Abstract
Iron (Fe) deficiency in crop production is a worldwide problem which often results in chlorosis in grapevines, particularly in calcareous soils. Siderophores secreted by microorganisms and Strategy II plants can chelate Fe and other metals in soil solution, and siderophore-Fe complexes can then be utilized by plants and microbes. Plants may also shift rhizosphere conditions to favor siderophore-producing microbes, which can increase plant available Fe. Between-row cover crops (barley, rye, wheat, wheat/vetch) were planted as living mulch to address grapevine chlorosis by enhancing soil health in two vineyards in central Washington. The objectives of the current study were to (1) enrich for siderophore-producing organisms from within the indigenous rooting zone community of ‘Concord’ grapevines, and (2) perform comparative genomics on putative siderophore producing organisms to assess potentially important Fe acquisition-related functional domains and protein families. A high-throughput, chrome azurol S (CAS)-based enrichment assay was used to select siderophore-producing microbes from ‘Concord’ grapevine root zone soil. Next-generation whole genome sequencing allowed the assembly and annotation of ten full genomes. Phylogenetic analysis revealed two distinct clades among the genomes using the 40 nearest neighbors available in the public database, all of which were of the Pseudomonas genus. Significant differences in functional domain abundances were observed between the clades including iron acquisition and metabolism of amino acids, carbon, nitrogen, phosphate, and sulfur. Diverse mechanisms of Fe uptake and siderophore production/uptake were identified in the protein families of the genomes. The sequenced organisms are likely pseudomonads which are well-suited for iron scavenging, suggesting a potential role in Fe turnover in vineyard systems.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chlorosis, or leaf yellowing, in grapevines is a global issue which negatively impacts production, as it may induce leaf loss, poor berry quality, low yield, and accelerated vine death [1]. Although deficiency/toxicity of several nutrients may induce chlorosis in grapevines, iron (Fe) deficiency is commonly responsible in calcareous soil [2,3,4,5]. While Fe is abundant in soil, it is primarily in a non-bioavailable form (Fe3+), which makes Fe deficiency a major agricultural problem in much of the world [3, 6]. When soil pH is high and Fe availability is low, grapevines and other Strategy I plants release protons or reductants into the rhizosphere to increase the solubility of iron in solution [7]. From a management perspective, it is possible to reduce the severity of chlorosis through addition of synthetic Fe chelates, such as Fe-EDDHA [8, 9]. Unfortunately, application of Fe-EDDHA or any other synthetic chelators are not economically viable for irrigated fields and low-economic return crops, as the effects are short-lived [10,11,12].
Microorganisms and Strategy II plants have both evolved to obtain Fe from the environment through the production of low molecular weight compounds called “siderophores” [13]. Siderophores chelate Fe and other metals in soil solution and can be taken up by plants and microbes as bioavailable Fe sources [14, 15]. Plants such as red clover can recruit specific rhizosphere microbial communities with greater capacities for siderophore secretion [16], and microbial siderophore production is often associated with increased Fe acquisition in plants grown in calcareous soils, where the majority of Fe is in an unavailable form [17, 18].
Some of these siderophores are more effective than synthetic chelators at increasing Fe and metal availability [5]. In fact, application of siderophores isolated from pure microbial cultures and management practices promoting growth of siderophore-producing microorganisms in situ have both been suggested as transformative future technologies for enhancing Fe availability [19, 20]. Concurrently, new technologies and genomics applications are rapidly revealing novel soil microbial genomes capable of encoding previously undescribed siderophores, and other secondary metabolites that represent a relatively untapped resource for sustainable crop production and antimicrobials [21]. Siderophores also serve many other roles in complex microbial communities, in terms of competition, communication, and even antagonistic pleiotropy forcing cooperation [6, 22, 23]. Now recognized as both kin recognition molecules and as antagonistic agents [22,23,24], siderophores have been implicated in many microbial interactions, including intense competition and even “cheating,” where cheating is defined as “capitalizing on the ‘public goods’ secreted by” other organisms into the extracellular environs without significant contributions in return [25, 26].
The most recent work on microbiome roles in grapevine chlorosis has discovered distinct bacterial communities between healthy and chlorotic vines, without evidence of any role of pathogenicity [27]. Therefore, while microbiome interactions with grapevines and associated soils is a nascent field of study, the role and potential mechanisms of siderophore-producing microorganisms in grapevine iron chlorosis has not been investigated. The objectives of this study were to (1) enrich siderophore-producing organisms from within the indigenous root zone community of ‘Concord’ grapevines by planting between-row cover crops to enhance soil health in two vineyards in central Washington which consistently have chlorotic grapevines and (2) construct and analyze the genomes of these organisms for potentially important biosynthetic pathways and protein families controlling Fe nutrition and microbial community dynamics.
Materials and Methods
Study Site and Experimental Design
Rooting zone soil samples were collected in June 2016, from two 20-year-old vineyard sites located in Grandview and Sunnyside, WA, located between − 119° 50′ 28″ W and − 119° 41′ 1″ W latitude and 46° 24′ 78″ N and 46° 15′ 53″ N longitude. The soils of both vineyards were Warden silt loam soils (coarse-silty, mixed mesic superactive mesic Xeric Haplocambid) with very low clay content (< 3%) and typically between 60 and 65% silt [1]. Moisture content of the study sites fluctuates due to the application of drip irrigation [28]. Soil pH is between 7.8 and 8.3 (USDA NRCS https://websoilsurvey.sc.egov.usda.gov/App/WebSoilSurvey.aspx), and the area receives approximately 150-mm mean annual precipitation with mean annual temperature around 12 °C (Ag Weather Net http://weather.wsu.edu/).
