Abstract
This study aimed to gain insight into the microbial quality, safety and bacterial community composition of black soldier fly larvae (Hermetia illucens) reared at different facilities on a variety of organic waste streams. For seven rearing cycles, both on laboratory-scale and in large-scale facilities at several locations, the microbiota of the larvae was studied. Also samples of the substrate used and the residue (= leftover substrate after rearing, existing of non-consumed substrate, exuviae and faeces) were investigated. Depending on the sample, it was subjected to plate counting, Illumina Miseq sequencing and/or detection of specific food pathogens. The results revealed that the substrates applied at the various locations differed substantially in microbial numbers as well as in the bacterial community composition. Furthermore, little similarity was observed between the microbiota of the substrate and that of the larvae reared on that substrate. Despite substantial differences between the microbiota of larvae reared at several locations, 48 species-level operational taxonomic units (OTUs) were shared by all larvae, among which most belonged to the phyla Firmicutes and Proteobacteria. Although the substrate is assumed to be an important source of bacteria, our results suggest that a variety of supposedly interacting factors-both abiotic and biotic-are likely to affect the microbiota in the larvae. In some larvae and/or residue samples, potential foodborne pathogens such as Salmonella and Bacillus cereus were detected, emphasising that decontamination technologies are required when the larvae are used in feed, just as for other feed ingredients, or eventually in food.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Every year, an increasing volume of solid waste is generated worldwide. A large fraction of this waste exists of organic material, such as pre- and postconsumer food waste and animal manure [1]. In Europe alone, approximately 100 million tonnes of food products remain unused annually [2]. Furthermore, if left untreated, livestock waste products such as manure cause pollution to water bodies through eutrophication, to air through ammonia and greenhouse gas emissions (and thus contributing to global warming) and to soil through nutrient accumulation [3]. One method for valorisation of organic waste products consists of their use as a feeding substrate for mass-rearing of edible insects [4,5,6]. Some insect species, such as the black soldier fly (Hermetia illucens (Diptera: Stratiomyidae), further referred to as ‘BSF’), can be reared on a variety of organic side streams such as food waste [6,7,8], livestock manure [9,10,11] and faecal sludge [12, 13]. The larvae as well as specific compounds isolated from the larvae (e.g. protein, fat and chitin) show a large potential to be used in aquaculture [14, 15], livestock feed [8, 16], human food [17] or other applications such as biofuel [11, 18] and bioactive coatings [19].
So far, little attention has been paid to microbial dynamics associated with the rearing of BSF larvae on waste streams. Research has suggested that the gut microbial community in insects may be greatly influenced by the feeding substrate [20,21,22]. For BSF, such an effect was already demonstrated for the bacterial microbiota of larvae reared on either food waste, cooked rice or calf forage [7] and for the mycobiota of BSF larvae reared on chicken feed and/or vegetable waste [23]. As the microbial safety of the larvae is of great importance for their use as feed ingredient [17], the selection of the substrate can be an important factor in assuring food/feed safety [22]. Indeed, food pathogens that may be present in the substrate may be transferred to the larval intestinal tract and subsequently cause illness in the traditional farm animals given a BSF-based feed or in people consuming the derived animal products [17, 24, 25]. On the other hand, multiple studies show that BSF larvae possess antimicrobial capacities and are able to reduce pathogenic bacteria such as Salmonella and Escherichia coli in their substrate [1, 8, 12, 24,25,26]. In addition to the importance of the BSF microbiota for food and/or feed safety, microorganisms present in the rearing environment may have a potential towards optimising growth performance of the insect and insect-derived antimicrobial strategies [27, 28]. However, the variability in the microbiota of BSF larvae reared in different facilities, each with their own rearing methods and on different organic waste streams, is still unexplored. Therefore, this study aimed to gain insight into the variability in microbial quality, safety and bacterial community composition of BSF larvae reared at different facilities and in relation to the rearing substrate used. To this end, samples were taken during the rearing process of BSF larvae at laboratory scale as well as in three external facilities in Belgium, the Netherlands and Switzerland. As a consequence of considering different locations, larvae were cultivated on different organic waste streams, using slightly different practices and in slightly different environmental conditions. Samples of the larvae as such, but also of the substrates (i.e. the waste streams) and the residues (i.e. the mixture of non-consumed substrate, faeces or frass and exuviae) were analysed for their intrinsic parameters, microbial numbers and bacterial community composition (using high-throughput Illumina sequencing of 16S rRNA genes). In addition, larvae and residues from the three external facilities were also assessed for their microbial safety (through detection of a selection of food pathogens).
Methods
Laboratory Rearing Cycles
BSF larvae were reared at laboratory scale on four different waste streams (Table 1 and Online Resource 1). For each waste stream, one rearing cycle was conducted that consisted of three batches. Fruit/vegetable waste (LAB 1), consisting of a mixture of strawberries, apples, lettuce, cucumbers, red bell peppers, broccoli, carrots and chicory, was obtained from the local supermarket (Colruyt group, Geel, Belgium) and homogenised using a home-type blender (Espressions, Eindhoven, The Netherlands) and stored at − 21 °C. Supermarket and restaurant waste (consisting of vegetable and animal products; LAB 2) was obtained from a local waste management company (Renewi, Mol, Belgium), where it had been collected, unpacked and mixed into a slurry. Upon arrival at the laboratory, the slurry was stored at − 21 °C. Poultry blood (LAB 3) was obtained from a local poultry slaughtering facility (Pluvera-Klaasen & Co, Ravels, Belgium) and stored at − 21 °C. Finally, poultry manure (including shavings; LAB 4) was obtained from a local broiler farm (Proefbedrijf pluimveehouderij, Geel, Belgium) and stored at 3 °C. Substrates were obtained maximally 1 week prior to the start of the rearing cycle, except for the fruit/vegetable waste which was obtained 6 months before and kept frozen. No further treatments except from cooled/frozen storage were applied to the substrates prior to administering them to the larvae. Frozen substrates were thawed for 1 to 3 days at 3 °C, and all substrates were placed at room temperature for 4 h before administering to the larvae. At the start of each rearing cycle (day 0), 0.2 g of BSF eggs were placed in an open plastic cup (200 ml) with 15 g of apple slices and 15 g of commercial chicken starter feed (startmeel voor kuikens 259, AVEVE, Leuven, Belgium) and incubated at 30 °C (= nursery phase). On day 3, the same quantity of apple slices and chick feed was added. On day 4, the larvae, including the residue, were transferred into a larger plastic beaker without lid (1 l) containing 150 g of chick feed moisturised with 150 ml of tap water and placed in a large insect-rearing room (i.e. phase I, from day 4 to 6). On day 7, the larvae were placed in larger plastic container (20 l, 36 × 26 × 28 cm), and the specific side stream was added (phase II). Chick feed and/or water were also added depending on the cycle (Online Resource 2), in order to maintain a suitable moisture content of approximately 70%. That moisture content was chosen to allow proper larval growth and at the same time efficient drying of the residue towards the end of the cycle [29]. Larvae were harvested, by manually picking them out of the residue using sterile forceps, at day 14.
