (Kirkwood and Buff 1951; Shulgin and Ruckenstein 2005).
At one side when the thickness δ l of the local domain and the sample composition are known, from the value of the exchange equilibrium constant K the G pj term can be calculated by:
where V s is the volume of the solvation layer and n j,s is the number density of the j-th species inside V s. On the other side, the water-urea term has been calculated in the approximation of unitary activity coefficients (Shulgin and Ruckenstein 2005) as G wu = k B T k T −(n w + n u)ν w,b ν u, being k T the isothermal compressibility of the mixed solvent. Note that a direct relationship between Γpj , x w and K was derived by some of us (see Eq. 14 in Sinibaldi et al. 2007), taking the limit n p → 0. Another relevant parameter that can be easily determined is the excess solvation number N pj = n j G pj , which represents the number of displaced molecules j when a protein molecule is introduced into the mixture (Shimizu 2004).
Results
SANS and SAXS results are shown in Figs. 2 and 3, respectively. Scattering curves are ordered according to protein concentration (c), urea molar content, and sample deuteration grade. It is important to note that in each experimental condition the three dimensional structure of BSA resulted globular and compact, as shown by a few Kratky plots reported by way of example in Fig. 4. Therefore, even at the highest urea concentration considered, BSA maintains its native structure, as suggested by previous denaturation works (Ahmad and Qasim 1995).
SANS experimental data obtained at different experimental conditions. The corresponding best fitting curves are superimposed to the experimental points. The three columns correspond to samples prepared at the same urea molarity, while each row corresponds to different protein concentration, as indicated. Numbers reported close to each curve indicate the sample deuteration grades, which vary because urea and protein are not deuterated. For the sake of clarity, each curve has been scaled by a factor multiple of 0.5 cm−1
SAXS experimental data obtained at different experimental conditions. The corresponding best fitting curves are superimposed to the experimental points. The curves are organized as indicated in Fig. 2, using the same scale factor of 0.5 cm−1 to shift each curve
It appears also evident that at the highest investigated protein concentrations the position of the interference peak shows a remarkable sensitivity to the presence of urea. From a qualitative point of view, this means that small amounts of urea, which are not able to unfold the BSA, induce large modifications in the interactions between the proteins. This is a very important point: indeed, this is the reason that, in the global fit analysis, the interaction parameters were not considered constant for all the investigated experimental conditions. It should be also observed that changes in protein charges were already detected in lysozyme dissolved in water/urea mixtures (Bhattacharya et al. 2004; Ortore et al. 2008).
Best fit curves obtained by simultaneously analysing all the data are reported in Figs. 2 and 3. As they are superimposed to experimental points, the good quality of the fitting results can be directly appreciated. Fitted parameters (both common and related to each curve) are shown in Tables 1 and 2.
Concerning protein–protein interaction, results are reported in terms of protein effective charge as a function of the urea content of the solution in Table 2, and in terms of the J and d pairs for each experimental condition in Table 1. The BSA charge slightly increases with addition of urea in solution, showing a certain trend close to, but outside from, the calculated errors. Concerning the attractive potential, the depth decreases while increasing protein concentration, in agreement with previous literature results (Zhang et al. 2007; Javid et al. 2007); moreover, the depth also decreases with increasing urea content. The whole results lead to the conclusion that urea noticeably modifies protein-protein interactions, increasing the repulsive potential and decreasing the attractive one (Niebuhr and Koch 2005).
It should be noticed that to evidence eventual correlations between the attractive and the repulsive potentials, several tests have been performed by means of different fitting attempts. In any case no evident correlations between the two terms have been found. As an example, fixing the parameters related to the attractive potential, the effective protein charge qualitatively changes in a similar way as reported in Table 2, but the fit quality was worse.
The other fitted parameters common to all the experimental curves are also reported in Table 2. The protein volume changes simply testify that small amount of urea slightly modify the BSA surface architecture (Ravindra and Winter 2004): the fitted protein volumes, as well as the protein charges, suggest that weak modifications should happen at the protein surface in the presence of urea, without that relevant shape changes are induced, since the protein form factor well reproduces experimental curves in every conditions. Concerning the other parameters, it can be observed that the local domain thickness agrees with previous results obtained with other proteins, and it is in full agreement with radial distribution functions recently obtained by molecular dynamics for urea on protein surfaces, which show a first minimum at a distance greater than the thickness of the first water hydration layer (Baynes and Trout 2003). Moreover, the molecular volume of water in the local domain results to be smaller with respect to the water volume in the bulk and appears comparable to that found in previous works (Svergun et al. 1998; Sinibaldi et al. 2007).