The experimental design at each site was a randomized complete block with five cover crop treatments replicated across six blocks. Each block was a single row and the cover crops were planted in the inter-rows on both sides of the vines, with eight vines per plot and two vines as buffers between plots. The cover crop treatments were barley, rye, wheat, wheat/vetch, or control (resident vegetation). Soil samples were collected from the area within the vine rows (in row) or in the area between the vine rows and under the cover crops (between row).
Soil and Plant Tissue Sampling and Characterization
Soil samples were collected using a 2.5-cm traditional soil probe (AMS, American Falls, Idaho), to a depth of 30 cm from ten locations in each plot, (5 from within the row and 5 from under the cover crop) composited within plot, according to sampling location (VR—vine row, or CC—cover crop) in zip-sealed plastic bags, mixed thoroughly and placed on ice, in the dark for transport to Washington State University (WSU) for analyses. All sampling equipment was disinfected with 70% ethanol between plots. Basic soil and plant tissue characterization was performed according to Lewis et al. (2018).
At bloom, the leaf opposite the basal cluster was sampled from the center of six vines in each plot from both sides of the canopy. Two leaves were collected from each side of the vine, resulting in 24 leaves per plot sample [29]. Leaf tissues were dried and ground with a Wiley Mill (Thomas Scientific, Swedesboro, NJ). Samples were digested in nitric acid/hydrogen peroxide and analyzed for tissue nutrient concentration via inductively coupled plasma mass spectroscopy (ICP-MS; Thermo 6500 DUo ICP, Thermo Fisher Scientific, Waltham, MA). Leaf tissue N was determined by dry combustion [30] on a Carlo Erba NA-1500 elemental analyzer (CA Elantech, Inc., Lakewood, NJ). Visual assessment of chlorosis was made on both sides of the canopy and the vine ranked on a scale from 0 to 2 in increments of 1, with 0 representing a completely healthy vine with no visual signs of chlorosis, and 2 representing a severely chlorotic vine. All vines in the plot (8 total vines) were evaluated on both sides of the canopy and then the average of both sides was taken. In total, there were 16 evaluations per plot, which were made at bloom.
High-Throughput CAS-Fe Enrichment Assay
To rapidly screen many soil samples with several technical replicates for the presence of siderophore-producing microorganisms, a high-throughput microtiter plate (microplate) assay was adapted from the traditional chrome azurol S (CAS) assay of Schwyn and Neilands [31]. Deionized, 0.22 μm filtered water (double-deionized water or DDI H2O) was used to prepare all media and reagents. Glassware and other plastic materials were acid-washed with HCl and HNO3 (100 mM:100 mM) for at least 2 h. Iron-deficient modified M9 medium (MM9) medium was prepared from the following solutions: solution 1—12.8 g Na2PO4.7H2O, 0.3 g KH2PO4, 0.5 g NaCl, and 1 g NH4Cl per 200 mL; solution 2—14.7 g CaCl2.2H2O per 100 ml; solution 3—18 g MgSO4.7H2O per 100 mL; solution 4—20% glucose (20 g/100 mL); and solution 5—PIPES buffer (prepared by dissolving 6.048 g of PIPES into 156 mL Milli-Q water with stirring). The pH of the buffer was adjusted to 6.8 with 5 M NaOH. To prepare the MM9 medium, all solutions were autoclaved separately, and solutions 1–4 were added in a sterile hood as follows: 40 mL of solution 1, 20 μL of solution 2, 266 μL of solution 3, and 4 mL of solution 4. The solution was gently mixed by hand. The CAS-Fe agarose was prepared from the following solutions: solution 6—add 15.12 g PIPES buffer in 375 mL H2O while stirring, adjust pH to 6.8 with 5 M NaOH, add 5 g agarose, add 75 mL H2O, transfer to 1 L bottle; solution 7—0.0365 g HDTMA to 20 mL H2O, place at 37 °C until dissolved; solution 8—add 25 mL H2O to 250 mL bottle, add 5 mL 1 mM FeCl3/10 mM HCl while stirring, add 0.302 g CAS while stirring, slowly add 20 mL solution 7 while gently stirring. Autoclave solutions 6 and 8 separately (with stir bars). After autoclaving, stir CAS-Fe/HDTMA (solution 8) to ensure suspension; then, stir solution 6 and slowly add solution 8 while stirring (this should turn bright, royal blue). The final solution is kept in a 45 °C water bath until cooled and while being used in the following steps and should be used relatively quickly as CAS-Fe precipitation can occur.
From each composite soil sample (120 total), 0.5 g was added to 5.0 ml of sterile MM9 media and placed on a reciprocating shaker (Model 6010, Eberbach Corportation, St. Belleville, MI, USA) at low speed for 30 min. From this slurry, a 10−3 soil dilution in MM9 broth was created and incubated at 28 °C for 24 h. After initial incubation, 100 μl of each 10−3 soil sample dilution was added to six wells along one column of a sterile, 96-well microplate. Immediately, 100 μl of CAS-Fe agarose (45 °C) was transferred to each well and mixed thoroughly with the soil dilution. Sterile water was added to one column (6 wells), as a control for sterility within the plate, and all perimeter wells were filled with sterile water to reduce evaporative losses from the experimental wells. The microplate was then covered with a lid and sealed with parafilm to allow gas exchange without contamination. The microplate assay was performed twice for all soils leading to a total of 1440 microplate wells (120 soils × 6 wells × 2 assays). Color change of the microplate wells was assessed visually, and absorbance was recorded at 420 nm after 15 days of incubation at 28 °C. All 1440 microplate wells changed color, indicating siderophore production in all the enrichment cultures. Of those 1440 wells, 16 were selected based off visual assessment for sequencing DNA isolation and sequencing. Sample characteristics of those 10 wells which yielded whole genome assembles are present in Table 1.