External Rearing Facilities
Three external, large-scale rearing facilities specialised in the cultivation of BSF larvae for commercial or research purposes contributed on this study (Table 1 and Online Resource 1). These facilities were located in Belgium (Millibeter, EXT-BE), The Netherlands (Bestico B.V., Koppert Biological Systems, EXT-NL) and Switzerland (FiBL Research Institute of Organic Agriculture, EXT-CH). Each rearing facility was studied with respect to its own specific rearing infrastructure and methods. Here, all rearing cycles could also be divided into two phases during each of which a different substrate was administered. Similar to the laboratory cycles, one cycle was conducted by each facility, during which samples were taken from three batches of larvae. Briefly, at EXT-BE, larvae were supplemented during phase I with chick feed that was mixed with the phase II substrate (see further; ratio not determined by the rearer) and with supplementation of methylparaben (0.1%). Methylparaben was included in order to prevent moulding of the substrate, which can have detrimental effects on larval development as experienced by that rearer. The other rearers did not make use of methylparaben. The whole cycle was completed in crates (50 l), and no separation of the larvae from the residue took place after phase I. After a 3-day phase I period, phase II substrate was added to the crates (solely phase II substrate, without chick feed and methylparaben), which consisted of a mixture of dried distilled grains with solubles (DDGS, 20%), an apple waste stream from apple juice production (60%) and water (20%). Those ingredients were stored at room temperature for 2 months prior to homogenising them into a mixture, after which they were administered without further treatment to the larvae. The larvae were kept in a temperature-controlled, ventilated room during the complete cycle, and were harvested at day 14 (batches 1 and 2) or at day 21 (batch 3), depending on the larval development. At EXT-NL, larvae were grown for the first 7 days (= phase I) on a substrate of fine wheat bran (30%) and water (70%) in 50 l crates, after which they were separated from the bran by automated sieving and transferred into new crates of the same volume. In phase II, larvae were grown on a mixture of fermented potato peels (40%), yeast concentrate (40%) and wheat flour (20%). These ingredients were homogenised into a mixture which was stored for maximally 6 weeks before administering to the larvae without further treatment. Both phases took place in a temperature-controlled room (Online Resource 1). Larvae were considered harvest-ready at day 14. At EXT-CH, larvae were reared on feed for laying hens (34%) supplemented with water (66%) in phase I. During this phase (days 0–7), larvae were kept in 10 l plastic crates, which were placed in a climate chamber (days 0 to 4) or rearing room (days 4 to 7; Table 1 and Online Resource 1). After 7 days, larvae were separated from phase I substrate by manual sieving and transferred into larger containers (550 l) containing phase II substrate, and then housed in another room. The phase II substrate consisted of a homogenised mixture of fruit and vegetable wastes (40%), brewer’s spent grains (30%) and off-specification, pre-cooked ravioli/tortellini pasta (30%), which were kept for a maximum of 2 weeks in a barn at temperatures below 10 °C (approaching outside temperatures during winter in Switzerland). After homogenisation and before administering, the mixture was brought to rearing temperature without further treatment. In this cycle, larvae were harvested at day 19. More details on the rearing practices and feeding regimes of the cycles are shown in Online Resources 1 and 2.
Sampling and Sample Pre-Treatment
For each rearing cycle, samples were taken from each of the three batches per cycle. From these batches, samples were taken of both larvae and residues (one sample per batch, randomly collected from different places in each rearing crate) at harvest day. In addition, phase I and phase II substrates were sampled in triplicate for each rearing cycle. These samples were taken immediately before administering, after homogenising them with a sterile spoon and as they were added to the rearing crates (i.e. brought to room temperature after storage for LAB cycles and EXT-CH). For the cycles at the large-scale facilities (EXT-BE, EXT-NL and EXT-CH), samples were also taken from larvae and residues from each batch at subsequent sampling moments during the rearing phase. After sampling, larvae were washed with running tap water on a sieve (1 mm mesh size) for 1 min in order to remove remaining residue from the larval surface. This procedure was shown in a preliminary experiment to be sufficient to report reliable counts for the interior microbiota of the larvae, excluding microorganisms from their outer surface. In that experiment, three samples of BSF larvae were subjected to the rinsing procedure as described above (‘rinsed larvae’), while three other samples were subjected to the rinsing procedure and an additional disinfection protocol (‘rinsed + disinfected larvae’). The protocol existed of 5 g of larvae being subjected to three washing steps in 100 ml of 70% ethanol followed by three washing steps in 100 ml sterile distilled water. Each step was performed during 1 min at 200 rpm on a laboratory shaker (Unimax 1010, Heidolph, Germany). This experiment was repeated for two batches. The results showed that average microbial counts between rinsed and rinsed + disinfected larvae per batch maximally differed 0.6 log unit for any count, and thus that counts obtained for larvae that were only rinsed are representative for the interior larval microbiota. Because the larval gut was not dissected, it cannot be concluded that only the gut microbiota was included in our analysis, as other organs may also harbour microorganisms.
For all cycles under study, larval weights at harvest were determined prior to further analysis and the mean was calculated from three times ten larvae from each batch. All samples were stored at 3 °C for a maximum of 24 h until analyses. After storage, larvae were homogenised prior to analysis according to Stoops et al. [30]. Substrate and residue samples were analysed without rinsing or homogenisation.
Analyses
Intrinsic Parameters
Water activity was measured using a water activity meter (LabMaster aw, Novasina, Lachen Switzerland), until water activity and temperature (25 °C) were stable for 15 and 5 min, respectively. The moisture content was determined by calculating the difference in weight of 5 g of the initial sample before and after oven drying for 17 h at 105 °C. The pH was measured using a digital pH meter (Portamess 911, Knick, Berlin, Germany with SI analytics electrode, Mainz, Germany). For phase I substrate and residue samples, demineralised water was added prior to pH measurement in a 1:1 or 1:2 ratio (sample:water).
Microbial Counts
All samples were subjected to microbial analyses via plate count methods as described by Dijk et al. [31]. Total viable aerobic counts (TVC) were determined on Plate Count Agar (PCA, Biokar Diagnostics, Beauvais, France) and incubated at 30 °C for 72 h. Enterobacteriaceae were determined on Violet Red Bile Glucose agar (VRBG, Biokar Diagnostics) after incubation at 37 °C for 24 h. Lactic acid bacteria were determined on De Man, Rogosa and Sharpe agar (MRS, Biokar Diagnostics) and incubated at 30 °C for 72 h. Aerobic bacterial endospores were determined by giving the 10−1 dilution a heat shock (10 min at 80 °C), followed by a tenfold serial dilution, plating onto PCA and incubation at 37 °C for 48 h. Fungi were determined on Dichloran Rose Bengal Chloramphenicol agar (DRBC, Biokar Diagnostics) and incubated at 25 °C for 6 days.
Pathogen Detection
Larvae and residue samples taken at harvest from the large-scale rearing cycles (EXT-CH, EXT-BE and EXT-NL) were investigated for a selection of food pathogens. For samples of EXT-BE and EXT-NL, the presence of Salmonella spp. and Listeria monocytogenes was investigated using ISO methods ISO 6579 B″ and AFNOR BRD 07/4-09/98 B″. For samples of EXT-CH, enrichment was performed according to ISO 6579 (Salmonella spp.) and ISO 11290-1 (L. monocytogenes), respectively, and detection was for both pathogens performed using real-time PCR. Coagulase-positive staphylococci were determined according to AFNOR 3M-01/9-04/03 B (EXT-BE/NL) or ISO 6888-2 (EXT-CH). Bacillus cereus colonies were enumerated according to ISO 7932 (EXT-BE/NL/CH).
Metagenetic Analyses
For each rearing cycle, the bacterial community composition of the phase I and II substrates (two biological replicates before administration), as well as of the residues and larvae at harvest (three replicates, one of each of the three batches) was determined using Illumina MiSeq sequencing of partial 16S rRNA gene amplicons (V4 region, 250 bp). To this end, each sample was prepared as described above. Subsequently, DNA extraction, using the DNeasy Soil Kit according to the manufacturer’s instructions (Qiagen, Hilden, Germany), was performed in duplicate on each biological replicate, thus resulting in a total of four DNA extracts for the substrates (n = 2 × 2), and six for the larvae and residues (n = 3 × 2). PCR amplification (primer design shown in Online Resource 3), library preparation, sequencing, sequence processing and diversity analyses were performed as described by Wynants et al. [32]. Downstream diversity analyses used data rarefied to 7000 sequences per DNA extract. Unfortunately, all replicates from phase I substrate yielded too few sequence reads (most likely due to the lower microbial load) and were discarded from the analysis. However, a separate data analysis, rarified to 250 sequence reads and including only phase I substrates, was performed in order to gain insight into the most abundant community members of the phase I substrates (data not shown in manuscript). For the same reason, two DNA extracts from larval batches of EXT-NL were discarded (each belonging to a different biological replicate), resulting in four instead of six extracts. Sequences were clustered into operational taxonomic units (OTUs) based on a 97% similarity cut-off as a proxy for species. The taxonomic origin of each OTU was determined up to genus level with the SINTAX algorithm implemented in USEARCH based on the Silva Living Tree Project v123 (LTP v123) database (Fig. 1). Taxonomic assignments were considered reliable when bootstrap confidence values exceeded 0.80 (Online Resource 4). Sequences were deposited in the Sequence Read Archive (SAMN09425406 to SAMN09425515) under BioProject accession PRNJA476046.
