Abstract
The ecological interaction between microorganisms and seaweeds depends on the production of secondary compounds that can influence microbial diversity in the water column and the composition of reef environments. We adapted the 3H-leucine incorporation technique to measure bacterial activity in biofilms associated with the blades of the macroalgae Sargassum spp. We evaluated (1) if the epiphytic bacteria on the blades were more active in detritus or in the biofilm, (2) substrate saturation and linearity of 3H-leucine incorporation, (3) the influence of specific metabolic inhibitors during 3H-leucine incorporation under the presence or absence of natural and artificial light, and (4) the efficiency of radiolabeled protein extraction. Scanning electron microscopy showed heterogeneous distribution of bacteria, diatoms, and polymeric extracellular secretions. Active bacteria were present in both biofilm and detritus on the blades. The highest 3H-leucine incorporation was obtained when incubating blades not colonized by macroepibionts. Incubations done under field conditions reported higher 3H-leucine incorporation than in the laboratory. Light quality and sampling manipulation seemed to be the main factors behind this difference. The use of specific metabolic inhibitors confirmed that bacteria are the main group incorporating 3H-leucine but their association with primary production suggested a symbiotic relationship between bacteria, diatoms, and the seaweed.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The use of radiolabeled (3H or 14C) leucine has gained wider acceptance in the studies of microbial ecology since the 1980s [72]. This technique has the advantage of being simple, economical, and quick [87], and the leucine incorporation in protein content reveals the activity and carbon production of microorganisms [51]. Considering its efficiency in estimating energy fluxes, the leucine incorporation technique is still being adapted to different microbial habitats in marine, freshwater, and soil environments [4, 7, 8, 15, 16, 33, 42, 65]. In marine systems, the leucine uptake technique is widely used to measure bacterioplankton carbon production in oceanic and coastal environments [21, 95], such as estuarine silt [1], salt-marsh sand [90], coastal [44], and deep-sea sediments [24], as well as detritus and roots from estuarine macrophytes [98]. Despite its clear advantages, the leucine incorporation technique has not yet been updated to seaweed species. On the other hand, studies of bacteria–algae interactions using molecular techniques have revealed the community composition of microorganisms [2, 89, 92].
Biofilm supports functionally and phylogenetically diverse microbial communities that are involved in biogeochemical cycles of many elements [50]. Biofilm formation on plant surfaces is linked to active bacterial attachment processes and production of exopolymeric substances. Epiphytic bacteria on macroalgae benefit from organic compounds and oxygen furnished by the algae. In turn, bacteria mineralize exuded organic materials and provide CO2 and other metabolic compounds that favor algal growth [63]. On the other hand, antibiotics can be produced as secondary metabolites of both algae and bacteria, but many microbial populations are able to degrade them into the low concentrations usually found in natural environments [3]. On a small scale, the bacteria–algae interaction has an indirect influence on other organisms and in the composition of reef environments [70, 88].
The seaweed Sargassum is globally distributed and is a very important primary producer in reef environments, rocky shores, and the open ocean. Its photosynthetic activity may regulate microbial metabolism through production of secondary metabolites [2]. In the Cabo Frio upwelling region (Rio de Janeiro state, Brazil), Sargassum is ecologically very important as a food source and place of refuge for many organisms [27, 38, 59]. The presence of Sargassum in this region is seasonal, and its life cycle is associated with the upwelling period during spring/summer seasons [41].
The aim of this study was to adapt the 3H-leucine incorporation technique so as to measure the activity of epiphytic bacteria in biofilms associated with the blades of Sargassum. We used specific metabolic inhibitors to evaluate the activity of bacteria and other organisms in leucine uptake. We also did experiments in situ and under laboratory conditions to assay the influence of light in the bacteria–algae interaction. The presence and distribution of microorganisms were observed by scanning electron microscopy (SEM).
Methods
Seaweed Sampling and Processing
Specimens of Sargassum furcatum and other species were collected during 2006 and 2007 at different sites of Arraial do Cabo, RJ (23° S, 42° W), the main upwelling area on the Brazilian coast, in the Cabo Frio region [17, 94]. Samples were collected manually by SCUBA at 1–10-m depth and kept in ziplock bags with local water up until analysis in the laboratory. Blades were chosen randomly and collected with tweezers to avoid hand contact. Blades were incubated in the dark at 25 °C in 2.0-mL microcentrifuge tubes with 1.0 mL of local water filtered through 0.22-μm polycarbonate membranes.
