Abstract
Assessment of the diversity of algal assemblages in Antarctica has until now largely relied on traditional microbiological culture approaches. Here we used DNA metabarcoding through high-throughput sequencing (HTS) to assess the uncultured algal diversity at two sites on Deception Island, Antarctica. The first was a relatively undisturbed site within an Antarctic Specially Protected Area (ASPA 140), and the second was a site heavily impacted by human visitation, the Whalers Bay historic site. We detected 65 distinct algal taxa, 50 from within ASPA 140 and 61 from Whalers Bay. Of these taxa, 46 were common to both sites, and 19 only occurred at one site. Algal richness was about six times greater than reported in previous studies using culture methods. A high proportion of DNA reads obtained was assigned to the highly invasive species Caulerpa webbiana at Whalers Bay, and the potentially pathogenic genus Desmodesmus was found at both sites. Our data demonstrate that important differences exist between these two protected and human-impacted sites on Deception Island in terms of algal diversity, richness, and abundance. The South Shetland Islands have experienced considerable effects of climate change in recent decades, while warming through geothermal activity on Deception Island itself makes this island one of the most vulnerable to colonization by non-native species. The detection of DNA of non-native taxa highlights concerns about how human impacts, which take place primarily through tourism and national research operations, may influence future biological colonization processes in Antarctica.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
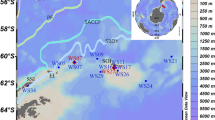
Deception Island, located in the South Shetland Islands, with a maximum altitude of 576 m a.s.l., is one of very few active volcanoes in the Antarctic Treaty area (above latitude 60° S) and one of only two in the region that has had human-witnessed eruptions [1]. It is a relatively young volcanic island, with an age less than 100 kya [2] and is still undergoing colonization. Deception Island is horseshoe-shaped with a diameter of ca. 15 km. Its inner flooded caldera forms Foster Bay, and about 57% of its land surface is currently covered by ice [1, 3]. Two national Antarctic research stations are presently active on the island (the Argentinean Decepción and Spanish Gabriel de Castilla), and historically Chile and the UK also operated stations on the island. The Chilean station was destroyed and the British station heavily damaged during the most recent series of eruptions on the island in the late 1960s [3]. Whalers Bay (62° 59′ S, 60° 34′ W) was also the site of an active whaling station in the early twentieth century, and the station’s remains are now designated as a historic site. Whalers Bay is also one of the most visited sites in Antarctica by tourists, with more than 80,000 tourists visiting the island in the summers of 2007–2010 [4]. The unique geology, history, biota, and esthetic values, as well as the activity of multiple national operators, underlie the designation of the entire island as an Antarctic Specially Managed Area (ASMA 4). In addition, Deception Island includes two Antarctic Specially Protected Areas (ASPAs), designated as ASPA 140 (terrestrial, formed of multiple subsites) and 145 (marine).
Terrestrial biological communities on Deception Island have received some research attention. The flora includes a number of Bryophyta (57 species) [5] and Marchantiophyta (6 species) [6], some associated with geothermally heated sites [3] and not found elsewhere in the Antarctic Treaty area, though also occur in similar geothermal habitats on the maritime Antarctic South Sandwich Islands [7, 8]. No flowering plants are cited in the management plan for the island [9], but both native Antarctic species are known to occur. Similarly, these sites also host a number of terrestrial micro-arthropods not found elsewhere in the treaty area [10,11,12]. As well as having specific geothermally active sites, the entire island is thought to be under slight geothermal influence and is thought to be particularly vulnerable as a result to the colonization (by natural means or human-assisted) of non-native species of plants and invertebrates [11, 13].
Traditionally the study of soil algal diversity in Antarctica has relied heavily on morphology alone, including two methods, direct observation using microscopy and cultivation [14,15,16,17,18,19,20]. Soil microalgal assemblages on Deception Island were investigated by Fermani et al. [17], who sampled across 18 sites that reflected the island’s geological and edaphic variability and reported the presence of 140 taxa, of which 26 were representatives of Chlorophyta. However, due to the extreme environmental conditions typical of Antarctica, microalgae and other taxa can exhibit significant morphological variation due to local environmental conditions [21]. In addition, cultivation methods can be selective towards generalist rather than rare or specialist species [15], with some species simply not being currently cultivable. Thus, traditional culturing methods fail to adequately represent the range of microalgal diversity in nature [22, 23].