DNA Isolation and Sequencing
After 15 days of incubation, 100 μL of each CAS-reactive well exhibiting strong color changes in the microplate assay were collected by sterile, disposable needle, and samples were transferred to sterile 0.2-mL tubes and preserved at − 80 °C. Sample suspensions were made by mixing 100 μL molecular-grade water with 100 μL of the sample (1:1). DNA was extracted from the samples using the DNeasy® PowerSoil® kit (QIAGEN, Hilden, Germany) according to the manufacturer’s directions. Genomic DNA libraries were prepared with the Nextera DNA Library Prep kit and the Nextera Index kit (Illumina, Inc., San Diego, CA, USA). Whole genome sequencing was performed using an Illumina MiSeq with the MiSeq Reagent Kit v3 (Illumina, Inc.), to run 600 cycles, for 2 × 300 bp (base pairs) paired-end reads. The MiSeq Reagent Kit v3 uses the standard Illumina primers, which target sequences added during library preparation with the Nextera Index kit.
Sequence Analysis in KBase
Basic prokaryotic genome assembly and annotation were performed using KBase, adapted from Allen et al. [32]. DNA sequences were paired and trimmed using the Trimmomatic app (version 0.36) [33]. Metagenomes were assembled using the metaSpades app (version 3.11.1) with a contig cutoff length of 600 bp [34]. After assembly, full genomes were binned from 10 samples using MaxBin2 (version 2.2.3) with a probability threshold of 0.8, the bacterial marker gene set, and a minimum contig length of 600 bp [35]. CheckM (version 1.0.8) [36] was used to assess the genome quality, and genomes estimated to be > 90% complete were extracted using BinUtil (version 1.0.0). These binned genomes were further assessed for quality using the QUAST [37] app (version 0.0.4) and then annotated using the Annotate Microbial Assembly app (version 0.0.12), which uses Rapid Annotation of microbial genomes using Subsystems Technology (RAST) [38], with the Domain set to “Bacteria” and the Genetic Code set to “Archaea, most bacteria, most Virii, and some Mitochondria”. Phylogenetic trees were then constructed from the 10 genomes along with the 40 nearest neighbors using the Insert Genome into Species Tree app (version 2.1.10), which utilizes FastTree 2 [39]. A pangenome was constructed Build Pangenome with OrthoMCL app (PangenomeOrthomcl v0.0.7) using a close relative of the newly sequenced genomes, Pseudomonas koreensis [GC 001605965.1]. Pangenomes were visualized using the Pangenome Circle Plot app (v1.2.0). Genomes were further compared by annotating the functional domains in the whole genome set (the 10 sequenced genomes) with DomainAnnotation (version 1.03) and viewing the results using the View Function Profile for Genomes app (version 1.0.1) with the domain namespace set to “SEED Roles.” Between the clades, relative frequency of functional domains was assessed for differences using a t test with a Bonferroni correction (α = 0.00035, 143 comparisons), and qualitative differences were assessed by manually browsing the contigs in KBase.
Results
Soil Sample Overview
Soil samples had a relatively narrow range of pH values (8.1–8.9) (Table 1). DPTA extractable Fe was also fairly consistent across samples (5–10 mg kg−1 soil), with the exception of one soil from the Grandview site having DTPA-Fe of 41 mg kg−1 soil. Soil moisture ranged from 5.3 to 16.3%.
Whole Genome Phylogeny
The novel high-throughput microplate assay was useful for identifying siderophore-producing bacteria from ‘Concord’ grapevine soils. All 1440 microplate wells changed color, indicating siderophore production in all the enrichment cultures. Of those 1440 wells, 16 were selected, based on visual assessment of pronounced color change, and targeted for sequencing, 10 wells of which yielded whole genome assemblies that are 96–100% complete (Table 2). The assemblies have an average N50 value of 81 kbp, with the lowest being 26 kbp and the highest being 160 kbp. The genome sizes are similar and ranged from 5.9 to 6.1 Mbp. The average number of contigs is 228 with six genomes having < 200 contigs.
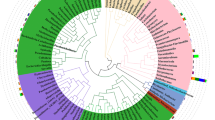
All ten taxa appear to be closely related pseudomonads with six taxa sharing a recent common ancestor and the other four sharing another recent common ancestor (Fig. 1). Sequenced whole genomes were separated into two clades with genomes 1–4 representing clade “A” and genomes 5–10 representing clade “B.” The phylogenetic construction of the two clades suggests two distinct lineages. Additionally, divergence events within each clade suggest sequencing of several distinct taxa. There were no clear correlations between clade identity and cover crop treatment, soil measurements, or chlorosis rank (Table 1).
Phylogenetic tree of the 10 newly constructed genomes with two distinct clades (A and B) located within the Pseudomonas genus. Generated by Insert Genome into Species Tree app in KBase (version 2.1.10) using the 40 closest relatives in the public database. Numbers in red are local support values. Asterisks represent genomes sequenced in this study
Pangenome Analysis
Genomes in clades A and B had a total of 29,252 and 39,476 genes, respectively (including the closest relatives) (Table S1). Of the genes in clade A, 26,363 are in homologous families among the pangenome, and 2889 are in singleton families, while in clade B, 36,475 genes are in homologous families and 3001 are in singleton families.
The clades identified in the phylogenetic analysis were also functionally distinct as determined via annotation of functional domains in the genomes using the SEED database. Protein families with significantly different relative abundance between the clades included those specifically associated with stress responses, iron acquisition, carbon metabolism, transport, metabolism of P, N, S, and amino acids (Table 3). The largest differences in functional potential of the clades (after controlling for differences in general relative abundance of each gene family) were observed in glutathionylspermidine and trypanothione; ABC transporter alkylphosphonate (TC 3.A.1.9.1); isoleucine degradation; methionine salvage; trehalose uptake and utilization; nitric oxide synthase; utilization of glutathione as a sulfur source; arginine and ornithine degradation; fermentations, mixed acid; and iron acquisition in Vibrio.