SEM
To observe the presence and distribution of microorganisms in biofilms formed on Sargassum blades, samples were fixed in buffered 4 % glutaraldehyde (0.05 M cacodylate, pH 7.4) and kept under refrigeration until SEM processing. Thereafter, blades were washed with buffered sterilized seawater (0.05 M cacodylate, pH 7.4) and postfixed in 1 % OsO4 for 1 h at 20 °C. After washing, the samples were submitted to a sequential dehydration in ethanol/water solutions (30, 50, 70, and 100 %). The samples were dried in critical-point drying equipment Bal-Tec SPD 030, mounted in aluminum stubs, and coated with a thin gold layer by using a sputter coater Bal-Tec SCD 050. Samples were analyzed and photographed with Zeiss EVO 40 using an accelerating voltage of 15 kV.
Blade Incubation Procedure
We implemented a series of technical evaluations prior to the main experiments. Firstly, we tested if epiphytic bacteria on blades were more active in detritus or in the biofilm. An experiment was done incubating (1) the intact blade, (2) the blade after 1 min of vortex, (3) the detritus released by the blade after vortex, (4) a scrubbed blade, and (5) its scrubbed material. Despite evidence of mechanical stress, the use of SDS or EDTA was avoided to reduce chemical influence on the live biofilm. During sampling, macroepibionts (filamentous algae, calcareous algae, and hydrozoans) were observed on the blades, and hence the influence of their presence on 3H-leucine incorporation was tested.
Blanks were prepared by addition of formaldehyde (5 % final concentration), and results were subtracted from samples killed with 5 % formaldehyde after 30 min of incubation. Extraction was done by filtration [16]. After 10 min in an ultrasonic bath, 5 % (final concentration) trichloroacetic acid (TCA) was added to precipitate the formed 3H-protein. After 30 min, samples were filtered through 0.22-μm membranes presoaked in unlabelled leucine (1,500 nM). Filters were washed two times with 5 % TCA and 80 % ethanol. After drying, membranes were immersed in a liquid scintillation cocktail for radioactivity measurement. Leucine incorporation rate (in moles per centimeter per hour) was calculated considering net disintegrations per minute (DPM), sample size, leucine concentration, and 3H-leucine specific activity (72 Ci mmol−1). Once there was no correlation between leucine incorporation and weight, size (width and height), or area of blade, we incubated 1 cm2 of blade in the following experiments. When the blade was larger than 1 cm2, we needed to cut the blade before incubation, but there was no significant change in results obtained with cut blades (n = 4, p = 0.49).
Substrate Saturation and Linearity of 3H-Leucine Incorporation
After the blade incubation procedure was determined, we carried out a series of experiments to assess leucine saturation concentration (15, 25, 35, 45, 55, 70, or 100 nM of final concentration) and the best incubation time (5, 15, 30, 45, 60, 90, or 120 min) with triplicates and two blanks. The samples were stored at −18 °C until protein extraction. All described experiments were done with two blanks and in triplicate adding 20 nM leucine. This concentration was used in previous bacterioplankton studies in the region [40]. The following experiments were done adding 40 nM leucine, and protein extraction was done after up to 15 days as longer storage periods lead to significant leucine breakdown by radiolysis, as shown by Miranda et al. [65] in freshwater periphyton samples.
Efficiency of Radiolabel Protein Extraction
We tested acid and alkaline extraction methods and also compared filtration [52] and microcentrifugation procedures [65] with minor modifications. The extraction methodologies were as follows:
-
1.
During the extraction by filtration [52], samples were first sonicated in an ultrasonic bath. The tested sonication times were 1, 5, and 10 min. Following sonication, 110 μL of 50 % trichloroacetic acid (final concentration equivalent to 5 % TCA) was added. After 30 min, the samples were filtered through 0.22-μm cellulose Millipore membranes presoaked in unlabeled (cold) leucine (1,000 nM). The filters were washed two times with 5 % TCA and two times with 80 % ethanol. After drying, the filters were placed in vials with 2 mL of scintillation cocktail (Cytoscint) and radioassayed by scintillation counting (TRICARB PACKARD 1600) for 30 min or after the accumulation of 10,000 counts. DPM values were obtained after quenching correction by using the external standard of the equipment. 3H-Leucine bacterial incorporation rates were calculated using DPM, the specific activity of 3H-leucine, and a conversion rate from DPM to 1 mol of leucine [51].