Recent developments in molecular biology have allowed considerable advances in the assessment of microbial diversity in environmental samples such as soil and water. DNA metabarcoding using high-throughput sequencing (HTS) represents a superior method for detection of rare species [23, 24], including their resting stages which are typically not detected in morphological surveys [25]. To date very few studies exist comparing morphological and molecular approaches for Antarctic algae. Rippin et al. [24] in a study on Livingston Island suggested that a molecular approach yielded richness estimates 11 times higher than a traditional morphological approach. Fraser et al. [26], working on continental Antarctica volcanoes, investigated the importance of geothermal areas for supporting life, including their possible role as refugia during glaciations. Garrido-Benavent et al. [27], also working on Livingston Island, used a more ecological approach to investigate the successional patterns of microorganisms, comparing assemblages of bacteria, fungi, and algae present in three different substrata (moraines, rocks, and soil). In the current study, we applied HTS for the first time to investigate Chlorophyta (Viridiplantae) diversity in assemblages present in soil samples from two sites on Deception Island, one from within a protected area (ASPA 140 subsite B) and the other under considerable visitor impact.
Materials and Methods
Taxonomic Definitions
We follow the definition of Viridiplantae of Leliaert et al. [28], as a monophyletic group comprising Chlorophyta and Streptophyta. Only green algae lineages are considered here, regardless of their systematic classification. We do not discuss the taxonomy of groups cited or nomenclatural validity, and geographical distributions are based on cited literature and Guiry and Guiry [29]. Taxa are referred to at genus and species level and higher hierarchical levels (e.g., families, orders) are omitted.
Study Sites and Sample Collections
Deception Island is located at 62° 57′ S and 60° 38′ W (Fig. 1). Two sites were selected to assess and compare the diversity of soil green algal communities on the island. The first was within a protected area (ASPA 140, subsite B, Crater Lake) (Fig. 1), which has a low degree of disturbance by both volcanic and human research activity (tourists are prohibited from entering). The area is distant from the shore, slightly raised elevation, and protected from direct exposure to sea spray by coastal hills. The second area was in Whalers Bay, which lies within the ASMA but beyond the limits of ASPA 140 and is a less protected and much more intensively visited site, both by tourists and national operator personnel, and was also more directly impacted by eruptions in the late 1960s [1, 3, 17]. The area is at sea level and its location close to the shore is more susceptible to sea spray.
During the austral summer of 2018/2019, soil samples (ca. 250 g each) were collected in both sites using a sterilized spatula and avoiding visibly vegetated areas (bare soil). Samples were kept in sealed sterile plastic bags (Whirl Pack®/US) and frozen (− 20 °C) until DNA extraction. Seven (non-composite) samples from each site were collected from the upper 5 cm of the soil profile for use in DNA studies, totaling 14 samples [30,31,32].
DNA Extraction and Data Analysis
DNA Extraction, Illumina Library Construction, and Sequencing
Total DNA was extracted from environmental samples using the QIAGEN Power Soil Kit (QIAGEN, Carlsbad, USA), following the manufacturer’s instructions. DNA quality was analyzed by agarose gel electrophoresis (1% agarose in 1 × Trisborate-EDTA) and then quantified using Quanti-iT™ Pico Green dsDNA Assay (Invitrogen). The internal transcribed spacer 2 (ITS2) of the nuclear ribosomal DNA was used as a DNA barcode for molecular species identification [33, 34] using the universal primers ITS3 and ITS4 [35]. Library construction and DNA amplification were performed using the Library kit Herculase II Fusion DNA Polymerase Nextera XT Index Kit V2, following Illumina 16S Metagenomic Sequencing Library Preparation Part #15044223 Rev. B protocol. Paired-end sequencing (2 × 300 bp) was performed on a MiSeq System (Illumina) by Macrogen Inc. (South Korea).
Data Analysis
Raw reads were interleaved and filtered using BBDuk version 38.34 (BBMap—Bushnell B.—sourceforge.net/projects/bbmap/) to remove Illumina adapters, known Illumina artifacts, and PhiX Control v3. Quality read filtering was carried out using Sickle version 1.33 [36] to trim ends 3′ or 5′ with low Phred quality score, and sequences smaller than 100 bp were also discarded. Remaining fastq sequences (Supplementary Table 1) were converted to FASTA format using reformat.sh from BBMap and compared against Viridiplantae ITS2 database (http://its2.bioapps.biozentrum.uniwuerzburg.de/) [37] using Nucleotide-Nucleotide BLAST 2.6.0+. Taxonomic assignments were determined by the maximal matching from BLASTn alignment using MEGAN V6.12.3 [38].