Relative abundance (%) of operational taxonomic units (OTUs) present in the samples of phase II substrates (a), residues (b) and larvae (c) per rearing cycle. Data are mean values of two extracts per replicate sample (n = 2 × 2 substrates, n = 2 × 3 for residues/larvae). Standard deviations varied between below 0.1 and 40.0%. Only OTUs represented by an average relative abundance of more than 5% of sequences in any sample are shown. OTUs with a mean relative abundance of less than 5% are grouped in ‘Other OTUs’
Statistical Analyses
Means for intrinsic parameters and plate counts between different rearing cycles for each sample type (Table 2), as well as means of larvae and of residues between sampling moments within one cycle for the external facilities (Table 3) were compared using one-way ANOVA in SPSS (v.23) followed by Tukey’s (in case of equal variances) or Games Howell’s (in case of unequal variances) post-hoc test. The same statistical approach was used for the diversity indices of samples subjected to metagenetic analyses (Table 4), which were calculated using Phyloseq (1.19-0): Chao1 index (representing the estimated species richness in the samples), equitability index (indicating the evenness in species abundances) and Shannon-Wiener index (a combined measure for species richness and relative abundances) [33,34,35]. Furthermore, Pearson correlation analyses were performed to detect pairwise correlations of average intrinsic parameters and average counts between phase I and II substrates, residues and larvae of all cycles (Online Resources 5 and 6). In all statistical analyses, a significance level of α = 0.05 was considered. In addition, the R-package vegan (v2.5-2) [36] was used to create a non-metric multidimensional scaling (NMDS) for larvae, phase II substrates and residues using the 200 most abundant OTUs found in the entire dataset (Fig. 2). The R-package was also used to conduct a cluster analysis (using the single linkage agglomeration method) on larvae from different rearing cycles based on all OTUs present in the larval samples. The clustering analyses were projected on top of the heat map in Fig. 3, which were constructed based on percentage abundances of each OTU in the larval samples (limited to OTUs present in at least 1% abundance in any larval sample). Both analyses were based on the Bray-Curtis similarity matrix.
Non-metric multidimensional scaling (NMDS) ordination (2D stress = 0.198), based on the Bray-Curtis similarity matrix, representing the bacterial community composition in the samples subjected to metagenetic analysis. The NMDS analysis was based on the 200 most abundant OTUs in this study. Different rearing cycles are depicted in different colours (LAB 1 = green, LAB 2 = dark blue, LAB 3 = orange, LAB 4 = yellow, EXT-BE = light blue, EXT-NL = purple, EXT-CH = black) and different sample types are depicted in different symbols (squares = phase II substrates, triangles = residues, dots = larvae)
Heat map of OTUs in larvae from different rearing locations. Only OTUs with an average relative abundance of at least 1% in any larval sample are shown. The middle value of 1.5% of the colour scheme represents the 75% percentile of relative abundances shown in the figure. OTUs depicted in white were not detected in that location. Cluster analysis, as shown by the dendrogram, was performed on all OTUs present at any abundance in the larvae
Results
Intrinsic Parameters
In the first place, the intrinsic parameters will be compared between all cycles and locations. These are all values obtained from the substrates before administering, and larvae and residues obtained at harvest (Table 2). For the cycles performed at large scale, also values during rearing were obtained from larvae and residues (Table 3) and these will be described in the second place. Results of larvae and residues at harvest are displayed in both tables, in order to be included in different comparisons (between cycles in Table 2 and between times within a cycle in Table 3). For all cycles, except for EXT-BE, the final sampling moment in Table 3 represents the larvae and residues at harvest as shown in Table 2. For EXT-BE - for which two batches were harvested at day 14 and one at day 21—results were represented at harvest in Table 2 and dependent on the sampling day in Table 3.
Table 2 demonstrates that both pH and moisture content of the phase I substrates, which were all grain-based, showed significant differences between cycles (and thus locations). The moisture content was the lowest for the laboratory cycles (ranging from 53.7 to 55.3%) and the highest for EXT-BE (94.5%). The pH value of phase I substrates was the lowest in the latter cycle (4.65), while the pH of other cycles ranged from 5.69 to 6.58. Water activity, on the other hand, did not differ significantly between cycles and was always higher than 0.95. For the phase II substrates, which were all organic waste streams, significant differences between cycles (and locations) were found for each intrinsic parameter. The pH ranged from 3.51 for the fruit/vegetable waste in LAB 1 to 7.29 for the poultry blood in LAB 3. Although the moisture content differed largely, ranging from 65.8 to 92.1%, the water activity remained higher than 0.95 for all phase II substrates. Also in the residue samples taken at harvest, the water activity was higher than 0.95 for all cycles/locations except for EXT-NL (aw = 0.83; Table 2), indicating a larger drying efficiency of the residue towards the end of that rearing cycle. The moisture content was also the lowest for that residue (23.2%), while the residues from other cycles showed a large variation and ranged from 46.3 to 73.8% on average. The average pH of the residue at harvest, ranging between 7.57 and 9.09, was higher than those of the administered substrates. An exception to this was cycle LAB 3, where the residue at harvest showed a lower pH of 5.80 than the near-neutral pH (7.29) of the substrate, being poultry blood. However, this pH of the residue showed a high standard deviation, indicating large differences between batches. For larval samples, the water activity and the moisture content at harvest respectively ranged from 0.96 to 0.97 and from 67.8 to 76.8%, between cycles. Remarkably, the pH of larvae at harvest from cycle EXT-CH (6.32) was significantly lower (p < 0.001) compared to all other cycles (averages ranging from 7.13 to 7.26). No correlations were detected between the intrinsic parameters of either phase I and II substrate and the residue or the larvae at harvest (Online Resource 5). One exception was the pH of the phase II substrate, which was negatively correlated to the pH of the residue (p = 0.026), although this result should be interpreted with caution given the high standard deviation for the residue pH of LAB 3.
Many intrinsic parameters showed prominent changes depending on the sampling moment, as is shown for the external rearing facilities (Table 3). This was expected for cycles which included a separation step after phase I (EXT-CH and EXT-NL), but it was also observed (although non-significant) for that cycle during which the residue contained-aside from faeces and exuviae—both the leftover of phase I substrate and also phase II substrate (EXT-BE). Also for the laboratory cycles, the pH of the residue at harvest (day 14) was on average 1.17 to 3.58 higher (depending on the cycle) when compared to the pH of the residue measured at day 9 (data not shown). Noteworthy, the pH of the larvae at EXT-CH, which showed a lower pH at harvest when compared to the other cycles with a value of 6.32, was 7.18 at the first sampling day (day 7, after feeding with the phase I substrate), thus showing a decrease in larval pH during rearing.