-
2.
The alkaline extraction procedure followed adaptations by Marxsen [62]. After thawing, the samples were centrifuged at 6,000 × g for 6 min. The supernatant was discarded, and 1 mL of 20 % acetic acid was added to remove carbonates. A new centrifugation was done (6,000 × g for 6 min), and samples received 0.2 mL of a solution composed of 80 mL of concentrated ethanol, 20 mL of distilled water, and 10 mg of cold leucine. After a new centrifugation (6,000 × g for 5 min) and addition of 0.2-mL alkaline solution (0.5 M NaOH, 25 mM EDTA, 1 % SDS), samples were heated to 100 °C for 2 h. After centrifugation (6,000 × g for 5 min), 0.5 mL from the supernatant was added into 4.5 mL of scintillation cocktail (Cytoscint) for beta radiation measurements.
-
3.
An adaptation was tested following Miranda et al. [65] for periphyton associated with macrophyte roots. Firstly, 1 cm2 of blade was incubated with 3H-leucine for 30 min in 1.0 mL of water from the studied site filtered at 0.22 μm; 60 μL of 100 % TCA stopped the incubation, and samples were frozen until protein extraction. Extraction started with a 5-min sonication bath, and the blade was removed. Samples were centrifuged at 2,500 × g for 15 min, and the supernatant was transferred to a new microcentrifuge tube. A new centrifugation was done at 13,000 × g for 10 min, and the supernatant was discarded; 1.5 mL of cold TCA (5 %) was added, and the centrifugation process was repeated. One milliliter of 80 % ethanol was added, and a new centrifugation was done. Finally, 1 mL of scintillation cocktail (Cytoscint) was added, and after an overnight period, the beta radiation was counted.
Influence of Specific Metabolic Inhibitors on 3H-Leucine Incorporation Under Dark as well as Natural and Artificial Light
The influence of light during the incubations was evaluated in two preliminary experiments using nine replicates and three blanks, one in the laboratory under artificial fluorescent light and another in the laboratory situated in a beach under natural light. We tested 5 % formaldehyde and 5 % TCA to stop incubation, but no difference was observed. Labeled proteins were extracted following Coelho-Souza et al. [16].
Thereafter, we tested the effect of specific metabolic inhibitors of heterotrophic bacteria, sulfate-reducing bacteria, yeasts, and primary producers. The experiments were done in field and laboratory environments, under dark and light conditions. We added 40 nM 3H-leucine and used two blanks and four replicates. We followed the method described by Miranda et al. [65] with some modifications (Fig. 7). Immediately after starting incubation in the field, the microcentrifuge tubes were put in 125-mL glass Winkler bottles with local water. To simulate dark conditions, the bottles were covered with aluminum foil. Bottles were put in open-end transparent acrylic tubes and incubated in situ (25 °C) for 75 min. Laboratory incubations were done under artificial fluorescent lamps as well as in the dark at 25 °C.
Specific inhibitors were added to samples 2 h before 3H-leucine addition. We tested three types of antibiotics (5 μg/L streptomycin, 100 units/L penicillin, and 0.2 nM chloramphenicol) and inhibitors of sulfate-reducing bacteria (20 mM sodium molybdate), photosynthesis (10 μM diuron), and eukaryotes—mainly yeasts (0.02 % cycloheximide).
In field experiments, we measured untreated controls, and samples were treated with combined antibiotics (5 μg/L streptomycin, 100 units/L penicillin, and 0.2 nM chloramphenicol) and with diuron, a photosynthesis inhibitor. In the laboratory, we compared untreated controls with samples treated with (1) 5 μg/L streptomycin, (2) 100 units/L penicillin, (3) 20 mM sodium molybdate, (4) 10 μM diuron, and (5) 0.02 % cycloheximide. All these experiments were carried under dark and light conditions.