Ecological Diversity Analysis
Rarefaction calculations were carried out using the rarefaction analysis command in the software MOTHUR, where we clustered sequences into OTUs by setting a 0.03 distance limit. The following diversity statistics were calculated: Fisher’s α, Shannon, Margalef, Simpson, and evenness, to assess alpha diversity. We also performed a diversity t test, for comparison of the Shannon and Simpson diversities in both areas using PAST 3.26 [39]. Non-metric multidimensional scaling (NMDS) ordination was performed to check the existence of differences in community composition at the sampled localities, using the Bray-Curtis index and also using PAST 3.26 [39]. For comparison between the two communities in relation to turnover and nestedness, we calculated beta diversity indexes using “beta-multi” option in the R package “betapart” using the Sørensen dissimilarity index [40]. As there are no studies available using the same approach, we used two lists of Chlorophyta extracted from publications by Broady [41, 42] in our statistical analyses for exploratory comparisons. Venn diagrams are based on Bardout et al. [43]
Results
A total of 4,097,406 reads were generated in the sequencing run, and 3,608,576 ITS2 reads remained after quality filtering (Suppl. Table ST1, Suppl. Fig. S1). The use of 97% similarity cutoff [44, 45] resulted in 67 ITS2 OTUs, representing 65 taxa of soil green algae according to the BLAST search. The calculated rarefaction curves approached a plateau, indicating that the reads gave an accurate representation of the local algal diversity in both sites (Suppl. Figs. S2 and S3).
Of the overall total of 65 taxa of soil green algae, 50 were present at ASPA 140 subsite B (Crater Lake) and 61 at Whalers Bay. These included 35 taxa previously unreported from Antarctica (Table 1). Abundance and distribution are shown in Table 2. The alpha diversity indices are presented in Table 3. The diversity t test was significant (p < 0.05), indicating differences in species composition between the two assemblages.
Alpha diversity indexes are presented in Table 3. The Shannon index values differed significantly between Crater Lake and Whalers Bay, with diversity being higher in Whalers Bay and that species assemblages differing between the two areas. The Simpson index was high in both Crater Lake and Whalers Bay and again significantly different. Equitability J (Pielou’s evenness) had values close to 0.6, suggesting that both species assemblages did not have similar patterns of abundance. Together, these indexes confirm that alpha diversity of these locations, even though numerically similar, were heterogeneous.
NMDS graphs (Suppl. Fig. S4), with stress value 0.09, and first and second axes explaining 80% and 17% of overall variation suggest little variation between the two assemblages, and both are equally heterogeneous. Beta diversity comparisons showed a dominance of the turnover component (βsim) for both assemblages (Table 4), suggesting that disturbances in the areas did not influence beta diversity.
Discussion
The present study corroborates previous findings that DNA metabarcoding using HTS is more effective in detecting soil microalgal diversity than traditional methods of cultivation or direct observation. For the two study sites examined here, ASPA 140 subsite B and Whalers Bay, algal richness detected by this means was about six times higher (65 taxa) than reported by Fermani et al. [17] from the same two locations based on cultivation (nine taxa). Consequently, the overlap in species diversity is high when comparing the two studies, as well as between sites based on the molecular data, since 46 of the 65 taxa (ca. 70%) were found in both ASPA 140 and Whalers Bay (Fig. 2). Of the taxa reported, Raphidonema nivale was the only one detected in both sites in both studies. About 53% (35 out of 65) taxa have not been previously recorded in Antarctica (Table 1), with previous records of some from areas remote from Antarctica (e.g., Caulerpa webbiana). We stress that detection of DNA does not confirm presence of the most similar organism whose sequence data are available in databases [46]; it means that a sample was somehow exposed to the organism or parts of it, even single cells a factor that needs to be taken into account when using molecular data to inform on conservation or management decisions, for instance when dealing with invasive species [47].
Venn diagram showing Chlorophyta taxa distribution and shared species between both study sites. Left, HTS data alone; right, HTS data plus culture data from Fermani et al. [17]
It is known that many factors, including extraction methodology, PCR, and primer bias, can affect the number of reads [25] and thus could lead to misinterpretation of abundance [48]. However, Giner et al. [49] concluded that such biases did not affect the proportionality between reads and cell abundance, implying that more reads are genuinely linked with higher abundance [50, 51]. Therefore, we used the number of reads as a proxy for relative abundance and for comparative purposes only.
