Abstract
The phytopathogen Pseudomonas syringae pv. syringae produces toxic cyclic lipodepsipeptides (CLPs): nona-peptides and syringopeptins. All CLPs inhibit the growth of many fungal species, including human pathogens, although different fungi display different degrees of sensitivity. The best studied CLPs are Syringomycin-E (SR-E), Syringotoxin-B (ST-B) and Syringopeptin-25A (SP-25A). Their biological activity is affected by membrane composition and their structural differences. We previously (Matyus et al. in Eur Biophys J 35:459–467, 2006) reported the molecular features and structural preferences of SR-E in water and octane environments. Here we investigate in atomic detail the molecular features of the two other main CLP components, ST-B and SP-25A, in water and octane by 200 ns molecular dynamics simulations (MD), using distance restraints derived from NMR NOE data (Ballio et al. in Eur J Biochem 234:747–758, 1995). We have obtained three-dimensional models of ST-B and SP-25A CLPs in different environments. These models can now be used as a basis to investigate the interactions of ST-B and SP-25A with lipid membranes an important further step towards a better understanding of the antifungal and antibacterial activity of these peptides.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pseudomonas syringae pv. syringae produces two groups of cyclic lipodepsipeptides (CLPs): the nonapeptides: Syringomycins (SRs) (Segre et al. 1989), Syringostatins (Fukuchi et al. 1991, 1992; Isogai et al. 1990), and syringotoxins (STs) (Ballio et al. 1990), and the more complex syringopeptins (SPs) (Ballio et al. 1991). They inhibit the growth of many fungal species with different degrees of efficiency (Sorensen et al. 1996) and have phytotoxic (Gross and Devay 1977) (Iacobellis et al. 1992) and antimicrobial effects (Lavermicocca et al. 1997). Syringopeptins have an antimicrobial spectrum of activity distinct from that of nonapeptides (Bender et al. 1999) and have stronger phytotoxic activity than the nonapeptides. This different antimicrobial-, hemolytic- and plant pathogenic activity might be explained by the different structures of these toxins, in particular the number of charges and the hydrophobicity of the different CLPs (Szabo et al. 2002). Their primary biological target is the plasma membrane (Dalla Serra et al. 1999a, b). The SPs have the highest pore-forming activity due to their high percentage of hydrophobic amino acids, while SRs are less active and STs have the lowest activity.
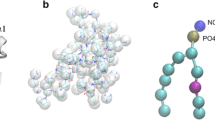
The chemical structures of the two lipodepsipeptides STs and SPs show definite differences. The primary structures of two prominent members of CLPs (ST-B and SP-25A) have been elucidated using NMR and mass spectroscopy and the absolute configurations at the chiral Cα atoms are known (Ballio et al. 1990, 1994). ST-B consists of a cyclic nonapeptide head, which contains four uncommon amino acids (Dab2: 2,4-diaminobutanoic acid, Dhb7: 2,3-dehydro-2-aminobutyric acid, (3-OH)Asp8: 3-hydroxy-aspartic acid, (4-Cl)Thr9: 4-chlorothreonone), closed by a lactone formed between the hydroxyl group of the Ser1 side chain and the carboxylate group of (4-Cl)Thr9. One hydrophobic 3-hydroxytetradecanoic fatty acid tail is linked via an amide bond to the Ser1 amino group (Fig. 1). The net charge of ST-B is +1.
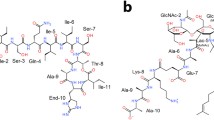
SP-25A contains 25 amino acids (Ballio et al. 1991). Eight of them (from 18 to 25) form a lactone ring via an esther bond between the hydroxyl group of allothreonine (Thr18) and the C-terminal tyrosine (Tyr25) residue. The N-terminal uncommon amino acid 2,3-dehydro-2-aminobutyric acid (Dhb1) is acylated by 3-hydroxydecanoic acid (R) (Fig. 2). The net charge of SP-25A is +2.