Microbial Counts
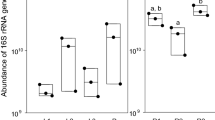
As shown in Table 2, the microbial counts-both total viable counts as well as the other microbial groups-differed to a large extent between cycles and locations. The phase I substrate of EXT-BE was higher for all microbial counts except for Enterobacteriaceae when compared to the phase I substrate of other cycles (all grain-based). Furthermore, even though methylparaben (0.1%) was administered with the phase I substrate, fungi were abundantly present (5.9 log cfu/g). Large differences were also observed between microbial counts of phase II substrates, which differed more in intrinsic parameters and likely also differed more in nutritional composition from each other than the phase I substrates did. Indeed, the lowest TVC was observed for the fruit/vegetable waste of rearing cycle LAB 1 (3.9 log cfu/g), whereas the highest TVC was observed for phase II substrate of EXT-BE, being a mixture of DDGS and an apple waste stream (8.6 log cfu/g). Similarly, also for larvae and residues, a large variability in microbial counts was observed between cycles, resulting in significant differences. Total viable counts of the larvae ranged on average from 8.0 to 9.8 log cfu/g, whereas those of the residue ranged from 8.5 to 10.2 log cfu/g. Other microbial counts showed even a larger variation between cycles for both residues and larvae. Furthermore, no correlations were observed between average microbial counts of the substrates on the one hand, and of the larvae or residue at the other hand (Online Resource 6). In contrast, a significant correlation was observed between the larvae and residues for the average number of fungi, lactic acid bacteria and endospores (p = 0.008, 0.005 and 0.016, respectively).
Also here, it should be noted that microbial counts for residues and larvae are dependent on the timing in a rearing cycle (Table 3). For instance, the larval microbial counts changed during the course of the cycles at EXT-CH, EXT-BE and EXT-NL. TVCs and fungal counts significantly decreased over the course of all cycles. The number of Enterobacteriaceae did not significantly change in any cycle, whereas for lactic acid bacteria and endospores either, no significant changes or significant decreases were observed.
Pathogen Detection
Listeria monocytogenes and coagulase-positive staphylococci were not detected in any cycle (although it should be noted that the detection limit for the latter at EXT-CH was set from 1000 to 10,000 cfu/g for the residues, due to the large background microflora during analysis). However, Salmonella enterica serovar Agona was present in the residue, but not in the larvae, of the one batch harvested at day 21 at EXT-BE. Additionally, Bacillus cereus was detected in quantities of 200 cfu/g in the residue of one batch of EXT-NL, as well as in all samples of larvae and residue from EXT-CH. In the latter samples, the bacterium was observed in quantities up to 6000 and 5000 cfu/g, respectively.
Metagenetic Analyses
High-throughput 16S rRNA gene sequencing was used to characterise the bacterial community composition of the substrate, residue and larvae samples. A total of 1306 OTUs was recovered from all samples (Online Resource 4). Relative OTU abundances and diversity indices were averaged over all replicate samples of phase II substrate, larvae and residue of each rearing cycle. Average sample coverage ranged from 81.2 to 99.5% (Table 4), indicating that the majority of the community members were recovered.
For the substrates, the richness, equitability and Shannon-Wiener diversity (Table 4) were the highest in cycles LAB 2 and 3, followed by LAB 1, which corresponded well to the large percentage of OTUs present in abundances of below 5% in these samples as seen in Fig. 1a. Furthermore, while the most abundant OTUs in rearing cycles LAB 1–3 were varying, the bacterial community of the other substrates (LAB 4, EXT-BE, EXT-NL and EXT-CH) showed more similarity with large abundances of OTUs belonging to the genus Lactobacillus (OTUs 6, 7, 9, 18, 26, 29, 41 and 57).
Similarly to the phase II substrate (Fig. 1a), the bacterial diversity of the residues (Fig. 1b) and of the larvae (Fig. 1c) differed largely between cycles and locations, as was also shown from the Chao1, Shannon-Wiener and equitability diversity indices (Table 4). In addition, NMDS analysis did not show clear clustering of phase II substrates, larvae and residues within rearing cycles (Fig. 2). Cluster analysis based on the larvae from different cycles also showed that, although LAB cycles 1–3 belonged to one cluster, and the external facilities to the other, LAB cycle 4 was more similar to the external facilities. The latter indicates that even within one location, vast differences in larval microbiota were observed (Fig. 3). Nevertheless, a total of 48 OTUs were in common for all larvae over all cycles, and most of these OTUs belonged to the phyla Proteobacteria (21 OTUs) and Firmicutes (21 OTUs) (Online Resource 7). However, none of them were present in abundances of more than 1% in all cycles. Figure 3 shows a heat map based on the OTUs that were present with a relative abundance of at least 1% in any larval sample, thus giving a more detailed overview as compared to Fig. 1 of the most prevalent OTUs present in the larvae. Also among these most abundant OTUs, some were present in larvae from all cycles. For instance, larvae from LAB 1 and LAB 2 showed a high abundance of a Morganella sp. (OTU 1), which was present in average abundances of 62.0 and 52.5%, respectively (Figs. 1c and 3). Also in the other cycles, that OTU was observed in abundances ranging from 0.5 to 2.1%. Other omnipresent OTUs in the larvae in this study were a Providencia sp. (OTU 23), ranging from less than 0.1 to 13.8% in abundance, and an Enterococcus sp. (OTU 11), with an abundance ranging from 0.9 to 9.9%. In addition, multiple OTUs corresponding to Pseudomonas sp. (e.g. OTUs 14, 32 and 46) were identified in larvae from all cycles, be it in abundances of below 2%. Furthermore, in all larvae (and residues) from cycles LAB 4, EXT-BE, EXT-NL and EXT-CH, members of Bacillaceae (OTUs 2, 4, 5, 15, 25, 33 and 110) were recovered in total abundances of more than 10%. A list of all 48 OTUs which were omnipresent in larvae from all cycles is given in Online Resource 7.
Some OTUs which were abundant in the larvae (Fig. 1c) were also abundant in the residue (Fig. 1b) of the same cycle. For instance, Morganella sp. (OTU 1) was also present in abundances higher than 10% in the residues of LAB 1 and 2. In addition, similar to the larvae of LAB 3, the residue of that cycle showed a high abundance of a Lactobacillus sp. (OTU 7). Residues of cycles LAB 4, EXT-BE, EXT-NL and EXT-CH were similar to the larvae of those cycles, characterised by a high abundance in members of Bacillaceae. Nevertheless, the overall bacterial communities of residues were still largely different from those of the larvae of the same rearing cycle.
Discussion
In this study, the microbiota of BSF larvae reared on a variety of waste streams at different locations, each with its own rearing methods and infrastructure, was studied. The aim of this study was (1) to characterise (part of) the variability in waste streams and rearing methods applied at different facilities, (2) to assess the variability in microbiota of BSF larvae reared in different facilities and (3) to study the correlation (if any) between the substrates used on the one hand, and the larvae and residues on the other. To this end, samples were taken from substrates, as well as from larvae and residues of seven different rearing cycles. As only one rearing cycle was studied per waste stream, it should be noted that inter-cycle variability for one location is not included in this study. More research is needed in order to assess the consistency in the microbiota when using the same substrate and rearing protocol in different cycles.
Microbial Characterisation of Waste Streams as BSF Rearing Substrates
A common aspect in all rearing cycles studied is the administration of a phase I substrate containing a grain-based product, such as laying hen/chick feed or wheat bran. The provision of a nutritionally dense substrate of known quality, such as chick feed or laying hen feed, during the first days of the cycle is a common practice in BSF rearing, in order to promote optimal growth during the first life stage. For most cycles, phase I substrates existed of this grain-based product, moisturised with water immediately before administration to the larvae. However, for cycle EXT-BE, the chick feed was already mixed in with the phase II substrate. Because of this, the moisture content was the highest and the pH was the lowest of all phase I substrates (likely due to the apple waste stream ingredient (pH = 3.78 ± 0.16) and DDGS ingredient (pH = 4.88 ± 0.00, data not shown; Table 2). Because of the deviating composition of this phase I substrate compared to the others, most microbial counts were higher (Table 2). Nevertheless, all phase I substrates showed a water activity of at least 0.96 and their pH ranged between 5.59 and 6.58. These properties make them highly opportune matrices for microbial growth [37], especially combined with the temperature in the rearing environment (on average between 24.9 and 29.1 °C; Online Resource 1) which was close to the optimal growth temperature for many microorganisms.