The calculated ecological indices indicate that both study sites hosted moderate algal diversity (Fisher’s α) and richness (Margalef) with low dominance (Simpson’s), with Whalers Bay being more diverse than ASPA 140 subsite B. In the absence of comparable published studies of molecular diversity of Antarctic algae against which to compare our data, we used data extracted from the studies of Broady [41, 42] (Table 3) using culture approaches, while accepting that the latter study sites are remote from Deception Island in Antarctica and not closely similar habitats. The use of HTS unveiled a cryptic algal community present in the soils of Deception Island which was more diverse, richer, and with low dominance when compared with Broady’s studies [41, 42], but we accept that the latter study sites are much further south (at least 15° of latitude) and also at around 3000 m a.s.l. Recent studies by Fraser et al. [17] and Garrido-Benavent et al. [27] had distinct aims and used different taxonomic ranks (genus and order, respectively), reducing the ability to make direct comparisons with the current study. However, comparing diversity data across studies (Table 4), the Shannon and Simpson indices differed little in the microalgal assemblages of ASPA 140 and Whalers Bay between the current study and those calculated by Garrido-Benavent et al. [27], although evenness was higher in the latter study. Garrido-Benavent et al. [27] described similar overall patterns for Chlorophyta, suggesting that local disturbances had not affected the assemblage composition.
Given the lack of modern molecular studies of the diversity of Antarctic microalgal communities, it is likely that many taxa reported for the first time from Antarctica, as in the data obtained here, will have much larger distributions than currently known. It is also important to note the caveat that studies, such as the current, of eDNA found in soil do not generate voucher material or cultures of the taxa identified (Table 5).
Some of the taxa found are common symbionts with lichen-forming fungi, such as the genera Asterochloris, Diplosphaera, and Trebouxia, and therefore, their geographical distributions are likely to be much wider. Gloetilopsis planctonica is a species of freshwater phytoplankton, whose presence in these soil samples is likely to be related to the close proximity of freshwater lakes to the sampling sites. The few marine species recorded (Caulerpa webbiana, Desmochloris halophile, Elliptochloris marina, Kornmannia leptoderma, Pseudendoclonium commune, P. submarinum) were found mostly in Whalers Bay, except P. submarinum which was present only in ASPA 140, which may be explained by the much closer proximity to the coast in the former location. Several of these species currently have very disjointed distributions globally: Pseudendoclonium commune is a photobiont found on coastal rocks in the North and Irish Seas associated with Pelvetia canaliculata (Phaeophyceae/Fucales) beds [52]; E. marina lives in symbiosis with sea anemones of the temperate Pacific Ocean in the Northern Hemisphere; D. halophile is an epibiont of a marine species of Cladophora in Wisconsin; and K. leptoderma is widely distributed in the Arctic, North Atlantic, Alaska, and Asia.
Pellizzari et al. [53] identified the seaweed Monostroma grevillei on Deception Island. This species has a biogeographic distribution similar to K. leptoderma, and both are considered cryptic monostromatic chlorophyceans. Pellizzari et al. [53] suggested that M. grevillei may have been introduced to Deception Island during whaling activity early in the twentieth century, in which ships of North Atlantic origin played a large role. K. leptoderma might represent a similar case, demanding further and detailed investigation. Most of the marine taxa present were recorded with very low abundance. However, C. webbiana displayed a higher abundance (2206 reads), which is potentially of great concern as this taxon is one of the most invasive macroalgae [54, 55]. Although currently considered to be restricted to warmer waters, it has been reported as far south as New Zealand (Norfolk Island), and the volcanic activity in Deception Island, causing water heating within Port Foster, may provide suitable conditions for the establishment of this organism that has, so far, not been reported in Antarctic waters [53].
Members of the genus Desmodesmus have been reported as potentially causing human infections [56]. The taxon was present at both current study sites, but with higher abundance in Whalers Bay. The widespread Stichococcus mirabilis was also very abundant especially in Whalers Bay and has been reported in South America as close as Argentina [57]. Taxa such as Chlorella and Stichococcus may be capable of intercontinental dispersal [57], as may also be the case for many Chlorophyta due to their small size and high resistance towards environmental stresses. Deception Island hosts a diversity of bird species, with species such as Larus dominicanus, Catharacta antarctica, and Chionis alba migrating between and common within the maritime Antarctic, southern South America, and the sub-Antarctic islands.
Human influence can also not be ruled out as a possible agent dispersing green algae to Antarctica. Many of the taxa reported here are also known from European locations, the origin of many tourist and national operator visitors to Deception Island. As a relatively young volcanic island still undergoing colonization, Deception Island is an important natural laboratory in which to study the taxonomy, ecology, and evolution of both resident and non-native species under extreme conditions.