A total of 44 siderophore-related protein families were identified in clades A and B with 6 families unique to clade B, and 32 families shared between the clades (Table 4). Protein families distinct to particular genomes included a ferrichrome-iron receptor family, an outer membrane ferripyoverdine receptor family, a ferripyochelin-binding protein family, and two Fe siderophore receptor families (Table 4). Clade B had six distinct siderophore-related protein families, including those involved in transport and sensing iron. Siderophore-related protein families shared across the clades included siderophore biosynthesis, transport, and regulatory protein families, and were generally found in each of the closely related Pseudomonas ancestors. Pyoverdine-related protein families comprised 16 of the 32 shared families among the clades, and included the following proteins from the Pvd protein family: PvdN, PvdE, PvdO, PvdM, PvdP, PvdS, and PvdA. Additionally, a pyoverdine non-ribosomal synthetase module gene, outer membrane ferripyoverdine receptor gene, outer membrane pyoverdine efflux protein, and three ABC transporter genes in the pyoverdine gene cluster were also identified. Protein families involved in aerobactin biosynthesis, enterobactin regulation, and Fe (III) dicitrate transport were detected in all ten genomes, and were identified as N6-hydroxylysine O-acetyltransferase, two-component response regulator, and ferric-dicitrate transport protein FecA, respectively.
Lastly, functional overlap was found between the two clades for a ferric siderophore transport protein family that was assigned to all genomes in both clades, as well as an additional protein family in Pseudomonas sp. RCNY00000000 from clade A, and several other protein families specific to clade B. From the pangenome analysis, six siderophore-related protein families were found in the newly sequenced genomes but not Pseudomonas koreensis.
Additionally, 61 heme-related protein families were discovered in the pangenome analysis, where 36 protein families were shared by each of the 10 newly constructed genomes and 25 protein families were uniquely distributed across the clades (Table 5). The majority (57%) of heme-related protein families were also shared among the clades and P. koreensis (Table 5). Other iron-related protein families were found either only in the clades or in conjunction with P. koreensis (Table 6). Protein families that were shared by all organisms from both clades were numerous and included protein families involved in iron binding (e.g., 4Fe-4S ferredoxin/iron-sulfur binding and outer membrane receptor for ferric coprogen and ferric-rhodotorulic acid.), transport (e.g., ferric iron ABC transporter/iron-binding protein, low-affinity iron permease, etc.), and utilization (heme iron utilization protein) (Tables 5 and 6). Of the protein families unique to clade B, four protein families were associated with the tripartite periplasmic protein Efe (EfeO, EfeB, and EfeU) (Table 6).
Discussion
Enrichment Limitations
Ten individual draft genomes of siderophore-producing pseudomonads were isolated from ‘Concord’ vineyard soils where vines consistently exhibit chlorosis and decreased vine health. These genomes were observed to be part of two distinct phylogenetic clades within the Pseudomonas genus and had different functional capacities, including siderophore biosynthesis, recognition, and transport. Importantly, each clade also had the capacity to produce at least one of each of the major functional groups of siderophores. We did not observe a clear connection between isolated genomes and cover cropping treatments and other field/plant parameters; however, the microbial enrichment process likely selected for similar sets of organisms, suited to the medium in addition to the above-ground treatments and vine health status. Additionally, samples were chosen for sequencing based on visual assessments of siderophore production, by CAS color-change reactivity, and many of the sequenced wells were of similar color, indicating similar siderophore functional type [40, 41]. The microplate assay was useful for isolating siderophore-producing bacteria and the analysis pipeline in KBase provided a robust and rapid phylogenetic and functional genomic assessment of many whole bacterial genomes simultaneously.
Functional Ecology of Pyoverdine
Pangenomic analysis of the CAS-selected cultures revealed a variety of siderophore and other protein families related to Fe nutrition and transport. Members of clade B appear more closely related to the P. koreensis compared with clade A, which can also be observed in the pangenome circle plots (Fig. 2). A substantial functional overlap was identified between two phylogenetically distinct clades including the capacity to express proteins related to the biosynthesis, transport, and/or perception of the three major groups of siderophores (i.e., carboxylates, hydroxamates, and catecholates), as well as mixed-type siderophores which have two or more iron-binding functional groups [42]. The majority of the siderophore-related protein families found in the ten sequenced genomes belonged to the mixed-type category and included protein families related to pyoverdine and aerobactin biosynthesis, utilization, and regulation.
Pangenome analyses of clades A (a) and B (b) performed using the Build Pangenome with OrthoMCL app (PangenomeOrthomcl version 0.0.7). Red indicates base singletons, sky blue represents non-core genes with respect to Pseudomonas koreensis [GC 001605965.1] compared with the respective pangenomes. Gray represents non-core genes within the pangenome. Dark blue represents core genes
Pyoverdine is a fluorescent siderophore produced by many species of pseudomonads but has been primarily studied in P. aeruginosa and P. fluorescens [43, 44]. Previously, in silico genomic analysis of P. fluorescens identified 31 genes putatively involved in pyoverdine synthesis, transport, or regulation [44]. In the current study, 16 different protein families related to pyoverdine biosynthesis, transport, and regulation were identified, with all protein families belonging to all ten sequenced genomes and P. koreensis. Pyoverdine production by pseudomonads has proven to be highly efficient in capturing ferric iron due to its high affinity for Fe(3+), as well as the high specificity of the pyoverdine outer membrane receptors [45, 46]. In a study comparing iron uptake capabilities between P. fluorescens C7R12 and a C7R12 pyoverdine minus mutant, it was found that the mutant was significantly more susceptible to iron deficiency compared to the wild-type strain [47]. Importantly, the C7R12 mutant had the capacity to produce other unknown siderophores, but Fe acquisition was still inhibited, suggesting a dominant role of pyoverdine in Fe nutrition within the cell. Additionally, the work of Cunrath et al. (2016) [48] and Baldi et al. (2016) [49] also found that pyoverdine was critical to maintenance of the balance of metal ions (the metallome) in a polluted environment, significantly preventing the accumulation of toxic metal within the cells of P. aeruginosa and P. putida.