The main result obtained by the global fit procedure is however the thermodynamic exchange constant, which in this case results K = 0.6 ± 0.1, in very good agreement with the value detected for lysozyme in urea/water mixtures (K = 0.52 ± 0.08, Ortore et al. 2008). The observed value, lower than one, clearly claims that in every experimental condition the local domain results enriched in urea with respect to the bulk: the BSA is preferentially solvated by urea. Therefore, despite the very small variations of BSA charge and volume induced by urea, it can be suggested that the observed effects are definitively related to the preferential interaction of urea with the protein surface.
Discussion
The results described in this work can be divided into two main classes: those regarding the analysis of the protein solvation shell and the ones related to the protein–protein interactions.
The results concerning urea preferential binding to the protein surface fully agree with literature (Timasheff and Xie 2003; Zhang et al. 1996) and with previous unpublished results (Ortore et al. 2008). However, a further step to obtain a deeper comparison with achieved results is possible. As previously described for a protein dissolved in a mixed solvent (Sinibaldi et al. 2007), we developed a method that combines SAS results obtained at relatively high concentrations with experimental thermodynamical data related to infinite dilution conditions, studied according to Kirkwood–Buff (KB) theory. The preferential binding coefficient is the most common and used parameter to describe the accumulation or diminution of components near or at the surface of a protein (Schellman 2005).
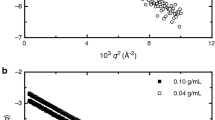
The comparison between preferential binding coefficients determined in this work by the thermodynamic constant and the ones already reported in literature (Zhang et al. 1996) is shown in Fig. 5. It can be directly observed that data are in agreement with each other. Note that data from Zhang and coworkers (Zhang et al. 1996) were derived from vapour pressure osmometry performed in quite similar buffer conditions, but in light water. Figure 6 shows the excess solvation number as a function of solvent composition. It can be observed that at all the investigated conditions the local domain is enriched in urea: note however that, up to a certain urea concentration, the local domain contains more water molecules than an equal volume of bulk: this is due to the higher density of water molecules in contact with the protein surface. As a conclusion, data on preferential binding (Figs. 5, 6) show that urea preferentially expels water from the protein surface, hence local composition of cosolvent is higher in respect to bulk.
Dependence on urea composition of the aqueous solution of the preferential binding coefficients Γpu (upper frame) and Γpw (bottom frame). The points reported in the upper frame are literature data as obtained by vapour pressure osmometry (Zhang et al. 1996)
The change in protein–protein interaction due to the presence of urea is another clearly visible result of this study. Although it was already suggested that the unfolding process induced by urea could be related to protein charge modifications (Caballero-Herrera et al. 2005), there is no precise description of this phenomenon. Now, the present results clearly suggest a denaturation mechanism in which the main process is the urea localization on the surface of the protein. The contact between urea with charged residues can lead to a repulsion between them, and hence induce an “outside in” denaturation process that finally change the protein surface architecture, and as a consequence, the total protein charge. It is evident that we are not able to state that this effect is the premise for the consequent full denaturation of the protein and/or that this effect can be widely associated to urea action on all proteins, but it merits to be further investigated. We point out that observations of charge modifications induced by small amount of urea in proteins solutions have been already proved by other experimental techniques (Bhattacharya et al. 2004). As already observed for lysozyme (Niebuhr and Koch 2005) we found that the attractive potential is modified by urea. However, we now demonstrate that for BSA the attractive contribution is also dependent on protein concentration. Since the attractive interaction term includes van der Waals as well as hydration or hydrophobic or other nonspecific contributions, it is difficult to assess the origin of the observed changes.
Conclusions
The goals of this study can be summarized in three points. Firstly, it is shown that a set of SAS experiments can provide quantitative information about the composition of protein solvation shell in a mixture if a solvation model is considered. Secondly, the same data set is useful to derive a complete description of protein–protein interactions. Thirdly, the quantitative data related to the first two points are achieved just using a global fit procedure in the analysis of experimental curves. The original methodology to determine a thermodynamic equilibrium constant responsible for the composition of protein solvent interphase from a neutron scattering experiment and to relate it to thermodynamic data, has been recently published (Sinibaldi et al. 2007). This further study both confirms the validity of the methodology to another protein in a different environment and opens the perspective to be adopted in SAXS experiments. The efficiency of this analysis is strictly related to the experimental conditions investigated. This methodology can be in fact successfully applied if it succeeds previous numerical simulations (Sinibaldi et al. 2007), which, taking into account the different scattering density of cosolvents and the protein form factor, set the best experimental conditions necessary to evidence the small differences expected in SAS results. Finally, we underline that our results have been achieved performing a global fit on both SANS and SAXS data, confirming the validity of the method for two different experimental techniques.