The conformation of SP-25A obtained by computer simulation in vacuo, using experimental NOE data, includes three different structural regions: a loop from 2 to 6, a left-handed helicoidal zone from 8 to 15, and the lactone ring from residues 18 to 25 (Ballio et al. 1995). In the first region two turns (one inverse γ-turn and one type II β-turn) are detected which are stabilized by hydrogen bonds (H-bond). The helicoidal zone is stabilized by three i · i + 4 H-bonds; a putative fourth one (between Ala9 and Ala13) is not detected. The two turns of the lactone ring give a structure similar to that found in the structure of ST-B (Ballio et al. 1995). These turns are stabilized by three H-bonds involving the i · i + 3 residues. The fatty chain is not involved in any of the structured region. It is flexible, despite the presence of the hydrogen bond between the OH of the fatty acid chain and the NH of Dhb1.
The present paper describes the structure and conformational flexibility of ST-B and SP-25A lipodepsipeptides in hydrophilic and hydrophobic environments during 200 ns molecular dynamics (MD) simulations. The MD simulations in water and octane environments predict structural preferences of both peptides, in particular, the importance of side-chain interactions in determining peptide structure. We also compared the SP-25A structure in vacuo, water and octane environments, taking into account available experimental information from Ballio et al. (1991, 1990, 1995).
We highlight the conformational similarity of nonapeptides and syringopeptin. The resulting structures will be used in future simulations of the interactions between CLPs and lipid bilayers in order to understand their pore forming mechanism.
Material and methods
Initial models of ST-B and SP-25A were created using InsightII (Accelrys), based on the experimental chemical structure and stereo chemistry (Ballio et al. 1990, 1991). The starting structure of the CLPs are arbitrary extended conformations which were solvated in a periodic cubic box (4.2 nm × 4.2 nm × 4.2 nm for ST-B, 5 × 5 × 5 for SP-25A) large enough to contain the peptide and 0.75 nm of solvent on all sides with either SPC water (Berendsen et al. 1981); or octane. The net charge on the ST peptide was +1, the net charge of the SP-25A was +2; the systems do not contain ions. The parameters of the uncommon 2,3-dehydro-aminobutyric acid (Dhb), and atoms (involved in the lactone bond) undefined in the force field were developed in our previous work (Matyus et al. 2006). Force constants for bonds and angles were taken from similar groups in the force field. Because bond lengths were constrained in the production runs their force constants are not critical. For a few atoms we estimated charges by comparison with the charges elsewhere in the force field for consistency.
Each system was energy minimized in 200 steps using the steepest descent algorithm. In all simulations, the temperature was 300 K and the pressure 1 bar, using the weak coupling method with τT = 0.1 ps and τP = 1 ps, respectively. The minimized ST-B and SP-25A models were simulated in water (W-ST, W-SP, resW-SP) and octane (O-ST, O-SP) for 200 ns, except O-SP was simulated for 100 ns. In resW-SP 82 short-range and 9 long-ranged NMR NOEs restraints were applied (Ballio et al. 1995), while in the W-SP, W-ST, O-SP and O-ST simulations no restraints were applied. Distances were restrained by adding a non-physical term to the potential function when the distance between specified pairs of atoms exceeds the experimentally determined distance. The potential form for distance restraints was quadratic between two specified upper bounds and linear beyond the largest bound (s = 0.25 + 0.05 nm, m/s = 0.3 + 0.05, m = 0.35 + 0.05, m/w = 0.38 + 0.05 nm, w = 0.4 + 0.05 nm). All simulations and the analyses of the resulting trajectories were done with the GROMACS-3.2.1. software package (Lindahl et al. 2001; van Maaren and van der Spoel 2001). For the unrestrained simulations the GROMACS ffgmx force-field and for the restrained simulation the GROMACS ffgmx2 force-field was used (Berendsen et al. 1995). The latter has the same parameters as ffgmx but includes explicit hydrogen sites for NOE restraints. Bonds were constrained using LINCS for the peptide (Hess et al. 1997) and SETTLE for water (Miyamoto and Kollman 1992). Electrostatic and Lennard–Jones interactions were calculated using a twin-range cut-off of 0.8/1.4 nm. Interactions within the short-range cut-off were updated every time step (2 fs), whereas interactions within the long-range cut-off were updated every ten steps together with the pair list. All atoms were given an initial random velocity obtained from a Maxwellian distribution at the desired initial temperature. Cluster analyses were carried out using the GROMOS clustering algorithm as described in Daura et al. (1999) with a cut-off of 0.3 nm. A maximum distance of 0.3 nm between hydrogen and acceptor and a cut-off angle of 60° between donor-hydrogen-acceptor were used as hydrogen bond criteria. The interproton distances were calculated for each trajectory separately using <r −6>−1/6 averaging. A cut-off value of 0.4 nm between charged groups was used to define salt bridges.