The varying composition of the phase II substrates clearly resulted in large differences in intrinsic parameters, in microbial counts and in composition and diversity of the bacterial community. Nevertheless, NMDS analysis (Fig. 2) showed the bacterial community of some substrates to be closely related to each other than to other substrates. For instance, phase II substrates of LAB 1, LAB 2 and EXT-CH, as well as substrates of LAB 4 and EXT-BE, were more similar in bacterial composition to each other than to the other substrates. The substrate of EXT-NL, in contrast, was the least similar to the other substrates, despite the fact that its community was highly abundant in the same Lactobacillus sp. (OTU 7) as the substrate of LAB 4. In general, it can be stated that various waste streams used to grow BSF larvae at different locations can highly differ in intrinsic parameters, microbial numbers as well as in bacterial community composition. It should also be noted that the results obtained from the substrates in this study cannot be extrapolated to other substrates which may differ in proportion of the ingredients used, in ingredient types or in the way they were treated, transported and/or stored.
Microbiota of Harvested Larvae from Different Rearing Cycles
Microbial numbers (Table 2) as well as bacterial communities (Figs. 1c and 3) and diversity indices (Table 4) of freshly harvested larvae differed to a large extent between rearing cycles. Given the observed changes in some of the microbial counts during the course of the rearing period (Table 3), it can be concluded that besides the selection of the substrate and the rearing methods, the timing of harvest likely influences the microbial numbers of the harvested larvae. Indeed, the timing of harvest determines the age and developmental stage of the harvested larvae, which in turn may affect their intrinsic parameters and microbiota. For instance, larval fat bodies are thought to represent a key tissue for insect humoral immunity and particularly for synthesis of antimicrobial peptides [38, 39], and it is also suggested that the fat content and accordingly the fat body sizes increase during BSF development [40]. As a consequence, larvae may express increasing levels of antimicrobial peptides as they mature. In addition, recent evidence exists for dietary effects on both BSF fat body metabolism [41] and antimicrobial peptide expression profiles [42]. Thus, both larval age as well as dietary changes may trigger feedback signals on temporal microbial dynamics.
Besides the possible influence of harvesting age, this study clearly shows that a large variability exists in the microbial quality of larvae reared at different facilities and on different substrates. The variability seen in this study is generally larger than the variability reported in two studies by Vandeweyer et al. [43, 44], who investigated intrinsic parameters, microbial counts and bacterial community composition of mealworms (Tenebrio molitor) and crickets (Acheta domesticus and Gryllodes sigillatus) from different facilities and different production batches. This observation can likely be explained by the fact that substrates and rearing procedures for the latter insect species are generally more comparable between production facilities than for BSF.
Remarkably, a total of 48 OTUs were in common for the larvae from all cycles (although none of them were present in abundances of more than 1% in every cycle; Fig. 3; Online Resource 7). Of these OTUs, Morganella sp. was also reported in other studies in BSF eggs [45] and BSF larvae grown on calf forage, food waste and cooked rice [7]. Both studies also mention the presence of a Providencia sp., while the latter also reports the presence of Enterococcus sp. on all three rearing substrates used. Similar to our study, Zheng et al. [45], who studied the microbiota in different BSF life stages, reported the presence of Pseudomonas sp. in BSF larvae, prepupae and adults, as well as the genus Bacillus in prepupae only. Except for Pseudomonas sp., the aforementioned OTUs detected in the larvae in this study were not detected in large abundancies in the phase II substrates (< 1%). However, it should be noted that Bacillaceae may have been present in the substrate as endospores, which are more difficult to detect through sequencing due to their resistance to DNA isolation techniques [46]. Possibly, the genera Morganella, Enterococcus, Pseudomonas and/or Providencia, as well as certain Bacillaceae sp. are part of a group of microorganisms often recurring in BSF larvae, regardless of substrates or other rearing conditions. In addition, NMDS analyses shows that larvae from different cycles to be positioned more closely together as compared to phase II substrates and residues (Fig. 2), suggesting that biotic and abiotic interactions in the larval gut may select for their bacterial community composition to become more alike. Whether a true ‘core microbiota’ is present, according to one of the definitions or approaches given in literature, such as for instance by Astudillo-Garcia et al. [47], remains to be established. Some genera could also be part of a so-called ‘house flora’ present in rearing facilities, but this cannot be stated with certainty since the microbiota of the production environment in rearing facilities has never been investigated so far, according to our knowledge. Further research may also focus on the exploitation of microorganisms abundantly present in the BSF gut as a probiotic to enhance biomass production and/or immunity [28]. For instance, research already showed that specific Bacillus subtilis strains could be isolated from BSF larvae, and when added to the substrate, they were shown to enhance larval growth, likely due to their aid in substrate digestion [27, 48]. As for Providencia, this genus was shown to attract females for oviposition [49] and is likely transmitted vertically through the haemolymph, although more research on this hypothesis is advised [28]. Thus, the microbial community of eggs and hatchlings may already differ between rearing facilities, before they even had contact with the substrate, depending on their parental origin. Such hypothesis was also suggested for mealworm rearing [50]. Although it is difficult to determine the historical origin of each BSF strain studied, each facility has been using a single strain for multiple years and hence multiple generations, before the rearing cycles in this study were conducted. Therefore, it is very likely that BSF reared at different locations harboured a location-specific microbiota. Moreover, specific interactions between the insect’s genotype on the one hand, and its microbiota on the other hand, may possibly affect key life history performances such as nutritional physiology and immune defences [51,52,53,54]. Furthermore, there is emerging evidence showing that BSF larvae comprise a vast genetic diversity, and possibly even a cryptic species complex (i.e. two or more distinct species classified as a single species [55]; Sandrock, personal communication). Concomitantly, variation in overall larval microbial composition may also be fuelled by interactions with the host’s genetic background, an aspect that clearly deserves further research. However, even within the same BSF strain that was used for cycles LAB1–4, large variation exists in microbial community between rearing cycles. Indeed, cluster analysis showed that LAB cycle 4 was even more similar to the external rearing facilities than to LAB 1–3 (Fig. 3), than to the other three lab-scale rearing cycles. The latter indicates that even when those larvae were reared in the same location, using the same rearing methods, the use of a different substrate and possibly other contributing factors (e.g., slightly different environmental conditions …) will likely have impacted the larval microbiota. Thus, although a given BSF strain could possess a characteristic innate microbiota, it is suggested that the microbial dynamics in the larval gut during rearing are largely determined by other biotic and abiotic factors in the rearing environment. The latter likely encompasses feed composition and quality, other microorganisms present in the rearing system, and the overall responses of microorganisms to relevant biotic and abiotic changes triggered by the larvae themselves (see also ‘Relation Between Rearing Substrates, Residues And Larvae’ section). As such, even within one location using the same rearing methods, differences in microbiota of the larvae may exist. It is thus reasonable to assume that even when the same substrate and rearing techniques are used in a facility, one rearing cycle may even differ from another when conducted at a different point in time.
Relation Between Rearing Substrates, Residues and Larvae
It is reasonable to assume that both intrinsic parameters as well as the microbial composition of the substrate affect the microbial dynamics during the rearing phase. However, despite the high variability in the microbial load of the substrates, the leftover residues (being the (partially) digested substrates, larval faeces and exuviae) showed an average TVC of at least 8.5 log cfu/g in all rearing cycles. In general, most other residue counts were also higher compared to the substrates, indicating the rearing system (i.e. environmental conditions, interactions with larvae and the nutrient composition of the substrates) to be highly suitable for microbial growth. However, no correlations were observed between microbial counts or intrinsic parameters between the phase I and II substrates on the one hand, and the residue on the other hand. In all cycles, except for LAB 3, the residue at harvest showed a higher pH value when compared to the substrates administered. Previous studies also detected increases in pH in the residue [1, 24, 56, 57]. The rise in pH can be explained in the first place by the production of ammonia during the digestion of proteins by the larvae, but the production of ammonia by the indigenous microflora of the substrate also has been hypothesised [1, 24]. The ammonia produced might have an antimicrobial effect on certain bacteria such as Salmonella sp. [24]. Nevertheless, even the residue of LAB 4, with pH 9.09, contained an average TVC of 8.9 log cfu/g and showed the highest Shannon-Wiener diversity. Further research is needed in order to unravel the interplay between substrate pH, larvae and the present microbiota.