Other recent work with pyoverdine biosynthesis establishes this siderophore as a “public good” that can be shared among cells, exploited by cheating organisms [25] and therefore implicates the production of this siderophore as playing a crucial role in cooperation and competitive co-evolution in complex microbial communities [50]. The organisms excreting pyoverdines into the extracellular environment not only have an advantage in iron scavenging abilities, but also promote the growth of non-siderophore producers, and provide intraspecific benefits to other beneficial microorganisms that would otherwise be at a disadvantage [26, 50]. However, pyoverdine structure varies across the Pseudomonas genus and some pyoverdines can effectively inhibit the growth of non-producers, often due to the specificity of their cognate receptors [25]. Future research should determine the extent and mechanisms of cheating in pseudomonads in these soils.
Rarity of Aerobactin in Soils
Buyer et al. (1991) was reportedly the first group to discover aerobactin production in a bacterium that did not belong to the family Enterobacteriaceae and found a halophilic pseudomonad capable of producing the siderophore [51]. Aerobactin is categorized as a mixed-type siderophore having two hydroxamates and one α-hydroxy carboxylate ligand, and has been primarily studied as a virulence factor in pathogenic species of bacteria, such as Escherichia coli and Salmonella spp. [52,53,54]. However, aerobactin production has been observed in select non-pathogenic bacteria including members of the Pseudomonas and Citrobacter genera [55], and has recently been observed widely distributed across the Vibrionaceae family [56]. Our results suggest aerobactin production may also be observed in pseudomonads from vineyard soils, as well as, other pseudomonads (the relatives).
Highly Diverse Membrane Receptors
Along with pyoverdine and aerobactin biosynthetic protein families, a large number of diverse membrane receptor protein families for a variety of iron chelates were also present in the newly sequenced genomes. Carboxylate-type siderophores chelate ferric iron only through α-hydroxyl and carboxylate functional groups, with perhaps the most common siderophore of this type being citrate [52]. Essential to the uptake of ferric-citrate is the outer membrane protein FecA which was identified in all ten assembled genomes, with an additional FecA encoding protein family found in Pseudomonas sp. RAZO00000000. With pKa values ranging from 3.5 to 5, carboxylate-type siderophores are preferred by microorganisms in acidic environments; however, they tend to be outcompeted by catecholates at near neutral pH levels [57]. Enterobactin, the most extensively studied catecholate type siderophore, has a pKa of approximately 6.5–8 and a high affinity for Fe at neutral pH [58]. In the current study, the enterobactin-responsive activator PfeR was found in each of the ten assembled genomes and is required for synthesis of the ferric enterobactin receptor PfeA in Pseudomonas aeruginosa [59]. According to a study by Michel et al. (2007), P. aeruginosa has also been found to produce ferripyochelin (a catecholate type siderophore) in Fe-limiting conditions [60]. In this context, a ferripyochelin-binding protein family was identified in each of the ten sequenced genomes suggesting their potential to utilize extracellular ferripyochelin in the soil environment. Hydroxamate type siderophores have pKa values that typically range from 8 to 10 making them effective chelators in alkaline soils, such as the soils analyzed in this study [61]. Among the most common and historically significant siderophores of this type is ferrichrome. Ferrichrome production has mainly been identified in species of fungi, and to our knowledge has yet to be identified in any Pseudomonas species. However, research with P. aeruginosa has shown that they are able to express ferrichrome outer membrane receptors to transport iron-bound ferrichrome inside their cells [62,63,64]. Furthermore, the energy necessary for ferrichrome uptake by P. aeruginosa is supplied by the inner membrane protein TonB [64]. The results suggest that the sequenced pseudomonads possess similar capabilities due to the identification of several ferrichrome-iron receptor protein families in addition to multiple TonB-binding protein families. Of note, TonB-dependent transporters have also been associated with mechanisms to utilize xenosiderophores, allowing uptake of virtually any siderophore produced by another organism, trans-kingdom, and even previously unrecognized siderophore-Fe complexes, by the TonB expressive organism [65].
Importance of Protein Efe
An interesting distinction between the clades was identified by the annotation of the three protein families EfeU, EfeO, and EfeB in clade B and P. koreensis, which are essential to the production of the tripartite periplasmic protein Efe. Studies with Escherichia coli have shown EfeUOB system is induced under low pH conditions to transport both ferric and ferrous iron into bacterial cells, and has a higher affinity for ferrous iron with optimal activity in aerobic and acidic conditions [66, 67]. Studies with Bacillus subtilis 168 revealed the EfeUOB system has a high affinity for both ferrous and ferric iron [58]. The EfeUOB system has received far less attention in the Pseudomonas genus, but orthologs have been identified, along with an additional putative component EfeM [66, 68]. Additionally, Efe-like proteins (Algp7) encoded in Sphingomonas sp. actively bind both ferric and ferrous Fe, as well as zinc and copper [69]. We observed orthologs of EfeU, EfeO, and EfeB in some pseudomonads isolated from vineyard soils, which suggests they could be involved in Fe cycling in these soils. It is possible the organisms composing clade B could capitalize on associations with strategy I plants, like grapevines, which rely on acidification of the rhizosphere and root exudates to increase Fe bioavailability [7]. This, however, suggests inter-kingdom iron piracy and cheating on the part of the pseudomonads composing clade B, and merits further investigation. Additionally, these data, along with those presented in Table 3, suggest there are fundamental biological differences in these two distinct clades ranging across various metabolic pathways.