Results
Figure 3 shows the root mean square deviation (RMSD) of the backbone and the whole peptide with respect to the starting structure as function of time for all five simulations (W-ST, O-ST, W-SP, resW-SP, O-SP). The backbone RMSD of both ST systems increases initially from the starting model, and then fluctuates around 0.24 nm during the entire trajectories, reflecting the rigidity of the ST backbone. The higher fluctuation of the whole peptide RMSD around 0.55 nm for W-ST and O-ST is due to side chain flexibility. The MD simulation of SP-25A started from a fully extended chain configuration. The RMSD of W-SP quickly rises at 0.75 nm during 25 ns as the peptide rapidly coils. The restrained system reaches the coiled structure sooner and the RMSD value levels off at 0.4 nm. The RMSD values are higher in octane solvent than in water system, from the same starting structure, with a rapid initial rise to 1.4 nm. In the second half of the trajectory its value decreases from 1.7 to 1.5 nm. This modification is due to conformational changes of the loop (residues from Pro2 to Val6) and of the lactone ring (data not shown).
The radius of gyration R g, a measure of the compactness averaged over whole trajectory, of the peptide backbone is ∼0.4 nm for both W-ST and O-ST, and ∼0.7 nm for W-SP, resW-SP and O-SP (Fig. 4). The SP-25A backbone is more compact in resW-SP than in the W-SP because about half of the distance restraints (short-range NMR NOE) imposed in the resW-SP simulation involve the amino acid backbone.
To identify the structures corresponding to the different RMSD levels and to identify the structural differences corresponding to the different radii of gyration in water and octane we used cluster analysis; structures from our 200 ns trajectories were clustered into related conformations based on the whole peptide RMSD. Figure 5 shows the dominant structures for each of the simulations.
A, B The central structures of the major clusters of ST-B conformations in the W-ST simulation; C, D the central structures of the major clusters of ST-B conformations in the O-ST simulation; E, F the central structures of the major clusters of SP-25A conformations in the resW-SP simulation; G, H, I the central structures of the major clusters of SP-25A in the O-SP simulation
Two ensembles of structures characterize the W-ST trajectory. The A and B conformers (Fig. 5A, B) are the central structures from the first and second largest clusters, with a population of 60 and 17%, respectively. The most significant difference between the two most populated conformers is the position of the alkyl chain, which is elongated next to the backbone in the B conformer and folded towards the peptide backbone in the A conformer. The residues from Orn5 to Asp8 are involved in a type III β-turn (Gao et al. 2002) in both conformers (with ϕ6 = −71°, ψ6 = −58°; ϕ7 = −70°, ψ7 = −63°; ϕ6 = −75°, ψ6 = −58°; ϕ7 = −58°, ψ7 = −50°, respectively), which is stabilized by a hydrogen bond in both clusters between the Asp8 carboxyl oxygen and the Orn5 amino hydrogen. The side chains of these two amino acids are not involved in salt bridge formation with one another. The Asp8 side chain approaches the Dab2 side chain and forms a salt bridge with it in the B conformer.
Two conformations characterize the ST-B structure in octane. Figure 5C, D shows the central structures of the most populated clusters (cluster C, 95% of total population, and cluster D, 4.5% of total population). In the O-ST the lactone ring is stabilized by the same hydrogen bond between Orn5 and Asp8 as observed in W-ST, and Asp8 forms a salt bridge with Dab2 throughout the trajectory. In the D conformer the hydroxyl group of Asp8 is involved in a second hydrogen bond with the amino group of Dab2. This conformer is stabilized by a third hydrogen bond between the hydroxyl group of Ser4 and the carbonyl group of Thr9. The residues from Orn5 to Asp8 are not involved in any known β-turn. A γ-turn is present, involving Thr6 and Asp8 in both conformers with ϕ7 = 69°, ψ7 = −82° and ϕ7 = −65°, ψ7 = −55°, respectively.
A detailed description of the mobility of the lactone ring in W-ST and O-ST can be obtained by calculating the root mean square fluctuation (RMSF) of the atomic position of the respective residues. The flexibility profile of residues from Dab2 to Ser4 is significantly different in hydrophobic and hydrophilic environments due to hydrogen bonding and the salt bridge between the side chains of Dab2 and Asp8. In addition, the hydroxyl group of Ser4 forms a H-bond with the carbonyl group of the Thr9 in the hydrophobic environment which is not detected in the hydrophobic environment. The relative rigidity of Asp8 is due to the presence of a salt bridge with Dab2 and an H-bond with Orn5 and Dab2 in hydrophobic solvent (Fig. 6).