Previous studies have indicated that the substrate can affect the bacterial [7] and fungal [23] composition of BSF larvae. However, in our study, also for the larvae, no correlations were observed between intrinsic parameters and microbial counts of the substrates and those of the larvae, respectively. The bacterial communities of the substrates (Fig. 1a) are clearly represented by a distinct set of bacteria compared to those of the larvae (Fig. 1c) and residues (Fig. 1b). Although most of the OTUs which are abundant in the larvae are also present in the phase II substrate, they are generally present in very low abundances (below 1%) in the latter. Similarly, very little of the OTUs abundant in the phase II substrates reach a high prevalence in the larvae or residues. Yet, multiple OTUs are also present in each cycle that are observed in the larvae but were not at all recovered from the substrate. Some of these OTUs may originate from the phase I substrate. Even though phase I substrates were discarded from data-analysis due to a too low number of sequence reads, a separate data-analyses of these substrates (rarefied to 250 sequence reads, data not shown) revealed that the most abundant OTUs (more than 5% of sequences) for the laboratory cycles belonged to genera Erwinia, Pedobacter, Parabacteroides and Hafnia, and to Microbacteriaceae. The wheat bran of EXT-NL was most abundant in an Erwinia sp., a Massilia sp. and a Parabacteroides sp. The laying hen feed from EXT-CH was highly colonised by an Erwinia sp. and two Pseudomonas sp. These genera are generally not found in high abundances in the residues or larvae. It should be noted, however, that, given the low number of sequence reads in these analyses, the results should be interpreted with caution. The phase I substrate of EXT-BE, which consisted of chick feed supplemented with methylparaben (0.1%) mixed into the phase II substrate, was as expected highly similar in bacterial composition to the phase II substrate alone. Yet, also here, no correlating patterns could be discerned between substrates and larvae/residues. It is therefore assumed that other factors besides substrate type also contribute to the bacterial composition of larvae and residues [28, 45]. First, bacteria from both the environment inside as well as the environment outside the rearing facility may enter the rearing containers through the air or via personnel [58]. If those bacteria are suited to the environment inside the crates, they might colonise the system and compete with other microorganisms for nutrients. Additionally, the environmental conditions in the rearing containers can be different from those in substrate storage (often in a cool environment), and they were also shown to change during rearing (e.g. pH and water activity). Although microorganisms themselves are likely among the causative drivers for these environmental changes [1, 24], these changes in turn can cause shifts in the microbial composition of the substrate post-administration. Furthermore, some bacterial species may survive the conditions in the larval gut and even multiply [20, 21]. Subsequently, they may be excreted into the residue in high numbers (as suggested by Wynants et al. [32] for lesser mealworms). This hypothesis may also explain the correlations observed between larvae and residues for some of the counts (lactic acid bacteria, endospores and fungi; Online Resource 6), as well as the occurrence of some OTUs abundant in both residues and larvae. Indeed, although NMDS analysis did not show any clustering of substrate, larvae and/or residues within rearing cycles, the residues and larvae of the same cycle were generally positioned slightly closer to each other as compared to the substrate (Fig. 2). Finally, the presence of the larvae themselves likely affects the microbiota of the rearing system, as was suggested in previous studies where the number of Salmonella sp. and E. coli were reduced in the presence of BSF larvae [1, 8, 12, 24,25,26]. BSF larvae (and/or members of their gut microbiota) likely produce antimicrobial components, which may influence the microbial community of the matrix by targeting specific bacteria [1, 28, 59, 60]. There can even exist interactions between environmental conditions, such as pH, and the stability of these antimicrobial compounds from BSF larvae in the rearing environment [28].
Microbial Safety Aspects
It is unclear whether Salmonella sp., which was revealed to be present in the residue—but not in the larvae—of the one batch that was harvested at day 21 at EXT-BE originated from one (or more) of the substrate components, or from the rearing environment. Even though the pathogen was not detected in any of the three larval samples (absent in 25 g), there is no guarantee of its absence in all larvae. As suggested in literature, a heat treatment to eliminate all possible Salmonellae is advised prior to processing of the larvae into feed and other products [17, 28]. Other decontamination technologies alternative to heat treatment (for instance high hydrostatic pressure or irradiation) may well be suitable too, provided the processing conditions necessary to kill Salmonella sp. in the particular matrix of the larvae are well established. Bacillus cereus, which was detected in one residue sample of EXT-NL as well as in larvae and residue samples of EXT-CH, is widely spread in soil, in water and in plants [61]. It could have contaminated the rearing environment via the substrate, as both cycles contained a vegetable component and both substrates from phase I and II contained endospores (Table 2). Indeed, Bacillus cereus has been identified in edible insects in previous studies [62, 63]. As shown from Illumina sequencing (Fig. 1), many Bacillaceae sp. were among the most abundant OTUs in the residues and larvae of rearing cycles LAB 4, EXT-BE, EXT-NL, and EXT-CH, confirming their wide-spread origins in BSF rearing. It is unknown whether the B. cereus cells detected for EXT-NL and EXT-CH were present in the larvae and/or residues as spores or vegetative form. However, given the large quantities recovered at EXT-CH combined with a pH not low enough to prevent spore germination, it can be assumed that the beneficial temperature and water activity and the nutrient-rich matrix may have encouraged endospores to germinate and multiply [64]. On the other hand, the progressive nutrient depletion during the course of the rearing cycle both by larvae and by microorganisms may have triggered sporulation during the rearing [65]. The specific mechanisms leading to the high numbers of (B. cereus) spores in the rearing system should be further established. Nevertheless, the presence of endospores and vegetative B. cereus cells may imply risks regarding the microbial safety of BSF larvae to be used in feed or eventually in food. First, spores are in general very resistant to heat treatments and/or other processing steps [64]. Second, in the current study, B. cereus counts of up to 3.8 log cfu/g were observed. Although the threshold cell density for production of the heat-resistant toxin cereulide is generally considered to be 4 to 5 log cfu/g, some studies also report even lower densities for toxin production [61]. As a consequence, the possible production of cereulide cannot completely be excluded. Attention should thus be paid to the presence B. cereus, both as endospore or in vegetative state, before and during the processing of BSF larvae into feed/food or other products [62].
Conclusions
The results from this study unravel a great variability in microbial quality and bacterial community composition when using different waste streams as a substrate for BSF larvae at laboratory and large scale. Although it was not disproved in this study that the substrates are an important source for bacterial species for the larvae and the residue, they are colonised by a different set of OTUs within each cycle. The results from this study indicate that the microbial quality and community composition of BSF larvae cannot be related based on the microbial composition of the substrate. Furthermore, it is clear that the microbiota of the larvae, both in numbers as well as the bacterial composition, also largely differs between rearing locations. Future research, however, should be dedicated to unravel the inter-batch variability within one location. Differences are likely caused by a multitude of factors, including differences in substrate type and rearing methods, interactions with other microbial community members and with the larvae and parental origin of the larvae. Nevertheless, a number of OTUs were present in more than one rearing cycle in the current study, be it in varying abundances. Some of these OTUs, such as a Morganella sp., a Enterococcus sp., Pseudomonas spp., a Providencia sp. and Bacillaceae, were also reported in BSF in literature. The wide-spread presence of these genera in different BSF larvae from different locations suggest the possible existence of a core microbiota in BSF larvae, although the abundance of these species seems highly variable depending on the abiotic and biotic factors in the rearing system, and possibly even the BSF strain used.