In conclusion, our results confirm that DNA metabarcoding was able to unveil a hidden algal community present in the soils of Deception Island, identifying a much higher level of diversity than previously recorded using traditional culture methods. In addition, although only two locations were sampled, there appeared to be important differences between the protected and the strongly human-impacted site in terms of diversity, richness, and abundance. As the western Antarctic Peninsula region is among the parts of the planet most affected by recent climatic changes, the detection of DNA of taxa from multiple different parts of the world highlights concerns about the potential impact of tourism and/or scientific activities on the future biological colonization of Antarctica.
Data Availability
All soil samples analyzed in this paper are stored in the Laboratory of Microbiology at Universidade Federal de Minas Gerais.
References
Environment Protocol (2005) Management Plan for Antarctic Specially Protected Area No. 140. http://www.ats.aq/documents/recatt/Att291_e.pdf. Accessed 18 Feb 2020
Smellie JL (2001) Lithostratigraphy and volcanic evolution of Deception Island, South Shetland Islands. Antarct Sci 13:188–209. https://doi.org/10.1017/S0954102001000281
Smith RIL (2005) The thermophilic bryoflora of Deception Island: unique plant communities as a criterion for designating an Antarctic Specially Protected Area. Antarct Sci 17(1):17–27. https://doi.org/10.1017/S0954102005002385
Roura R (2012) Being there: examining the behaviour of Antarctic tourists through their blogs. Polar Res 31:1–23. https://doi.org/10.3402/polar.v31i0.10905
Ochyra R, Smith RIL, Bednarek-Ochyra H (2008) The illustrated moss flora of Antarctica. Cambridge University Press, Cambridge, p 685
Bednarek-Ochyra H, Vána J, Ochyra R, Smith RIL (2000) The liverwort flora of Antarctica. Polish Academy of Sciences, Institute of Botany, Cracow 236p
Convey P, Smith RIL (2006) Geothermal bryophyte habitats in the South Sandwich Islands, maritime Antarctic. J Veg Sci 17:529–538. https://doi.org/10.1111/j.1654-1103.2006.tb02474.x
Convey P, Smith RIL, Hodgson DA, Peat HJ (2000) The flora of the South Sandwich Islands, with particular reference to the influence of geothermal heating. J Biogeogr 27:1279–1295. https://doi.org/10.1046/j.1365-2699.2000.00512.x
Management Plan for Antarctic Specially Protected Area N° 140 - Parts of Deception Island, South Shetland Islands (2005). Available at: https://www.ats.aq/devph/en/apa-database/45. Accessed 20 May 2020
Downie RH, Convey P, McInnes SJ, Pugh PJA (2000) The non-marine invertebrate fauna of Deception Island (Maritime Antarctic): a baseline for a comprehensive biodiversity database. Polar Record 36(199):297–304. https://doi.org/10.1017/S0032247400016788
Greenslade P, Patopov M, Russell D, Convey P (2012) Global Collembola on Deception Island. J Insect Sci 12(1):111–116. https://doi.org/10.1673/031.012.11101
Pugh PJA, Convey P (2000) Scotia Arc Acari: antiquity and origin. Zool J Linnean Soc 130:309–328. https://doi.org/10.1111/j.1096-3642.2000.tb01633.x
Smith RIL, Richardson M (2011) Fuegian plants in Antarctica: natural or anthropogenically assisted immigrants? Biol Invasions 13:1–5. https://doi.org/10.1007/s10530-010-9784-x
Adams BJ, Bardgett RD, Ayres E, Wall DH, Aislabie J, Bamforth S, Bargagli R, Cary C, Cavacini P, Connell L, Convey P, Fell JW, Frati F, Hogg ID, Newsham KK, O'Donnell A, Russell N, Seppelt RD, Stevens MI (2005) Diversity and distribution of Victoria land biota. Soil Biol Biochem 38:3003–3018. https://doi.org/10.1016/j.soilbio.2006.04.030
Broady PA (1996) Diversity, distribution and dispersal of Antarctic terrestrial algae. Biodivers Conserv 5:1307–1335. https://doi.org/10.1007/BF00051981.pdf. Accessed 18 Feb 2020
Cavacini P (2001) Soil algae from northern Victoria Land (Antarctica). Polar Biosci 14:45- 60. https://www.researchgate.net/publication/252601941_Cavacini_P_Soil_algae_from_northern_Victoria_Land_Antarctica_Polar_Biosci_14. Accessed 18 Feb 2020