Thirty-five NOE violations >0.1 nm are found in the starting structure of SP25A (Electronic supplementary material Table 1). Averaging the distances over the last 100 ns of the resW-SP trajectory 20 violations are observed. Thirteen violations are localized around lactone ring closure in the starting structure, but after 100 ns MD only seven violations are detected. The helicoidal zone contains the highest number of violations after 100 ns MD, because with eight violations (down from ten initially). No violations are found in the loop. Three violations are found in the alkyl chain in the starting structure, all of which disappear during the MD simulation. Because of the better agreement between the calculated and measured NOEs after 200 ns than between the calculated and measured NOEs of the starting structure, we assume that the long MD simulations provide a substantially improved description of the SP-25A structure and dynamics.
In water, the restrained SP-25A adopts two dominant conformations with populations of 81% (conformer E) and 18% (conformer F). Conformer F is found between 120 and 150 ns; the conformer E is predominant in the remaining parts of the trajectory (Fig. 5). The first structured region of SP-25A is considerably different in the two conformers. The loop is characterized by a distorted γ-turn between Pro2 and Ala4 in the E conformer where the ϕ3, ψ3 values at the Val3 residue differ appreciably from the ideal values for the i + 1 residue, being 116°, -113° instead of ϕ3 = 75°, ψ3 = −64° (typical values for the type γ-turn).In the F conformer one distorted type II β-turn, involving the Val3 to Val6 residues is detected with the ϕ4 = −61°, ψ4 = 125°, ϕ5 = 104°, ψ5 = −92°. The type II β-turn displays a strong Ha(4)-5N and a weak 4N-5N NOE, where the corresponding distances are 0.21 and 0.44 nm [0.22 and 0.45 are the theoretical values (Schuman et al. 2003)] (ESM Table 1). The two hydrogen bonds (between Pro2 and Ala4; Val3 and Val6) predicted (Ballio et al. 1995) for this region occurred less than 50% of the whole trajectory in the water environment. The backbone amide NH of Val3 forms a hydrogen bond with the CO group of Leu7, which characterizes the whole trajectory with 95% occurrence in conformer E and 98% in conformer F.
The helicoidal region is characterized by two and three hydrogen bonds in conformer E and F, respectively. One of them is i · i + 4 type between the CO group of Ala8 and the NH of Dhb12 in both conformers. Ala8 forms a second H-bond with Val11 in both conformers. The third one between CO of Ala13 and NH of Ala15 is found only in the F conformer. All hydrogen bonds occur in more than 50% of the trajectory. This region ends with a γ-turn involving the Ala15 and Dhb17 with ϕ16 = 84, ψ16 = −146 in both conformers. The NOEs characteristic of the γ-turn are shown in ESM Table 1.
For both clusters, the lactone ring of the central structure is very similar to the starting structure (the RMSD of the lactone ring central structure relative to the starting structure is 0.08 nm). Residues from Ala22 to Tyr25 are involved in a distorted type II β-turn which is stabilized by one hydrogen bond between OH of Tyr25 and NH of Ala22 in the E conformer. The calculated NOEs characteristics of a type II β-turn (for the whole trajectory) (ESM Table 1) correspond to theoretical values (Schuman et al. 2003). Residues from Ser19 to Ala22 form a distorted type II′ β-turn stabilized by a hydrogen bond but its occurrence is less than 50% of the trajectory. However, the calculated NOEs characteristics correspond to theoretical values (ESM Table 1).
The alkyl chain is not involved in any of the structured regions. It is flexible, despite the presence of hydrogen bonds between the hydroxyl group of the second carbon of the chain and the CO of Val6, and CO of the alkyl chain and NH of Ala16 in the E conformer (Fig. 5). These two hydrogen bonds are not found in conformer F, where the alkyl chain is more elongated than in conformer E.
In octane, the SP-25A adopts three dominant conformations with populations of 36% (conformer G, 65–100 ns), 25% (conformer H, 10–33 ns) and 12% (conformer I, 43–60 ns) (Fig. 5). The structures of these three conformers are very different.