Statistical Tests
Data were not normally distributed even when transformed. Therefore, we used the Mann–Whitney nonparametric test to compare data from experiments with two treatments and the Kruskal–Wallis test if there were more treatments [101]. Experiments with the addition of specific metabolic inhibitors were statistically tested using bifactorial ANOVA [93]. Therefore, we considered the following factors as fixed: (1) light/dark conditions and (2) the different inhibitors nested in factor 1. For all tests, we considered a 0.05 significance level.
Results
Microorganism Distribution on Blades
The presence of bacteria and other microorganisms on Sargassum blades was confirmed by SEM images. The surface of the seaweed blades was fouled to different degrees, and the microbial community varied in structure (Fig. 1). Blade coverage varied from a smooth detritus or bacteria coating (Fig. 1a, b) to a complex coating (Fig. 1c–f) heterogeneously formed by bacteria, diatoms (mainly Cocconeis), and other cells immersed in a matrix of extracellular polymeric substance (EPS) (Fig. 1c–f). A three-dimensional network with bacteria and diatoms was registered (Fig. 1d). A very dense EPS matrix was observed around diatoms (Fig. 1e). The high abundance of diatoms near criptostoma, an algae structure responsible for nutrient uptake, was also noted.
SEM images of S. furcatum blades showing a the presence of detritus on blades, bar = 2 μm; b attachment of bacteria to blade surface, bar = 1 μm; c biofilm formation and the initial colonization of blade surface by bacteria, bar = 5 μm (insert figure shows bacteria in detail, bar = 1 μm); d a portion more densely occupied with bacteria and diatoms, bar = 4 μm; e an abundant EPS matrix forming a three-dimensional structure, where some diatoms are immersed, bar = 9 μm; and f the presence of diatoms (Cocconeis) close to the criptostoma, bar = 120 μm, (insert figure shows a detail of diatom immersed on EPS, bar = 4 μm)
Blade Incubation
The high variability in distribution of microorganisms on blades observed on SEM images was corroborated by 3H-leucine incorporation data. The leucine incorporation experiment indicated that bacteria were mainly associated not only with the biofilm but also with epiphytic detritus (Fig. 2). We observed that measurements of bacterial activity were clearly influenced by the incubation procedure (p = 0.03). Intact blades tended to have the highest bacterial activity (up to seven times higher than brushed blades) and the lowest data variability. Bacterial activity was similar in incubations of extracted detritus and of blades without detritus. The brushing of blades might destroy the biofilm structure and influence the bacteria–algae interaction. Biofilm extracted by brushing also presented low bacterial activity and is probably associated with a low brushing efficiency. Brushing also resulted in higher activity in blanks (91 % of DPMsamples), as observed when incubating detritus removed by vortexing (70 %). The blank average was 43 % in detritus samples, while it was around 7 % when blades were incubated.
The variability of 3H-leucine incorporation is not only related to heterogeneity of microorganism distribution in biofilms but also to fouling by macroorganisms. The presence of macroepibionts decreased bacterial activity by almost three times (Fig. 3), and the presence of hydrozoans resulted in the lowest 3H-leucine incorporation (p = 0.02). Therefore, we used blades without visible macroepibionts in the following experiments.
Substrate Saturation and Linearity of 3H-Leucine Incorporation
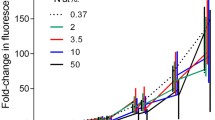
3H-leucine incorporation increased due to higher substrate concentration but tended to stabilize around 40–50 nM (Fig. 4). Applying the Michaelis–Menten equation, maximum velocity (V max) was an order of magnitude higher than our results, and K m (substrate concentration that yields a half-maximal velocity) was four orders higher than the leucine concentrations used herein. Therefore, leucine millimolar concentrations were needed to obtain substrate saturation. We decided to use 40 nM of leucine in the next experiments to decrease the possibility of leucine incorporation by algae and avoid an aberrant substrate concentration. Also, the variance was not high within this concentration range. The time-course curve indicated the highest 3H-leucine incorporation after 120 min, but it was linear between 15 and 60 min (Fig. 5).