References
Ahmad N, Qasim MA (1995) Fatty acid binding to bovine serum albumin prevents formation of intermediate during denaturation. Eur J Biochem 227:563–565
Baynes BM, Trout BL (2003) Proteins in mixed solvents: a molecular-level perspective. J Phys Chem B 107:14058–14067
Bhattacharya J, Ghoshmoulick R, Choudhuri U, Chakrabarty P, Bhattacharya PK, Lahiri P, Chakraborti B, Dasgupta AK (2004) Unfolding of hemoglobin variants—insights from urea gradient gel electrophoresis photon correlation spectroscopy and zeta potential measurements. Anal Chim Acta 522:207–214
Bloom JD, Labthavikul ST, Arnold CROFH (2006) Protein stability promotes evolvability. Proc Natl Acad Sci USA 103:5869–5874
Caballero-Herrera A, Nordstrand K, Berndt KD, Nilsson L (2005) Effect of urea on peptide conformation in water: molecular dynamics and experimental characterization. Biophysical Journal 89:842–857
Courtenay ES, Capp MW, Anderson CF, Record MT Jr (2000) Vapor pressure osmometry studies of osmolyte–protein interactions: implications for the action of osmoprotectants in vivo and for the interpretation of “osmotic stress” experiments in vitro. Biochemistry 39:4455–4471
Curry S, Mandelkow H, Brick P, Franks N (1998) Crystal structure of human serum albumin complexed with fatty acid reveals an asymmetric distribution of binding sites. Nat Struct Biol 5:827–835
Dill KA, Ozkan SB, Weikl TR, Chodera JD, Voelz VA (2007) The protein folding problem: when will it be solved? Curr Opin Struct Biol 17:342–346
Farnum M, Zukoski C (1999) Effect of glycerol on the interactions and solubility of bovine pancreatic trypsin inhibitor. Biophys J 76:2716–2726
Feigin LA, Svergun DI (1987) Structure analysis by small-angle X-ray, neutron scattering. Plenum Press, New York
Gekko K, Timasheff SN (1981) Thermodynamic and kinetic examination of protein stabilization by glycerol. Biochemistry 20:4667–4676
Hansen JP, McDonald IR (1986) The theory of simple liquids. Academic Press, London
Jacrot B (1976) The study of biological structures by neutron scattering from solution. Reg Prog Phys 39:911
Javid N, Vogtt K, Krywka C, Tolan M, Winter R (2007) Protein–protein interactions in complex cosolvent solutions. Chem Phys Chem 8:679–689
Karen R, Tang S, Bloomfield VA (2002) Assessing accumulated solvent near a macromolecular solute by preferential interaction coefficients. Biophys J 82:2876–2891
Kirkwood JG, Buff FP (1951) The statistical mechanical theory of solutions. J Chem Phys 19:774–777
Lide DR (1996) Handbook of chemistry and physics, 77th edn. CRC Press, Cleveland
Narayanan J, Liu XY (2003) Protein interactions in undersaturated and supersaturated solutions:a study using light and X-ray scattering. Biophys J 84:523–532
Niebuhr M, Koch MHJ (2005) Effects of urea and trimethylamine-n-oxide (tmao) on the interactions of lysozyme in solution. Biophys J 89:1978–1983
Ortore MG, Spinozzi F, Carsughi C, Mariani P, Bonetti M, Onori G (2005) High pressure small-angle neutron scattering study of the aggregation state of β-lactoglobulin in water and water/ethylene glycol solutions. Chem Phys Lett 418:338–342
Ortore MG, Sinibaldi R, Spinozzi F, Carsughi F, Clemens D, Bonincontro A, Mariani P (2008) Sans study on the molecular action of denaturants: the lysozyme-urea case (submitted)
Pagan DL, Gunton JD (2005) Phase behavior of short-range square-well model. J Chem Phys 122:184515
Pellicane G, Costa D, Caccamo C (2004) Theory and simulation of short-range models of globular protein solutions. J Phys Condens Matter 16:4923–4936
Ravindra R, Winter R (2004) Pressure perturbation calorimetry: a new technique provides surprising results on the effects of co-solvents on protein solvation and unfolding behaviour. Chem Phys Chem 4:566–571
Schellman JA (2005) Destabilization and stabilization of proteins. Quart Rev Biophys 38:351–361
Schellmann JA (2003) Protein stability in mixed solvents: a balance of contact interaction and excluded volume. Biophys J 85:108–125
Shimizu S (2004) Estimating hydration changes upon biomolecular reactions from osmotic stress, high pressure, and preferential hydration experiments. Proc Natl Acad Sci USA 101:1195
Shulgin IL, Ruckenstein E (2005) A protein molecule in an aqueous mixed solvent: fluctuation theory outlook. J Chem Phys 123:054909