Fermani P, Mataloni G, Vijver BV (2007) Soil microalgal communities on an Antarctic active volcano (Deception Island, South Shetlands). Polar Biol 30:1381–1393. https://doi.org/10.1007/s00300-007-0299-6
Garraza GG, Mataloni G, Fermani P, Vinocur A (2011) Ecology of algal communities of different soil types from Cierva Point, Antarctic Peninsula. Polar Biol 34:339–351. https://doi.org/10.1007/s00300-010-0887-8
Mataloni G, Tell G, Wynn-Williams D (2000) Structure and diversity of soil algal communities from Cierva Point (Antarctic Peninsula). Polar Biol 23(3):205–211. https://doi.org/10.1007/s003000050028
Zidarova R (2007) Diversity and distribution of algae on Livingston Island, Antarctica. Comptes rendus de l'Académie bulgare des sciences: sciences mathématiques et naturelles 60(4):435–442. https://www.researchgate.net/publication/228490897_Algae_from_Livingston_Island_S_Shetland_Islands_a_checklist. Accessed 01 Jan 2020
Huss V, Frank C, Hartmann EC, Hirmer M (1999) Biochemical taxonomy and molecular phylogeny of the genus Chlorella sensu lato (Chlorophyta). J Phycol 35(3):587–598. https://doi.org/10.1046/j.1529-8817.1999.3530587.x
Rondon MR, August PR, Bettermann AD, Brady SF, Grossman TH, Liles MR, Loiacono KA, Lynch BA, MacNeil IA, Minor C (2000) Cloning the soil metagenome: a strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl Environ Microbiol 66(6):2541–2547. https://doi.org/10.1128/aem.66.6.2541-2547.2000
Ruppert K, Kline RJ, Rahman MS (2019) Past, present, and future perspectives of environmental DNA (eDNA) metabarcoding: a systematic review in methods, monitoring, and applications of global eDNA. Global Ecol Conserv 17:1–29. https://doi.org/10.1016/j.gecco.2019.e00547
Rippin M, Borchhardt N, Williams L, Colesie C, Jung P, Büdel B, Karsten U, Becker B (2018) Genus richness of microalgae and cyanobacteria in biological soil crusts from Svalbard and Livingston Island: morphological versus molecular approaches. Polar Biol 41:909–923. https://doi.org/10.1007/s00300-018-2252-2
Medinger R, Nolte V, Pandey RV, Jost S, Ottenwalder B, Schlotterer C, Boenigk J (2010) Diversity in a hidden world: potential and limitation of next-generation sequencing for surveys of molecular diversity of eukaryotic microorganisms. Mol Ecol 19(1):32–40. https://doi.org/10.1111/j.1365-294X.2009.04478.x
Fraser CI, Connell L, Lee CK, Cary SC (2018) Evidence of plant and animal communities at exposed and subglacial (cave) geothermal sites in Antarctica. Polar Biol 41:417–421. https://doi.org/10.1007/s00300-017-2198-9
Garrido-Benavent I, Pérez-Ortega S, Durán J, Ascaso C, Pointing SB, Rodríguez-Cielos R, Navarro F, de los Ríos A (2020) Differential colonization and succession of microbial communities in rock and soil substrates on a maritime Antarctic glacier forefield. Front Microbiol 11. https://doi.org/10.3389/fmicb.2020.00126
Leliaert F, Smith DR, Moreau H, Herron MD, Verbruggen H, Delwiche CF, De Clerck O (2012) Phylogeny and molecular evolution of the green algae. Crit Rev Plant Sci 31:1–46. https://doi.org/10.1080/07352689.2011.615705
Guiry MD, Guiry GM (2020) AlgaeBase. World-wide electronic publication, National University of Ireland, Galway. http://www.algaebase.org. Accessed 01 Jan 2020
Archer SDJ, Lee KC, Caruso T, Maki T, Lee CK, Cary SC, Cowan DA, Maestre FT, Pointing SB (2019) Airborne microbial transport limitation to isolated Antarctic soil habitats. Nat Microbiol 4:925–932. https://doi.org/10.1038/s41564-019-0370-4
Borruso L, Sannino C, Selbmann L, Battistel D, Zucconi L, Azzaro M, Turchetti B, Buzzini P, Guglielmin M (2018) A thin ice layer segregates two distinct fungal communities in Antarctic brines from Tarn Flat (Northern Victoria Land). Sci Rep 8:6582. https://doi.org/10.1038/s41598-018-25079-3
Cerqueira AES, Silva TH, Nunes ACS, Nunes DD, Lobato LC, Veloso TGR, De Paulo SO, Kasuya MCM, Silva CC (2018) Amazon basin pasture soils reveal susceptibility to phytopathogens and lower fungal community dissimilarity than forest. Appl Soil Ecol 131:1–11. https://doi.org/10.1016/j.apsoil.2018.07.004
Chen S, Yao H, Han J, Liu C, Song J, Shi L, Zhu Y, Ma X, Gao T, Pang X, Luo K, Li Y, Li X, Jia X, Lin Y, Leon C (2010) Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS One 5(1):e8613. https://doi.org/10.1371/journal.pone.0008613