The G conformer is stabilized by two hydrogen bonds with occurrences higher than 50% of the whole trajectory. The hydroxyl group of the alkyl chain is involved in a hydrogen bond with NH of Ala16. The second structured region is characterized by one i · i + 4 hydrogen bond between CO of Ala9 and NH of Ala13. Around Leu7, not involved in the helicoidal region, loop, or lactone ring, a γ-turn is formed with ϕ7 = 20°, ψ7 = −87. The second region ends with an inverse γ-turn involving Ala15 and Dhb17 with ϕ16 = −53°, ψ16 = 151°, and the characteristic NOEs (ESM Table 1). In the lactone ring, the predicted γ-turn between Dab23 and Tyr25 is distorted (the ϕ24 = 134°, ψ24 = −132° values differ from ideal values) which is evident in the calculated NOEs too. Between Ser19 to Ala22 residues one type-VIa2 β-turn is found.
Four hydrogen bonds and five turns characterize the H conformer. The helicoidal zone is characterized by one i · i + 4 hydrogen bond between NH of Ala15 and CO of Val11. Another three interactions (CO Leu7 and NH Ser19; CO Val13 and NH Val21; CO Dab24 and NH Ala4) connect the lactone ring with the loop and helicoidal zones, determining a different molecular shape observed in conformer G. The loop is involved in an inverse γ-turn (Pro2 and Ala4) where the ϕ3 = −126°, ψ3 = 73 for the i + 1 residue and with characteristic NOEs (ESM Table 1); and a distorted type VIa2 β-turn between residues from Val3 to Val6. It ends with an inverse γ-turn involving Val6 and Ala8 with ϕ7 = −55°, ψ7 = 95°. Residues from Dab23 to Tyr25 are involved in a distorted γ-turn (the Dab24 residue differs appreciably from the ideal values for the i + 1 residue, being 134°, −132°). The last turn, type VIa1 β-turn, is included in the third region from Ser19 to Ala22.
In conformer I the loop is characterized by a distorted inverse γ-turn between Pro2 and Ala4 where the ϕ3 = −93°, ψ3 = 125° for the Val3 residue and is stabilized by one hydrogen bond between CO of Ala4 and NH of Val6. The loop region ends with an inverse γ-turn involving the Val6 and Ala8 with ϕ7 = −78°, ψ7 = 71°; the helicoidal zone ends with Ala15 and Dhb17 with ϕ16 = −62°, ψ16 = 203°. The third region is stabilized by a hydrogen bond between CO of Ala 20 and NH of Dab24 and a type I β-turn is detected between residues Ser19 to Ala22.
The alkyl chain is flexible despite the presence of hydrogen bonds between the CO of the chain and NH of Thr18.
Discussion
It was shown in earlier studies that syringopeptins have stronger phytotoxic activity than the nonapeptides (Lavermicocca et al. 1997). SP-25A has the highest pore forming activity, while SRE has less activity and ST-B has the weakest activity on planar bilayers and red blood cells (Agner et al. 2000a; Szabo et al. 2002). SR-E and ST-B show temperature-dependent pore inactivation, but SP-25A-pores did not inactivate (Agner et al. 2000b; Szabo et al. 2002). To understand the distinct biological effect of ST-B and SP-25A on membranes a detailed knowledge of their structures in different environments is required. This paper is focused on the structural features and preferences of ST-B and SP-25A in octane and water as a step towards understanding their membrane activity. We modeled ST-B and SP-25A structure in solution by long (200 ns) molecular dynamics simulations with and without explicit incorporation of NOE-based distance restraints. Analyses of the trajectories suggested two families of structures in all systems except O-SP in which three clusters were observed.
The most significant difference found among the ST-B conformers is the position of the alkyl chain, which is folded across the peptide backbone in the A conformer, while it is elongated along the backbone in the B and C conformers, and folded towards to the peptide backbone in the D conformer. The Orn5 amide group and the Asp8 carboxyl group form a hydrogen bond in the A conformer, a salt bridge in the B conformer and both in C and D conformers. The amino acid residues from Orn5 to Asp8 are involved in a type III β-turn in the A and B conformers, but not in the C and D conformers.
In our previous paper we highlighted the structural preferences of SR-E in hydrophobic and hydrophilic environments, in particular the importance of side-chain interactions in determining peptide stability (Matyus et al. 2006). Due to the similarities in the primary structure of ST-B and SR-E they have some similar conformational characteristics.