Two food pathogens, Salmonella sp. and Bacillus cereus, were identified in some of the residues and/or larvae. Their presence implies biological risks when BSF larvae are to be used in feed and maybe food. It is advised to apply an adequate heat treatment during processing of BSF larvae to reduce vegetative Salmonella and Bacillus cereus cells and to elaborate the time-temperature conditions to attain sufficient reduction. However, Bacillus cereus spores may eventually survive processing, as well as possible toxins produced during rearing. While one strategy to avoid this is to use only substrates not carrying Bacillus cereus, this would hinder the extensive use of food/feed side streams as substrate, in turn jeopardising the economic feasibility and sustainable nature of BSF rearing. Hence more research should focus on how to mitigate these risks to obtain microbially safe and toxin-free BSF larvae. The use of classical feed additives or fermentation of the substrate as strategies to control the microbial community during rearing may contribute to this aim. When exploring the potential of substrate fermentation, an ultimate innovation would be the development of a (mixture of) strain(s) as starter culture that not only secures microbial safety of the larvae but also promotes their growth.
References
Lalander CH, Fidjeland J, Diener S, Eriksson S, Vinnerås B (2015) High waste-to-biomass conversion and efficient Salmonella spp. reduction using black soldier fly for waste recycling. Agron Sustain Dev 35(1):261–271
European Commission (2015) Flash Eurobarometer 425. Report: Food waste and date marking. http://data.europa.eu/euodp/en/data/dataset/S2095_425_ENG. Accessed 10 August 2018
Martinez J, Dabert P, Barrington S, Burton C (2009) Livestock waste treatment systems for environmental quality, food safety and sustainability. Bioresour Technol 100:5527–5536
Diener S, Zurbrügg C, Tockner K (2009) Conversion of organic material by black soldier fly larvae: establishing optimal feeding rates. Waste Manag Res 27(6):603–610
van Huis A (2013) Potential of insects as food and feed in assuring food security. Annu Rev Entomol 58:563–583
Salomone R, Saija G, Mondello G, Giannetto A, Fasulo S, Savastano D (2017) Environmental impact of food waste bioconversion by insects: application of life cycle assessment to process using Hermetia illucens. J Clean Prod 140(2):890–905
Jeon H, Park S, Choi J, Jeong G, Lee S, Choi Y, Lee S (2011) The intestinal bacterial community in the food waste-reducing larvae of Hermetia illucens. Curr Microbiol 62:1390–1399
Nguyen TTX, Tomberlin JK, Vanlaerhoven S (2015) Ability of black soldier fly (diptera: stratiomyidae) larvae to recycle food waste. Environ Entomol 44(2):406–410
Sheppard DC, Newton GL, Thompson SA, Savage S (1994) A value added manure management system using the black soldier fly. Bioresour Technol 50(3):275–279
Myers HM, Tomberlin JK, Lambert B, Kattes D (2008) Development of black soldier fly (diptera: stratiomyidae) larvae fed dairy manure. Environ Entomol 37(1):11–15
Li Q, Zheng L, Cai H, Garza E, Yu Z, Zhou S (2011) From organic waste to biodiesel: black soldier fly, Hermetia illucens, makes it feasible. Fuel 90:1545–1548
Lalander C, Diener S, Magri ME, Zurbrügg C, Lindström A, Vinnerås B (2013) Faecal sludge management with the larvae of the black soldier fly (Hermetia illucens)—from a hygiene aspect. Sci Total Environ 45:312–318
Banks IJ, Gibson WT, Cameron MM (2014) Growth rates of black soldier fly larvae fed on fresh human faeces and their implication for improving sanitation. Tropical Med Int Health 19(1):14–22
Stadtlander T, Stamer A, Buser A, Wohlfahrt J, Leiber F, Sandrock C (2017) Hermetia illucens meal as fish meal replacement for rainbow trout on farm. J Insects Food Feed 3(3):165–175
Henry M, Gasco L, Piccolo G, Fountoulaki E (2015) Review on the use of insects in the diet of farmed fish: past and future. Anim Feed Sci Technol 203:1–22
Maurer V, Holinger M, Amsler Z, Früh B, Wohlfahrt J, Stamer A, Leiber F (2015) Replacement of soybean cake by Hermetia illucens meal in diets for layers. J Insects Food Feed 1:1–8
Wang YS, Shelomi M (2017) Review of black soldier fly (Hermetia illucens) as animal feed and human food. Foods 6(10):91
Zheng L, Hou Y, Li W, Yang S, Li Q, Yu Z (2012) Biodiesel production from rice straw and restaurant waste employing black soldier fly assisted by microbes. Energy 47:225–229
Elieh-Ali-Komi D, Hamblin MR (2016) Chitin and chitosan: production and application of versatile 653 biomedical nanomaterials. Int J Adv Res 4:411–427
Dillon RJ, Dillon VM (2004) The gut bacteria of insects: nonpathogenic interactions. Annu Rev Entomol 49(1):71–92
Engel P, Moran NA (2013) The gut microbiota of insects–diversity in structure and function. FEMS Microbiol Rev 37(5):699–735
EFSA Scientific Committee. (2015). Risk profile related to production and consumption of insects as food and feed. EFSA Journal 2015, 13(10), 60 pp. https://doi.org/10.2903/j.efsa.2015.425
Boccazzi IV, Ottoboni M, Martin E, Comandatore F, Vallone L, Spranghers T, Eeckhout M, Mereghetti V, Pinotti L, Epis S (2017) A survey of the mycobiota associated with larvae of the black soldier fly (Hermetia illucens) reared for feed production. PLoS One 12(8):e0182533
Erickson MC, Islam M, Sheppard C, Liao J, Doyle MP (2004) Reduction of Escherichia coli O157: H7 and Salmonella enterica serovar enteritidis in chicken manure by larvae of the black soldier fly. J Food Prot 67(4):685–690
Čičková H, Newton GL, Lacy RC, Kozánek M (2015) The use of fly larvae for organic waste treatment. Waste Manag 35:68–80
Liu Q, Tomberlin JK, Brady JA, Sanford MR, Yu Z (2008) Black soldier fly (Diptera: Stratiomyidae) larvae reduce Escherichia coli in dairy manure. Environ Entomol 37(6):1525–1530
Xiao X, Mazza L, Yu Y, Cai M, Zheng L, Tomberlin JK, Yu J, van Huis A, Yu Z, Fasulo S, Zhang J (2018) Efficient co-conversion process of chicken manure into protein feed and organic fertilizer by Hermetia illucens L.(Diptera: Stratiomyidae) larvae and functional bacteria. J Environ Manag 217:668–676
De Smet J, Wynants E, Cos P, Van Campenhout L (2018) Microbial community dynamics during rearing of black soldier fly larvae (Hermetia illucens) and impact on exploitation potential. Appl Environ Microbiol 84(9):e02722–e02717
Cheng JY, Chiu SL, Lo IM (2017) Effects of moisture content of food waste on residue separation, larval growth and larval survival in black soldier fly bioconversion. Waste Manag 67:315–323
Stoops J, Crauwels S, Waud M, Claes J, Lievens B, Van Campenhout L (2016) Microbial community assessment of mealworm larvae (Tenebrio molitor) and grasshoppers (Locusta migratoria migratorioides) sold for human consumption. Food Microbiol 53:122–127
Dijk, R., van den Berg, D., Beumer, R., de Boer, E., Dijkstra, A., Mout, L., … in ’t Veld, S. (2015). Microbiologie van voedingsmiddelen: Methoden, principes en criteria. (R. Dijk, D. van den Berg, R. Beumer, E. de Boer, A. Dijkstra, L. Mout, … S. in ’t Veld, Eds.) (5th ed.). Capelle aan den Ijssel, The Netherlands: MYbusinessmedia