Richardson RT, Lin C, Sponsler DB, Quijia JO, Goodell K, Johnson RM (2015) Application of ITS2 metabarcoding to determine the provenance of pollen collected by honey bees in an agroecosystem. Appl Plant Sci 3(1):1400066. https://doi.org/10.3732/apps.1400066
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic Press, New York, pp 315–322
Joshi NA, Fass JN (2011) Sickle: a sliding-window, adaptive, quality-based trimming tool for FastQ files (version 1.33) [software]. https://github.com/najoshi/sickle. Accessed 20 May 2020
Ankenbrand MJ, Keller A, Wolf M, Schultz J, Förster F (2015) ITS2 database V: twice as much. Mol Biol Evol 32:3030–3032. https://doi.org/10.1093/molbev/msv174
Huson DH, Beier S, Flade I, Górska A, El-Hadidi M, Mitra S et al (2016) MEGAN community edition - interactive exploration and analysis of large-scale microbiome sequencing data. PLoS Comput Biol 12:e1004957. https://doi.org/10.1371/journal.pcbi.1004957
Hammer, Ø, Harper DAT, Ryan PD (2001) PAST: paleontological statistics software package for education and data analysis. Palaeontol Electron 4(1):1–9. http://palaeo-electronica.org/2001_1/past/issue1_01.htm. Accessed 01 Jan 2020
Baselga A, Orme CDL (2012) “betapart”: an R package for the study of beta diversity. Methods Ecol. Evol. 3, 808–812. https://doi.org/10.1111/j.2041-210X.2012.00224.x
Broady PA (1983) Taxonomic and ecological investigations of algae on steam-warmed soil on Mt Erebus, Ross Island, Antarctica. Phycologia 23(3):257–271. https://doi.org/10.2216/i0031-8884-23-3-257.1
Broady PA (1989) Survey of algae and other terrestrial biota at Edward VII Peninsula, Marie Byrd Land. Antarct Sci 1(3):215–224. https://doi.org/10.1017/S0954102089000337
Bardou P, Mariette J, Escudié F, Djemiel C, Klopp C (2014) jvenn: an interactive Venn diagram viewer. BMC Bioinforma 15:293. https://doi.org/10.1186/1471-2105-15-293
Chen W, Zhang CK, Cheng Y, Zhang S, Zhao H (2013) A comparison of methods for clustering 16S rRNA sequences into OTUs. PLoS One 8(8):e70837. https://doi.org/10.1371/journal.pone.0070837
Koeppel AF, Wu M (2013) Surprisingly extensive mixed phylogenetic and ecological signals among bacterial operational taxonomic units. Nucleic Acids Res 41:5175–5188. https://doi.org/10.1093/nar/gkt241
Darling JA, Mahon AR (2011) From molecules to management: adopting DNA-based methods for monitoring biological invasions in aquatic environments. Environ Res 111:978–988. https://doi.org/10.1016/j.envres.2011.02.001
Comtet T, Sanionigi A, Viard F, Casiraghi M (2015) DNA (meta)barcoding of biological invasions: a powerful tool to elucidate invasion processes and help managing aliens. Biol Invasions 17:905–922. https://doi.org/10.1007/s10530-015-0854-y
Weber AA, Pawlowski J (2013) Can abundance of protists be inferred from sequence data: a case study of Foraminifera. PLoS One 8:e56739. https://doi.org/10.1371/journal.pone.0056739
Giner CR, Forn I, Romac S, Logares RC, Massana R (2016) Environmental sequencing provides reasonable estimates of the relative abundance of specific picoeukaryotes. Appl Environ Microbiol 82(15):4757–4766. https://doi.org/10.1128/AEM.00560-16
Deiner K, Bik HM, Mächler E, Seymour M, Lacoursière-Roussel A, Altermatt F, Creer S, Bista I, Lodge DM, de Vere N, Pfrender ME, Bernatchez L (2017) Environmental DNA metabarcoding: transforming how we survey animal and plant communities. Mol Ecol 26:5872e5895–5872e5895. https://doi.org/10.1111/mec.14350
Hering D, Borja A, Jones JI, Pont D, Boets P, Bouchez A, Bruce K, Drakare S, Hanfling B, Kahlert M, Leese F, Meissner K, Mergen P, Reyjol Y, Segurado P, Vogler A, Kelly M (2018) Implementation options for DNA-based identification into ecological status assessment under the European Water Framework Directive. Water Res 138:192–205. https://doi.org/10.1016/j.watres.2018.03.003
Cooley DR, Mullins RF, Bradley PM, Wilce RT (2011) Culture of the upper littoral zone marine alga Pseudendoclonium submarinum induces pathogenic interaction with the fungus Cladosporium cladosporioides. Phycologia 50(5):541–547. https://doi.org/10.2216/10-84.1