Both molecules have two dominant conformers, determined by the position of the alkyl chain both in hydrophobic and hydrophilic environments. The negatively charged Asp residue stabilizes the backbone of SR-E and ST-B molecules, forming hydrogen bonds and salt bridges. The shape of the backbone conformation has been described as the “seam of a tennis ball” and is in agreement with the results of Ballio et al. (1994). Furthermore, all CO and the side chains of polar groups (Dab2 and Orn5) are external to the backbone. Vaillo and coworkers predicted the presence of a β-turn in SR-E molecules by CD spectroscopy (Vaillo et al. 1992) which was confirmed by our simulations (Matyus et al. 2006). They suggested that due to the presence of Dhb and Phe residues the lactone ring possesses a turn which may be stabilized by the vicinal Asp residue. Our simulations reveal that the Dhb7 residue of ST-B molecule is also involved in a β-turn (water solvent) and a γ-turn (octane solvent) stabilized by hydrogen bonding of the Asp residue. The φ, ψ values at the Dhb7 residue differ from the ideal values because the Asp residue is deprotonated, forming a salt bridge with one of the positively charged residues (Dab2 residue). This would require a considerable degree of conformational change in the peptide backbone (Vaillo et al. 1992). The Orn5 has a less important role in stabilization compared to Arg in SR-E molecule however they have +1 charge and same position in the lactone ring of both molecules. The negatively charged Asp interacts favorably with another positively charged residue (Dab2), whose position is closer to Asp in ST-B than in SR-E.
The SP-25A molecule has a marked amphiphilic character and its primary structure is very different from nonapeptides. It contains a flexible fatty acid chain and a long peptide moiety with mixed chirality and a high content of Dhb residues that produce rigidity along the peptide chain. The last eight residues form a lactone ring with a +2 charge.
Ballio and co-workers simulated SP-25A in vacuo using NOE data and they found three structural regions. In the loop region two turns are detected with two hydrogen bonds. The helicoidal zone is stabilized by three i · i + 4 H-bonds; the fourth one (between Ala9 and Ala13) is not detected. The presence of two turns in the lactone ring results in a conformation resembling the seam of a tennis ball (Ballio et al. 1995). These turns are stabilized by three hydrogen bonds involving the i · i + 3 residues. Their CD spectrum studies on the spectrum of SP-24A showed the presence of a contribution from α-helical forms in aqueous solution. For the spectrum in 2,2,2-trifluoroethanol/H2O, the amount of left-handed α-helix was calculated to be 15% and of β-turns 11% were calculated (Ballio et al. 1995).
After 200 ns MD simulation of SP-25A in solvent the three structural characteristic features of the peptide (loop, helicoidal, and ring regions) determined in vacuum are also present. The structure of the loop region in solution is determined by a mixed chirality which produces conformational preferences for turns. The loop is characterized by one γ-turn involving the Pro2 and Ala4 residues in the E conformer and a β-turn involving the Val3 and Val6 residues in the F conformer. The hydrogen bonds stabilize the turns but they are present less than 50% occurrence in the whole trajectory. In octane, the formation of turns is favorable because the loop is involved in both an inverse γ-turn and a β-turn which are stabilized by hydrogen bonds in most of the trajectory. The Leu7 residue is not part of a structured region, probably because of the heterochiral junction with Val6 (Ballio et al. 1995). In octane it forms a γ-turn throughout the trajectory.
The presence of Ala residues promotes a helicoidal conformation. Visual inspection of the E and F conformers shows that this region is indeed formed. It is stabilized by two or three hydrogen bonds (with different occurrences in whole trajectory) but just one of them is an i · i + 4 type H-bond characteristic of alpha-helices (between the Ala8 and Dhb12 residues). In the hydrophobic environment this region is also characterized by one i · i + 4 H-bond. In case of the G conformer this is the predicted, but experimentally not detected H-bond between the Ala9 and Ala13 residues.
The third region, the lactone ring, is formed by eight residues and has structural similarities with both SR-E and ST-B. It contains two uncommon, positively charged Dab amino acids, as in SR-E. It is involved in a β-turn as in the nonapeptides. This turn (including residues Ser19 to Ala 22) was expected (Ballio et al. 1995) but not identified previously. Due to the presence of the second turn in the ring its shape might resemble the seam of a tennis ball, predicted by (Ballio et al. 1995).