Conclusions
Ten whole pseudomonad genomes, composing two distinct taxonomic and functionally distinct clades, were sequenced from ‘Concord’ vineyard soils. Based on the quantity and diversity of iron- and siderophore-related protein families observed in the genomes, the organisms in this study can either produce or utilize many different types of siderophores and other sources of iron. While the organisms can produce and uptake pyoverdine (a known “public goods” source of Fe), they are also capable of scavenging Fe by taking up siderophores generated by other organisms and various other forms of Fe, which is a trait of “cheating” organisms. The complex iron scavenging system present in these organisms suggests their potential role in attenuating soil Fe cycling and bioavailability, which could have implications for Fe limitation to ‘Concord’ grapevines and their associated microbial communities.
References
Davenport JR, Stevens RG (2006) High soil moisture and low soil temperature are associated with chlorosis occurrence in Concord grape. Hortscience 41:418–422
Pradubsuk S, Davenport JR (2010) Seasonal uptake and partitioning of macronutrients in mature ‘Concord’ grape. J Am Soc Hortic Sci 135:474–483
Abadia J, Vazquez S, Rellan-Alvarez R, El-Jendoubi H, Abadia A, Alvarez-Fernandez A, Lopez-Millan AF (2011) Towards a knowledge-based correction of iron chlorosis. Plant Physiol Biochem 49:471–482. https://doi.org/10.1016/j.plaphy.2011.01.026
Garcia-Mina JM, Bacaicoa E, Fuentes M, Casanova E (2013) Fine regulation of leaf iron use efficiency and iron root uptake under limited iron bioavailability. Plant Sci 198:39–45. https://doi.org/10.1016/j.plantsci.2012.10.001
Singh K, Singh Y, Upadhyay AK, Mori S (2011) Phytosiderophore-based molecular approach for enhanced iron acquisition to increase crop production under high pH calcareous soils. Indian J Agric Sci 81:679–689
Colombo C, Palumbo G, He JZ, Pinton R, Cesco S (2014) Review on iron availability in soil: interaction of Fe minerals, plants, and microbes. J Soils Sediments 14:538–548. https://doi.org/10.1007/s11368-013-0814-z
Kim SA, Guerinot ML (2007) Mining iron: iron uptake and transport in plants. FEBS Lett 581:2273–2280. https://doi.org/10.1016/j.febslet.2007.04.043
Lopez-Millan AF, Grusak MA, Abadia A, Abadia J (2013) Iron deficiency in plants: an insight from proteomic approaches. Front Plant Sci 4. https://doi.org/10.3389/fpls.2013.00254
Rajaie M, Tavakoly AR (2018) Iron and/or acid foliar spray versus soil application of Fe-EDDHA for prevention of iron deficiency in Valencia orange grown on a calcareous soil. J Plant Nutr 41:150–158. https://doi.org/10.1080/01904167.2017.1382523
Bavaresco L, Goncalves M, Civardi S, Gatti M, Ferrari F (2010) Effects of traditional and new methods on overcoming lime-induced chlorosis of grapevine. Am J Enol Vitic 61:186–190
Gamble AV, Howe JA, Delaney D, van Santen E, Yates R (2014) Iron chelates alleviate iron chlorosis in soybean on high pH soils. Agron J 106:1251–1257. https://doi.org/10.2134/agronj13.0474
Smith BR, Cheng LL (2006) Fe-EDDHA alleviates chlorosis in ‘Concord’ grapevines grown at high pH. Hortscience 41:1498–1501
Oburger E, Gruber B, Schindlegger Y, Schenkeveld WDC, Hann S, Kraemer SM, Wenzel WW, Puschenreiter M (2014) Root exudation of phytosiderophores from soil-grown wheat. New Phytol 203:1161–1174. https://doi.org/10.1111/nph.12868
Puschenreiter M, Gruber B, Wenzel WW, Schindlegger Y, Hann S, Spangl B, Schenkeveld WDC, Kraemer SM, Oburger E (2017) Phytosiderophore-induced mobilization and uptake of Cd, Cu, Fe, Ni, Pb and Zn by wheat plants grown on metal-enriched soils. Environ Exp Bot 138:67–76. https://doi.org/10.1016/j.envexpbot.2017.03.011
Saha M, Sarkar S, Sarkar B, Sharma BK, Bhattacharjee S, Tribedi P (2016) Microbial siderophores and their potential applications: a review. Environ Sci Pollut Res 23:3984–3999. https://doi.org/10.1007/s11356-015-4294-0
Jin CW, Li GX, Yu XH, Zheng SJ (2010) Plant Fe status affects the composition of siderophore-secreting microbes in the rhizosphere. Ann Bot 105:835–841. https://doi.org/10.1093/aob/mcq071
Masalha J, Kosegarten H, Elmaci O, Mengel K (2000) The central role of microbial activity for iron acquisition in maize and sunflower. Biol Fertil Soils 30:433–439
Desai A, Archana G (2011) Role of siderophores in crop improvement
Ahmed E, Holmstrom SJM (2014) Siderophores in environmental research: roles and applications. Microb Biotechnol 7:196–208. https://doi.org/10.1111/1751-7915.12117
Ahmed E, Holmstrom SJM (2015) Siderophore production by microorganisms isolated from a podzol soil profile. Geomicrobiol J 32:397–411. https://doi.org/10.1080/01490451.2014.925011
Crits-Christoph A, Diamond S, Butterfield CN, Thomas BC, Banfield JF (2018) Novel soil bacteria possess diverse genes for secondary metabolite biosynthesis. Nature 558:440–444. https://doi.org/10.1038/s41586-018-0207-y
Barber MF, Eldel NC (2015) Buried treasure: evolutionary perspectives on microbial iron piracy. Trends Genet 31:627–636. https://doi.org/10.1016/j.tig.2015.09.001
Popat R, Harrison F, da Silva AC, Easton SAS, McNally L, Williams P, Diggle SP (2017) Environmental modification via a quorum sensing molecule influences the social landscape of siderophore production. Proc R Soc B Biol Sci 284:20170200. https://doi.org/10.1098/rspb.2017.0200
Castro RO, Bucio JL (2013) Small molecules involved in transkingdom communication between plants and rhizobacteria. In: de Bruijn FJ (ed) Molecular microbial ecology of the rhizosphere. Wiley, Hoboken, pp 295–307
Butaite E, Baumgartner M, Wyder S, Kummerli R (2017) Siderophore cheating and cheating resistance shape competition for iron in soil and freshwater Pseudomonas communities. Nat Commun 8:414. https://doi.org/10.1038/s41467-017-00509-4
O’Brien S, Lujan AM, Paterson S, Cant MA, Buckling A (2017) Adaptation to public goods cheats in Pseudomonas aeruginosa. Proc R Soc B Biol Sci 284:20171089. https://doi.org/10.1098/rspb.2017.1089
Lewis RW, LeTourneau MK, Davenport JR, Sullivan TS (2018) ‘Concord’ grapevine nutritional status and chlorosis rank associated with fungal and bacterial root zone microbiomes. Plant Physiol Biochem 129:429–436. https://doi.org/10.1016/j.plaphy.2018.06.011
Davenport JR, Stevens RG, Whitley KM (2008) Spatial and temporal distribution of soil moisture in drip-irrigated vineyards. Hortscience 43:229–235
Singer SD, Davenport JR, Hoheisel G-A, Moyer M (2018) Vineyard nutrient management in Washington State.