Extraction Procedure
The sonication time was the first extraction parameter tested, and we concluded that 5-min sonication was adequate (data not shown). We found no significant difference in protein extraction efficiency between the acid (TCA) extraction followed by filtration and the alkaline extraction followed by centrifugation (Table 1; n = 10, p = 0.57). However, in this situation, the filtration method showed blank values five times higher than the centrifugation method.
Acid extraction is easier to use than alkaline extraction and showed a better cost/benefit. When the centrifugation and filtration methods were compared, the former yielded blanks with almost 20 times smaller DPM values. On average, the blank/sample ratio using centrifugation was 4 % while using filtration was 17 %. Therefore, we decided to apply acid extraction with centrifugation in further experiments.
Influence of Specific Metabolic Inhibitors Under Different Light Conditions
Previous experiments showed that under laboratory conditions, 3H-leucine incorporation tended to be higher under the influence of light. However, it was statistically different under natural light but not under fluorescent light (n = 9, p = 0.02 and p = 0.16, respectively). This indicated that light quality was an important factor regulating 3H-leucine incorporation, indicating that leucine incorporation was coupled with primary production. Therefore, experiments under light/dark conditions were carried out both in situ and in the laboratory, adding specific metabolic inhibitors to verify the role of different organisms in 3H-leucine incorporation. Then, we tested two treatment conditions (light influence and inhibitors action). Meanwhile, in the field, we just evaluated the action of antibiotics and diuron, while in the laboratory, we also tested the action of sulfate-reducing bacteria (SRB) and eukaryote inhibitors.
The influence of specific inhibitors in incorporation of 3H-leucine was dependent on the incubation procedure. Incorporation was higher and with smaller variance in in situ experiments than in laboratory ones (Fig. 6). In the former (Fig. 6a), the presence of antibiotics decreased 3H-leucine incorporation in both the presence and absence of light (n = 4, p = 0.02). An 85 % reduction was observed under light and 63 % under dark conditions. On the other hand, the addition of a photosynthesis inhibitor only reduced incorporation rates significantly in the dark, on average decreasing 47 %, while a 32 % reduction was observed under light. Meanwhile, the high variance should also be considered. For instance, there was no significant difference when the same treatments were used under light or dark conditions (n = 4, p = 0.83).
The effect in addition of specific metabolic inhibitors on 3H-leucine incorporation in experiments done under light or dark in a field and b laboratory conditions. Details in the text. One asterisk statistically different under dark treatments, two asterisks statistically different under light treatments
A wide range of inhibitor types were applied in the laboratory experiments and 3H-leucine incorporation rates tended to be higher in the dark (p = 0.73), except in the presence of antibiotics and of a eukaryotic inhibitor (Fig. 6b). As expected, the presence of antibiotics significantly decreased heterotrophic activity (p = 0.02), but the other inhibitors caused no significant reduction under both light and dark conditions. Specifically in the dark, the mean 3H-leucine incorporation rates decreased 96 % with streptomycin, 86 % with penicillin, 44 % with a eukaryotic inhibitor, 19 % with an SRB inhibitor, and 13 % with a photosynthetic inhibitor. Despite the lack of statistical significance, the reduction with eukaryotic inhibitor was high and suggested a eukaryote–prokaryote coupling in Sargassum biofilm. In the presence of artificial fluorescent light, incorporation rates decreased only under antibiotics, by around 90 % with streptomycin and 58 % with penicillin.
Discussion
The association between epiphytic bacteria and seaweeds is well described [9], including species of the genus Sargassum [47]. To verify the presence of bacteria and other microorganisms in our samples, biofilm formation was followed by scanning electron microscopy. The greatest advantage of this technique is to provide a three-dimensional image of the studied surface. However, a molecular study of the microbial composition in Sargassum biofilm is also recommended.
Different groups of bacteria were observed in Sargassum biofilms in many levels of complexity (Fig. 1). The abundance and growth of microorganisms are not related in natural systems, and cell death rates as well as grazing rates should be estimated in future studies. Bacteria such as Pseudomonas, Pseudoalteromonas, and Coccus as well as diatoms such as Cocconeis, Navicula, and Nitszchia were observed on Sargassum biofilms from the studied region (Baeta-Neves, personal communication). The importance of bacteria and diatoms in the biofilm stages is well established, with a high Cocconeis abundance in the first stage [19, 86]. Murray et al. [67] observed that exudation by diatoms stimulated bacterial DNA synthesis. However, the interaction between auto- and heterotrophic microorganisms can be positive or negative for both groups [18, 56, 66].