Wynants E, Crauwels S, Verreth C, Gianotten N, Lievens B, Claes J, Van Campenhout L (2018) Microbial dynamics during production of lesser mealworms (Alphitobius diaperinus) for human consumption at industrial scale. Food Microbiol 70:181–191
Chao A (1984) Nonparametric estimation of the number of classes in a population. Scand J Stat 11:265–270
Pielou EC (1966) The measurement of diversity in different types of biological collections. J Theor Biol 13:131–144
Shannon CE (1948) A mathematical theory of communication. Bell Syst Tech J 27:379–423 & 623–656
Development Core Team R (2013) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Adams MR, Moss MO (2008) Food microbiology3d edn. RSC Publishing, Cambridge
Park SI, Kim JW, Yoe SM (2015) Purification and characterization of a novel antibacterial peptide from black soldier fly (Hermetia illucens) larvae. Dev Comp Immunol 52(1):98–106
Zdybicka-Barabas A, Bulak P, Polakowski C, Bieganowski A, Waśko A (2017) Immune response in the larvae of the black soldier fly Hermetia illucens. ISJ 14:9–17
Liu X, Chen X, Wang H, Yang Q, ur Rehman K, Li W, Cai M, Li Q, Mazza L, Zhang J, Yu Z, Zheng L (2017) Dynamic changes of nutrient composition throughout the entire life cycle of black soldier fly. PLoS One 12(8):e0182601
Pimentel AC, Montali A, Bruno D, Tettamanti G (2017) Metabolic adjustment of the larval fat body in Hermetia illucens to dietary conditions. J Asia Pac Entomol 20(4):1307–1313
Vogel H, Müller A, Heckel DG, Gutzeit H, Vilcinskas A (2018) Nutritional immunology: diversification and diet-dependent expression of antimicrobial peptides in the black soldier fly Hermetia illucens. Dev Comp Immunol 78:141–148
Vandeweyer D, Crauwels S, Lievens B, Van Campenhout L (2017) Microbial counts of mealworm larvae (Tenebrio molitor) and crickets (Acheta domesticus and Gryllodes sigillatus) from different rearing companies and different production batches. Int J Food Microbiol 242:13–18
Vandeweyer D, Crauwels S, Lievens B, Van Campenhout L (2017) Metagenetic analysis of the bacterial communities of edible insects from diverse production cycles at industrial rearing companies. Int J Food Microbiol 261:11–18
Zheng L, Crippen TL, Singh B, Tarone AM, Dowd S, Yu Z, Wood TK, Tomberlin JK (2013) A survey of bacterial diversity from successive life stages of black soldier fly (Diptera: Stratiomyidae) by using 16S rDNA pyrosequencing. J Med Entomol 50(3):647–658
Filippidou S, Junier T, Wunderlin T, Lo CC, Li PE, Chain PS, Junier P (2015) Under-detection of endospore-forming Firmicutes in metagenomic data. Comput Struct Biotechnol J 13:299–306
Astudillo-García C, Bell JJ, Webster NS, Glasl B, Jompa J, Montoya JM, Taylor MW (2017) Evaluating the core microbiota in complex communities: a systematic investigation. Environ Microbiol 19(4):1450–1462
Yu G, Cheng P, Chen Y, Li Y, Yang Z, Chen Y, Tomberlin JK (2011) Inoculating poultry manure with companion bacteria influences growth and development of black soldier fly (Diptera: Stratiomyidae) larvae. Environ Entomol 40(1):30–35
Zheng L, Crippen TL, Holmes L, Singh B, Pimsler ML, Benbow ME, Tarone AM, Dowd S, Yu Z, Vanlaerhoven SL, Wood TK, Tomberlin JK (2013) Bacteria mediate oviposition by the black soldier fly, Hermetia illucens (L.) (Diptera: Stratiomyidae). Sci Rep 3:2563
Osimani A, Milanović V, Cardinali F, Garofalo C, Clementi F, Pasquini M, Riolo P, Ruschioni S, Isidoro N, Loreto N, Franciosi E, Tuohy K, Petruzzelli A, Foglini M, Gabucci C, Tonucci F, Aquilanti L (2018) The bacterial biota of laboratory-reared edible mealworms (T. molitor L.): from feed to frass. Int J Food Microbiol 272:49–60
Early AM, Shanmugarajah N, Buchon N, Clark AG (2017) Drosophila genotype influences commensal bacterial levels. PLoS One 12(1):e0170332
Dobson AJ, Chaston JM, Newell PD, Donahue L, Hermann SL, Sannino DR, Westmiller S, Wong CAN, Clark AG, Lazzaro BP, Douglas AE (2015) Host genetic determinants of microbiota-dependent nutrition revealed by genome-wide analysis of Drosophila melanogaster. Nat Commun 6:6312
Näpflin K, Schmid-Hempel P (2018) Host effects on microbiota community assembly. J Anim Ecol 87(2):331–340
Vorburger C, Perlman SJ (2018) The role of defensive symbionts in host–parasite coevolution. Biol Rev 93:1747–1764
Bickford D, Lohman DJ, Sodhi NS, Ng PKL, Meier R, Winker K, Ingram KK, Das I (2007) Cryptic species as a window on diversity and conservation. Trends Ecol Evol 22(3):148–155
Popa R, Green TR (2012) Using black soldier fly larvae for processing organic leachates. J Econ Entomol 105(2):374–378
Ma J, Lei Y, Rehman KU, Yu Z, Zhang J, Li W, Li Q, Tomberlin JK, Zheng L (2018) Dynamic effects of initial pH of substrate on biological growth and metamorphosis of black soldier fly (Diptera: Stratiomyidae). Environ Entomol 47(1):159–165
Schneider JC (2009) Principles and procedures for rearing high quality insects. Mississippi State University, Mississippi
Choi WH, Yun JH, Chu JP, Chu KB (2012) Antibacterial effect of extracts of Hermetia illucens (Diptera: Stratiomyidae) larvae against gram-negative bacteria. Entomol Res 42(5):219–226
Spranghers T, Michiels J, Vrancx J, Ovyn A, Eeckhout M, De Clercq P, De Smet S (2018) Gut antimicrobial effects and nutritional value of black soldier fly (Hermetia illucens L.) prepupae for weaned piglets. Anim Feed Sci Technol 235:33–42
Stenfors Arnesen LP, Fagerlund A, Granum PE (2008) From soil to gut: Bacillus cereus and its food poisoning toxins. FEMS Microbiol Rev 32(4):579–606
Fasolato L, Cardazzo B, Carraro L, Fontana F, Novelli E, Balzan S (2018) Edible processed insects from e-commerce: food safety with a focus on the Bacillus cereus group. Food Microbiol 76:296–303
Grabowski NT, Klein G (2017) Microbiology of processed edible insect products—results of a preliminary survey. Int J Food Microbiol 243:103–107
Wells-Bennik MHJ, Eijlander RT, Den Besten HMW, Berendsen EM, Warda AK, Krawczyk AO, Nierop Groot MN, Xiao Y, Zwietering MH, Kuipers OP, Abee T (2016) Bacterial spores in food: survival, emergence, and outgrowth. Annu Rev Food Sci Technol 7:457–482
van der Voort M, Abee T (2013) Sporulation environment of emetic toxin-producing Bacillus cereus strains determines spore size, heat resistance and germination capacity. J Appl Microbiol 114(4):1201–1210
Funding
The research that yielded these results was funded by the Belgian Federal Public Service of Health, Food Chain Safety and Environment through the contract RT 15/9 EDINCO.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Ethical Approval
All applicable international, national and/or institutional guidelines for the care and use of animals were followed.
Rights and permissions
About this article
Cite this article
Wynants, E., Frooninckx, L., Crauwels, S. et al. Assessing the Microbiota of Black Soldier Fly Larvae (Hermetia illucens) Reared on Organic Waste Streams on Four Different Locations at Laboratory and Large Scale. Microb Ecol 77, 913–930 (2019). https://doi.org/10.1007/s00248-018-1286-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-018-1286-x