Bremner J (1996) Nitrogen-total Methods of Soil Analysis Part 3—Chemical Methods: 1085–1121
Schwyn B, Neilands JB (1987) Universal chemical assay for the detection and determination of siderophores. Anal Biochem 160:47–56. https://doi.org/10.1016/0003-2697(87)90612-9
Allen B, Drake M, Harris N, Sullivan T (2017) Using KBase to assemble and annotate prokaryotic genomes. Curr Protoc Microbiol 46:1E.13.1–1E.13.18. https://doi.org/10.1002/cpmc.1037.
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. https://doi.org/10.1093/bioinformatics/btu170
Nurk S, Bankevich A, Antipov D, Gurevich AA, Korobeynikov A, Lapidus A, Prjibelski AD, Pyshkin A, Sirotkin A, Sirotkin Y, Stepanauskas R, Clingenpeel SR, Woyke T, McLean JS, Lasken R, Tesler G, Alekseyev MA, Pevzner PA (2013) Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J Comput Biol 20:714–737. https://doi.org/10.1089/cmb.2013.0084
Wu Y-W, Simmons BA, Singer SW (2015) MaxBin 2.0: an automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics 32:605–607
Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW (2015) CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome research: gr. 186072.186114.
Gurevich A, Saveliev V, Vyahhi N, Tesler G (2013) QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075
Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M (2013) The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res 42:D206–D214
Price MN, Dehal PS, Arkin AP (2010) FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. https://doi.org/10.1371/journal.pone.0009490
Perez-Miranda S, Cabirol N, George-Tellez R, Zamudio-Rivera L, Fernandez F (2007) O-CAS, a fast and universal method for siderophore detection. J Microbiol Methods 70:127–131
Sullivan TS, Ramkissoon S, Garrison VH, Ramsubhag A, Thies JE (2012) Siderophore production of African dust microorganisms over Trinidad and Tobago. Aerobiologia 28:391–401. https://doi.org/10.1007/s10453-011-9243-x
Aznar A, Dellagi A (2015) New insights into the role of siderophores as triggers of plant immunity: what can we learn from animals? J Exp Bot 66:3001–3010. https://doi.org/10.1093/jxb/erv155
Lamont IL, Martin LW (2003) Identification and characterization of novel pyoverdine synthesis genes in Pseudomonas aeruginosa. Microbiology 149:833–842
Moon CD, Zhang XH, Matthijs S, Schafer M, Budzikiewicz H, Rainey PB (2008) Genomic, genetic and structural analysis of pyoverdine-mediated iron acquisition in the plant growth-promoting bacterium Pseudomonas fluorescens SBW25. BMC Microbiol 8:7
Meyer JM, Abdallah MA (1978) The fluorescent pigment of Pseudomonas fluorescens: biosynthesis, purification and physico-chemical properties. J Gen Microbiol 107:319–328
Hohnadel D, Meyer JM (1988) Specificity of pyoverdine-mediated iron uptake among fluorescent Pseudomonas strains. J Bacteriol 170:4865–4873
Mirleau P, Delmorme S, Philippot L, Meyer JM, Mazurier S, Lemanceau P (2000) Fitness in soil and rhizosphere of Pseudomonas fluorescens C7R12 compared with a C7R12 mutant affected in pyoverdine synthesis and uptake. FEMS Microbiol Ecol 34:35–44
Cunrath O, Geoffroy VA, Schalk IJ (2016) Metallome of Pseudomonas aeruginosa: a role for siderophores. Environ Microbiol 18:3258–3267. https://doi.org/10.1111/1462-2920.12971
Baldi F, Gallo M, Battistel D, Barbaro E, Gambaro A, Daniele S (2016) A broad mercury resistant strain of Pseudomonas putida secretes pyoverdine under limited iron conditions and high mercury concentrations. Biometals 29:1097–1106. https://doi.org/10.1007/s10534-016-9980-y
O'Brien S, Hesse E, Lujan A, Hodgson DJ, Gardner A, Buckling A (2018) No effect of intraspecific relatedness on public goods cooperation in a complex community. Evolution 72:1165–1173. https://doi.org/10.1111/evo.13479
Buyer JS, DeLorenzo V, Neilands JB (1991) Production of the siderophore aerobactin by a halophilic pseudomonad. Appl Environ Microbiol 57:2246–2250
Sandy M, Butler A (2009) Microbial iron acquisition: marine and terrestrial siderophores. Chem Rev 109:4580–4595
Valvano MA, Silver RP, Crosa JH (1986) Occurrence of chromosome- or plasmid-mediated aerobactin iron transport systems and hemolysin production among clonal groups of human invasive strains of Escherichia coli K1. Infect Immun 52:192–199
Fischbach MA, Lin H, Liu DR, Walsh CT (2006) How pathogenic bacteria evade mammalian sabotage in the battle for iron. Nat Chem Biol 2:132–138
Chen LM, Dick WA, Streeter JG (2000) Production of aerobactin by microorganisms from a compost enrichment culture and soybean utilization. J Plant Nutr 23:2047–2060. https://doi.org/10.1080/01904160009382164