Studies with Ulva biofilms showed the presence, colonization, and competition between microorganisms as well as the importance of these interactions in the biofouling process [78, 79]. The synergic action due to bacteria interaction also resulted in higher antibiotic resistance [10]. These studies highlighted the competitive success of Pseudoalteromonas [77]. Pseudomonas aeruginosa is also common in distinct biofilm matrixes and showed positive as well as negative interactions with other microorganisms. Nadell et al. [68] observed that the same clone of P. aeruginosa generated distinct phenotypic lineages able to adapt their behavior to local conditions and chemical gradients, and their function structured the biofilm.
Bacterioplankton composition changes over a millimeter scale [57, 84], and no distribution pattern was observed in the Sargassum biofilm with the SEM images. Biofilm structure can determine the flux of compounds and consequently the bacterial metabolic and physiological activities [68]. These characteristics increase variance and make data interpretation difficult.
Blade Incubation Procedure
We tried to adapt the 3H-leucine incorporation technique to epilithic algal matrixes (EAM), including calcareous algae such as Jania sp. and Amphiroa sp. (data not shown). EAM are important benthic primary producers in the studied area [28]. The detritus associated with EAM incorporated more 3H-leucine than the algal matrix itself, and incubations done in the dark showed a higher 3H-leucine incorporation. However, when incorporation of EAM and Sargassum sp. was expressed on a weight basis, the biofilm associated with Sargassum blades presented 20 times higher 3H-leucine incorporation rates than the detritus associated with EAM. The presence of detritus with a highly variable bacterial activity was a common feature for both Sargassum and EAM.
Bacteria may use any available surfaces in order to survive under starvation and/or nutrient deprivation conditions. These surfaces could be both detritus [11] and biofilm [77]. Marine snow can be a source of benthic bacteria and originates from many sources such as plankton, suspended sediment, and macroalgae fragments, as well as feces from fish and other organisms [25]. In general, deep water is a source of diatoms in upwelling systems [40, 94] and the high abundance of diatoms was also observed in the biofilm SEM images.
As observed by SEM images, bacterial distribution and activity onto the blade may explain the wide variation reported in the 3H-leucine incorporation rates. We observed that the activity of bacteria could be associated with both detritus and biofilm on Sargassum blades and the structural complexity of both surfaces is a source of variance. The higher variance of bacterial activity in detritus than in the biofilm could be associated with the expected higher level of particulate inorganic materials in the former (Fig. 2). The highest measure of DPMblank/DPMsample observed was when detritus was incubated, suggesting abiotic incorporation throughout the 3H-leucine particle adsorption. Furthermore, the high rates of abiotic adsorption of the radiolabeled substrate in samples with high organic matter can interfere with the measurement of bacterial production [65].
The composition of the macroalgal biofilm is determined by a range of factors including the colonization rate by free floating bacteria, community growth, and the polymers that biofilms contain [82]. EPS are not only useful for attachment, locomotion, feeding, and protection but also act as a glue to bind other materials [99]. Ras et al. [80] showed that EPS size diversity was higher in mixed heterotrophic/autotrophic biofilms than heterotrophic ones. The distribution of microorganisms is also associated with the nature of substrate and with the water flow that transports dissolved and particulate organic matter [20]. Burke et al. [9] suggested that the competitive lottery model may explain the colonization pattern on algal surfaces. It has been shown that mechanical, nutritional, and metabolic signals, as well as quorum-sensing and host-derived signals, can shift biofilm development [53]. For instance, quorum-sensing signals play an important role in the ecological interaction between bacteria and their eukaryotic host [39].
Resources as well as species diversity change seasonally and influence microorganism interspecific competition [64]. All these factors could explain the variability observed in the 3H-leucine incorporation measured herein. This expressive variability had also been observed in biofilms from other environments, which could be associated with the spatial–temporal scale of each study [13, 16, 54].