Conclusions
Using long molecular dynamics simulations we have obtained three-dimensional structures for ST-B and SP-25A CLP molecules. Our results indicate that ST-B exists in two dominant structures in both water and octane. SP-25A has two dominant structures in water that are consistent with NMR NOE measurements, and three dominant structures in octane (All 3D structures of CLPs will be available from our home page: http://moose.bio.ucalgary.ca). The C conformer and the E conformer are more relevant to previous experiments. They can now be used as a basis to investigate the interactions of ST-B and SP-25A with lipid membranes, the mechanism of channel formation, and the structure of channels formed by CLPs.
References
Agner G, Kaulin YA, Gurnev PA, Szabo Z, Schagina LV, Takemoto JY, Blasko K (2000a) Membrane-permeabilizing activities of cyclic lipodepsipeptides, syringopeptin 22A and syringomycin E from Pseudomonas syringae pv. syringae in human red blood cells and in bilayer lipid membranes. Bioelectrochemistry 52:161–167
Agner G, Kaulin YA, Schagina LV, Takemoto JY, Blasko K (2000b) Effect of temperature on the formation and inactivation of syringomycin E pores in human red blood cells and bimolecular lipid membranes. Biochim Biophys Acta 1466:79–86
Ballio A, Bossa F, Collina A, Gallo M, Iacobellis NS, Paci M, Pucci P, Scaloni A, Segre A, Simmaco M (1990) Structure of Syringotoxin, a bioactive metabolite of Pseudomonas-Syringae pv. syringae. Febs Lett 269:377–380
Ballio A, Barra D, Bossa F, Collina A, Grgurina I, Marino G, Moneti G, Paci M, Pucci P, Segre A, Simmaco M (1991) Syringopeptins, new phytotoxic lipodepsipeptides of Pseudomonas-Syringae pv. syringae. Febs Lett 291:109–112
Ballio A, Collina A, Dinola A, Manetti C, Paci M, Segre A (1994) Determination of structure and conformation in solution of Syringotoxin, a lipodepsipeptide from Pseudomonas-Syringae Pv. syringae by 2D Nmr and molecular-dynamics. Struct Chem 5:43–50
Ballio A, Bossa F, Di Giorgio D, Di Nola A, Manetti C, Paci M, Scaloni A, Segre AL (1995) Solution conformation of the Pseudomonas syringae pv. syringae phytotoxic lipodepsipeptide syringopeptin 25-A. Two-dimensional NMR, distance geometry and molecular dynamics. Eur J Biochem 234:747–758
Bender CL, Alarcon-Chaidez F, Gross DC (1999) Pseudomonas syringae phytotoxins: mode of action, regulation, and biosynthesis by peptide and polyketide synthetases. Microbiol Mol Biol Rev 63:266–292
Berendsen HJC, Postman JPM, van Gusteren, Hermans J (1981) Intermolecular forces. D. Reidel Publishing Company, Dordrecht, pp 331–342
Berendsen HJC, van der Spoel D, van Drunen R (1995) Gromacs: a message-passing parallel molecular dynamics implementation. Comp Phys Comm 91:43–51
Chang CT, Wu CS, Yang JT (1978) Circular dichroic analysis of protein conformation: inclusion of the beta-turns. Anal Biochem 91:13–31
Dalla Serra M, Bernhart I, Nordera P, Di Giorgio D, Ballio A, Menestrina G (1999a) Conductive properties and gating of channels formed by syringopeptin 25A, a bioactive lipodepsipeptide from Pseudomonas syringae pv. syringae, in planar lipid membranes. Mol Plant Microbe Interact 12:401–409
Dalla Serra M, Fagiuoli G, Nordera P, Bernhart I, Della Volpe C, Di Giorgio D, Ballio A, Menestrina G (1999b) The interaction of lipodepsipeptide toxins from Pseudomonas syringae pv. syringae with biological and model membranes: a comparison of syringotoxin, syringomycin, and two syringopeptins. Mol Plant Microbe Interact 12:391–400
Daura X, Gademann K, Jaun B, Seebach D, van Gunsteren WF, Mark AE (1999) Peptide folding: when simulation meets experiment. Angew Chem Int Ed 38:236–240