Thode SK, Rojek E, Kozlowski M, Ahmad R, Haugen P (2018) Distribution of siderophore gene systems on a Vibrionaceae phylogeny: database searches, phylogenetic analyses and evolutionary perspectives. PLoS One 13:e0191860. https://doi.org/10.1371/journal.pone.0191860
Dertz EA, Raymond KN (2003) Siderophores and transferrins. In: Que L, Tolman WB (eds) Comprehensive coordination chemistry II. Elsevier, Ltd., Philadelphia
Miethke M, Marahiel MA (2007) Siderophore-based iron acquisition and pathogen control. Microbiol Mol Biol Rev 71:413–451
Dean CR, Neshat S, Poole K (1996) PfeR, an enterobactin-responsive activator of ferric enterobactin receptor gene expression in Pseudomonas aeruginosa. J Bacteriol 178:5361–5369
Michel L, Bachelard A, Reimmann C (2007) Ferripyochelin uptake genes are involved in pyochelin-mediated signaling in Pseudomonas aeruginosa. Microbiology 153:1508–1518
Cline GR, Powell PE, Szaniszlo PJ, Reid CPP (1982) Comparison of the abilities of hydroxamic, synthetic, and other natural organic acids to chelate iron and other ions in nutrient solution. Soil Sci Soc Am J 46:1158–1164
Llamas MA, Sparrius M, Kloet R, Jimenez CR, Vandenbroucke-Grauls C, Bitter W (2006) The heterologous siderophores ferrioxamine B and ferrichrome activate signaling pathways in Pseudomonas aeruginosa. J Bacteriol 188:1882–1891. https://doi.org/10.1128/jb.188.5.1882-1891.2006
Lee K, Lee KM, Go J, Ryu JC, Ryu JH, Yoon SS (2016) The ferrichrome receptor A as a new target for Pseudomonas aeruginosa virulence attenuation. FEMS Microbiol Lett 363. https://doi.org/10.1093/femsle/fnw104
Hannauer M, Barda Y, Mislin GLA, Shanzer A, Schalk IJ (2010) The ferrichrome uptake pathway in Pseudomonas aeruginosa involves an Iron release mechanism with acylation of the siderophore and recycling of the modified desferrichrome. J Bacteriol 192:1212–1220. https://doi.org/10.1128/jb.01539-09
Rudolf M, Stevanovic M, Kranzler C, Pernil R, Keren N, Schleiff E (2016) Multiplicity and specificity of siderophore uptake in the cyanobacterium Anabaena sp PCC 7120. Plant Mol Biol 92:57–69. https://doi.org/10.1007/s11103-016-0495-2
Grosse C, Scherer J, Koch D, Otto M, Taudte N, Grass G (2006) A new ferrous iron-uptake transporter, EfeU (YcdN), from Escherichia coli. Mol Microbiol 62:120–131. https://doi.org/10.1111/j.1365-2958.2006.05326.x
Cao J, Woodhall MR, Alvarez J, Cartron ML, Andrews SC (2007) EfeUOB (YcdNOB) is a tripartite, acid-induced and CpxAR-regulated, low-pH Fe2+ transporter that is cryptic in Escherichia coli K-12 but functional in E-coli O157 : H7. Mol Microbiol 65:857–875. https://doi.org/10.1111/j.1365-2958.2007.05802.x
Rajasekaran MB, Mitchell SA, Gibson TM, Hussain R, Siligardi G, Andrews SC, Watson KA (2010) Isolation and characterisation of EfeM, a periplasmic component of the putative EfeUOBM iron transporter of Pseudomonas syringae pv. Syringae. Biochem Biophys Res Commun 398:366–371. https://doi.org/10.1016/j.bbrc.2010.06.072
Temtrirath K, Okumura K, Maruyama Y, Mikami B, Murata K, Hashimoto W (2017) Binding mode of metal ions to the bacterial iron import protein EfeO. Biochem Biophys Res Commun 493:1095–1101. https://doi.org/10.1016/j.bbrc.2017.09.057
Acknowledgments
The authors wish to thank Kalyani Muhunthan for assistance in laboratory procedures and the members of the Davenport Lab for assistance with sampling and plant tissue analyses.
Funding
Funding was provided by the Washington State Concord Grape Research Council, by the Washington State University BioAg program, and by the USDA/NIFA through Hatch project 1014527.
Author information
Authors and Affiliations
Contributions
Ricky W. Lewis performed whole genome sequencing, processed and analyzed the sequencing data, and assisted with manuscript writing. Anjuman Islam gathered samples, performed the microplate siderophore production assay, extracted DNA, and assisted with manuscript writing. Lee Opdahl assisted with whole genome sequencing, interpretation of results, and manuscript writing. Tarah S. Sullivan and Joan R. Davenport conceived of the experimental design and assisted with sample gathering, interpretation of results, and manuscript writing.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Rights and permissions
About this article
Cite this article
Lewis, R.W., Islam, A., Opdahl, L. et al. Comparative Genomics, Siderophore Production, and Iron Scavenging Potential of Root Zone Soil Bacteria Isolated from ‘Concord’ Grape Vineyards. Microb Ecol 78, 699–713 (2019). https://doi.org/10.1007/s00248-019-01324-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-019-01324-8