Pellizzari F, Silva MC, Silva EM, Medeiros A, Oliveira MC, Yokoya NS, Pupo D, Rosa LH, Colepicolo P (2017) Diversity and spatial distribution of seaweeds in the South Shetland Islands, Antarctica: an updated database for environmental monitoring under climate change scenarios. Polar Biol 40(8):1671–1685. https://doi.org/10.1007/s00300-017-2092-5
Amat JN, Cardigos F, Santos RS (2008) The recent northern introduction of the seaweed Caulerpa webbiana (Caulerpales, Chlorophyta) in Faial, Azores Islands (north-eastern Atlantic). Aquat Invasions 3:417–422. https://doi.org/10.3391/ai.2008.3.4.7
Cardigos F, Monteiro M, Fontes J, Serrão R (2015) Fighting invasions in the marine realm, a case study with Caulerpa webbiana proliferation in the Azores. In: Canning-Clode J (ed) Biological invasions in changing ecosystems vectors, ecological impacts. Management and Predictions. De Gruyter Open Ltd, Warsaw/Berlin, pp 279–300
Westblade LF, Ranganath S, Dunne WM, Burnham CAD, Fader R, Ford BA (2015) Infection with a chlorophyllic eukaryote after a traumatic freshwater injury. N Engl J Med 372(10):982–984. https://doi.org/10.1056/NEJMc1401816
Hodač L, Hallmann C, Spitzer K, Elster J, Faßhauer F, Brinkmann N, Lepka D, Diwan V, Friedl T (2016) Widespread green algae Chlorella and Stichococcus exhibit polar-temperate and tropical-temperate biogeography. FEMS Microbiol Ecol 92(8):fiw122. https://doi.org/10.1093/femsec/fiw122
Acknowledgments
The authors thank the National Council for Scientific and Technological Development (CNPq), the Brazilian Antarctic Program (PROANTAR), Science and Technology National Institute of Cryosphere (INCT Criosfera II), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Fundação de Amparo a Pesquisa do Estado de Minas Gerais (FAPEMIG), National Fund for Scientific and Technological Development (FNDCT), Brazilian Navy, and Brazilian Congresswoman Jô Moraes and Instituto de Ciências Biológicas at University of Brasilia for their support for this research. PC is supported by NERC core funding to the BAS “Biodiversity, Evolution and Adaptation” Team. We thank Laura Gerrish, BAS Mapping and Geographic Information Centre, for preparing Fig. 1. This study also contributes to the SCAR “State of the Antarctic Ecosystem” international research program. Michael Stech for providing useful insights into the manuscript.
Funding
The research was funded by the PROANTAR, University of Brasilia Funds, and Brazilian Congresswoman Jô Moraes parliament fund.
Author information
Authors and Affiliations
Contributions
PEASC collected soil samples, optimized protocols for DNA extraction, and wrote the first version of manuscript. MCS contributed significantly in the later versions of manuscript. OHBP filtered the data and performed the metagenomic analysis once DNA information was available. ETA performed the ecological analyses. DKH helped optimizing laboratory protocols and revised all the manuscript versions. THS worked with protocol optimization and gave inputs to methodology. FP provided significant contributions to the manuscript discussion and revised all its versions. PC contributed to the result interpretations, gave important feedback to discussion and revised all manuscript versions. LHR collected the soil samples, provided the necessary infrastructure for DNA extraction, helped writing the first version of manuscript, and revised all versions.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Ethics Approval
The collections and studies performed in ASPA 140 were authorized by the Secretariat of the Antarctic Treaty and by PROANTAR.
Code Availability
Not applicable.
Rights and permissions
About this article
Cite this article
Câmara, P.E.A.S., Carvalho-Silva, M., Pinto, O.H.B. et al. Diversity and Ecology of Chlorophyta (Viridiplantae) Assemblages in Protected and Non-protected Sites in Deception Island (Antarctica, South Shetland Islands) Assessed Using an NGS Approach. Microb Ecol 81, 323–334 (2021). https://doi.org/10.1007/s00248-020-01584-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-020-01584-9