Fukuchi N, Isogai A, Suzuki A (1991) Stereochemistry of syringostatin, syringomycin and syringotoxin, phytotoxins of Pseudomonas syringae pv. syringae. Agric Biol Chem 55:625–627
Fukuchi N, Isogai A, Nakayama J, Takayama S, Yamashita S, Suyama K, Takemoto JY, Suzuki A (1992) Structure and stereochemistry of 3 phytotoxins, syringomycin, syringotoxin and syringostatin, produced by Pseudomonas-Syringae Pv. Syringae. J Chem Soc Perkin Trans 1:1149–1157
Gao F, Wang Y, Qiu Y, Li Y, Sha Y, Lai L, Wu H (2002) Beta-turn formation by a six-residue linear peptide in solution. J Pept Res 60:75–80
Gross DC, Devay JE (1977) Role of Syringomycin in holcus spot of maize and systemic necrosis of Cowpea caused by Pseudomonas-Syringae. Physiol Plant Pathol 11:1–11
Hess B, Bekker H, Berendsen HJC, Fraaije JGEM (1997) LINCS: a linear constraint solver for molecular simulations. J Comput Chem 18:1463–1472
Iacobellis NS, Lavermicocca P, Grgurina I, Simmaco M, Ballio A (1992) Phytotoxic properties of Pseudomonas syringae pv. syringae toxins. Physiol Mol Plant Pathol 40:107–116
Isogai A, Fukuchi N, Yamashita S, Suyama K, Suzuki A (1990) Structures of Syringostatin-a and Syringostatin-B, novel phytotoxins produced by Pseudomonas-Syringae Pv-Syringae isolated from Lilac Blights. Tetrahedron Lett 31:695–698
Lavermicocca P, Iacobellis NS, Simmaco M, Graniti A (1997) Biological properties and spectrum of activity of Pseudomonas syringae pv. syringae toxins. Physiol Mol Plant Pathol 50:129–140
Lindahl E, Hess B, van der Spoel D (2001) GROMACS 3.0: a package for molecular simulation and trajectory analysis. J Mol Model 7:306–317
Matyus E, Monticelli L, Kover KE, Xu Z, Blasko K, Fidy J, Tieleman DP (2006) Structural investigation of syringomycin-E using molecular dynamics simulation and NMR. Eur Biophys J 35:459–467
Miyamoto S, Kollman PA (1992) Settle––an analytical version of the Shake and Rattle algorithm for rigid water models. J Comput Chem 13:952–962
Schuman J, Campbell AP, Koganty RR, Longenecker BM (2003) Probing the conformational and dynamical effects of O-glycosylation within the immunodominant region of a MUC1 peptide tumor antigen. J Pept Res 61:91–108
Segre A, Bachmann RC, Ballio A, Bossa F, Grgurina I, Iacobellis NS, Marino G, Pucci P, Simmaco M, Takemoto JY (1989) The structure of Syringomycins A1, E and G. FEBS Lett 255:27–31
Sorensen KN, Kim KH, Takemoto JY (1996) In vitro antifungal and fungicidal activities and erythrocyte toxicities of cyclic lipodepsinonapeptides produced by Pseudomonas syringae pv. syringae. Antimicrob Agents Chemother 40:2710–2713
Szabo Z, Grof P, Schagina LV, Gurnev PA, Takemoto JY, Matyus E, Blasko K (2002) Syringotoxin pore formation and inactivation in human red blood cell and model bilayer lipid membranes. Biochim Biophys Acta 1567:143–149
Vaillo E, Ballio A, Luisi PL, Thomas RM (1992) The spectroscopic properties of the lipodepsipeptide, Syringomycin-E. Biopolymers 32:1317–1326
van Maaren PJ, van der Spoel D (2001) Molecular dynamics simulations of water with novel shell-model potentials. J Phys Chem B 105:2618–2626
Acknowledgments
The EM work was supported by Hungarian Scientific Research Fund, Grants F 043192 and D 048670. DPT is an Alberta Heritage Foundation for Medical Research Senior Scholar and Canadian Institutes of Health Research New Investigator. This work was supported by the Natural Science and Engineering Research Council (Canada).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Mátyus, E., Blaskó, K., Fidy, J. et al. Structure and dynamics of the antifungal molecules Syringotoxin-B and Syringopeptin-25A from molecular dynamics simulation. Eur Biophys J 37, 495–502 (2008). https://doi.org/10.1007/s00249-007-0242-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00249-007-0242-3