Shulgin IL, Ruckenstein E (2006) A protein molecule in a mixed solvent: the preferential binding parameter via the Kirkwood–Buff theory. Biophys J 90:704–707
Shulgin IL, Ruckenstein E (2007) Local composition in the vicinity of a protein molecule in an aqueous mixed solvent. J Phys Chem B 111:3990–3998
Sinibaldi R, Ortore MG, Spinozzi F, Carsughi F, Frielinghaus H, Cinelli S, Onori G, Mariani P (2007) Preferential hydration of lysozyme in water/glycerol mixtures: a small angle neutron scattering study. J Chem Phys 126:235101–235109
Smith PE (2006) Chemical potential derivatives and preferential interaction parameters in biological systems from Kirkwood–Buff theory. Biophys J 91:849–856
Spinozzi F, Carsughi F, Mariani P, Teixeira CV, Amaral LQ (2000) Sas from inhomogeneous particles with more than one domain of scattering density, arbitrary shape. J Appl Cryst 33:556–559
Spinozzi F, Gazzillo D, Giacometti A, Mariani P, Carsughi F (2002) Interaction of proteins in solution from small angle scattering: a perturbative approach. Biophys J 82:2165–2175
Spinozzi F, Carsughi F, Mariani P, Saturni L, Bernstorff S, Cinelli S, Onori G (2006) Met-myoglobin association in dilute solution during pressure-induced denaturation: an analysis at pH 4.5 by high-pressure small-angle x-ray scattering. J Phys Chem B (in press)
Stradner A, Sedgwick H, Cardinaux F, Poon W, Egelhaaf S, Schurtenberger P (2004) Equilibrium cluster formation in concentrated protein solutions and colloids. Nature 432:492–495
Stradner A, MThurston G, Schurtenberger P (2005) Tuning short-range attractions in protein solutions: from attractive glasses to equilibrium clusters. J Phys Condens Matter 17:S2805–S2816
Svergun D, Richard S, Koch MHJ, Sayers Z, Kuprin S, Zaccai G (1998) Protein hydration in solution: experimental observation by X-ray, neutron scattering. Proc Natl Acad Sci USA 95:2267–2272
Svergun DI, Petoukhov MV, Koch MHJ (2001) Determination of domain structure of proteins from X-ray solution scattering. Biophys J 80:2946–2953
Teixeira J (1992) Introduction to small angle neutron scattering applied to colloidal science. In: Chen S-H, Huang JS, Tartaglia P (eds) Structure and dynamics of supramolecular aggregates and strongly interacting colloids. Kluwer, Drodecht
Timasheff SN (1998) Control of protein stability and reactions by weakly interacting cosolvents: the simplicity of the complicated. Adv Protein Chem 51:355–432
Timasheff SN, Xie G (2003) Preferential interactions of urea with lysozyme and their linkage to protein denaturation. Biophys Chem 105:421–448
Valente J, Verma K, Manning M, Wilson W, Henry C (2005) Second virial coefficient studies of cosolvent-induced protein self-interaction. Biophys J 89:4211–4218
van Oss CJ (2003) Long-range and short-range mechanisms of hydrophobic attraction and hydrophilic repulsion in specic and aspecic interactions. J Mol Recogn 16:177–190
Yancey P, Clark M, Hand S, Bowlus R, Somero G (1982) Living with water stress: evolution of osmolyte systems. Science 217:1214–1222
Zhang W, Capp MW, Bond JP, Anderson CF, Record MT Jr (1996) Thermodynamic characterization of interactions of native bovine serum albumin with highly excluded (glycine betaine) and moderately accumulated (urea) solutes by a novel application of vapor pressure osmometry. Biochemistry 35:10506–10516
Zhang F, Skoda MWA, Jacobs RMJ, Martin RA (2007) Protein interactions studied by saxs: effect of ionic strength and protein concentration for bsa in aqueous solutions. J Phys Chem B 111:251–259
Acknowledgments
We thank Fondazione Cariverona for financial support.
Author information
Authors and Affiliations
Corresponding author
Additional information
Advanced neutron scattering and complementary techniques to study biological systems. Contributions from the meetings, “Neutrons in Biology”, STFC Rutherford Appleton Laboratory, Didcot, UK, 11–13 July and “Proteins At Work 2007”, Perugia, Italy, 28–30 May 2007.
Rights and permissions
About this article
Cite this article
Sinibaldi, R., Ortore, M.G., Spinozzi, F. et al. SANS/SAXS study of the BSA solvation properties in aqueous urea solutions via a global fit approach. Eur Biophys J 37, 673–681 (2008). https://doi.org/10.1007/s00249-008-0306-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00249-008-0306-z