Microbial biofilms control the fluxes of compounds in seaweed surfaces, protecting them against toxins and ultraviolet radiation. On the other hand, the presence of macroepibionts changes the shape and weight of surface, competing for resources and decreasing the macroalgae growth rates [97]. Therefore, macroepibionts should inhibit both microorganisms and macroalgae growth since they compete for space and resources, respectively. It could explain the decrease in leucine incorporation that we found (Fig. 3). Bacterial activity in relation to the blade portion and position could also be associated with the presence of macroepibionts and with seaweed photosynthetic activity. Gao and Umezaki [35] reported higher photosynthetic activity in apical blades and vesicles. Differences in 3H-leucine incorporation between apical and basal blades as well as different portions of the blades were not observed (data not shown). On the other hand, vesicles were not measured, and their importance in photosynthesis increases with the age of Sargassum [34], showing an adaptation process of the macroalgae since the abundance of macroepibionts on blades also increases with the age of the latter [75].
Substrate Saturation
The potential pitfalls of methods based on radiolabel incorporation are attributed to the paucity of information on substrate use and on the metabolism of heterogeneous natural populations, particularly the lack of information regarding potential variability in colonization species and their metabolic requirements [46]. As mentioned by Dixon and Turley [23], some of the problems in techniques using 3H when applied to sediments, or detritus, are (1) the difficulty in deriving an empirical factor to convert substrate incorporation rates to bacterial cell production, as an effect of slurring the sediment to disperse added precursors; (2) the abiotic adsorption of the radiolabeled product onto sediment particles that interferes with the signal-to-noise ratio; (3) the difficulties and uncertainties in extraction of the radiolabeled product from detritus; (4) the potentially high isotopic dilution; (5) the catabolism of tritiated label (e.g., 3H2O formation); and (6) how to relate the incorporation rates to accurate rates of bacterial production. However, the advantages of the approach compensate its pitfalls, and to our knowledge, no better tool has yet been developed to measure bacterial secondary production.
In this context, we avoided empirical factors, bacterial production estimates, and addition of high substrate concentrations to reduce isotopic dilution. The isotope dilution is the proportion in which an added radiolabeled substrate is incorporated in comparison with the exogenous concentration and the substrate biosynthesis [74]. If isotope dilution is not measured, substrate incorporation can be underestimated [6, 12]. Isotope dilution can be minimized by adding high leucine concentration [30]. To avoid underestimation of bacterial production and to minimize precursor degradation, we recommend to use saturated concentrations of leucine and short incubation times in future studies and also to evaluate if saturation curves vary among sites and habitats. On the other hand, incorporation rates are dependent upon the concentration of leucine that is added. As leucine concentration is increased, incorporation also increases up to a saturating concentration. Concentrations have to be high enough to repress the “de novo” synthesis of leucine by bacteria and to “swamp” the incorporation of unlabelled external leucine that might be present in the samples. Minimal concentrations meeting these requirements should be used. At higher concentrations, there could be some incorporation inhibition or incorporation by organisms other than bacteria or even adsorption to particles [37, 48]. Therefore, we decided to use unsaturated concentrations of leucine since it was more important to avoid Sargassum incorporation.
Usually, leucine incorporation rates are higher in eutrophic than oligotrophic systems [73]. The supply of organic carbon may determine how much leucine is incorporated directly into protein and how much is degraded to other amino acids [52]. In marine bacterioplankton, saturation of leucine is typically around 10–20 nM, and in the Cabo Frio upwelling system, it was 10 nM [14]. The concentration used to estimate bacterioplankton production in ultraoligotrophic systems is 40–50 nM [55], and using these concentrations, we observed a low associated error in our measurements. Tornblom and Sondegaard [91] reported that leucine saturation was 400 nM when 1 cm of Zostera marina leaf was incubated, and higher bacterial activity was observed in older leafs.
It is important to consider that algae could also be taking up leucine and turning the label over as dissolved organic matter, but Sargassum incorporation is more probable if saturated concentrations were applied (Fig. 4). Fleurence [31] observed that leucine represented 4–9 % of the proteins found in phaeophytes and that protein content corresponded to 3–15 % of their dry weight. In the Cabo Frio region, protein content made up to 16 % of Sargassum vulgare dry weight [61], and leucine represented 9 % of all protein content [60]. Leucine assimilation by Sargassum can be a technical artifact that needs to be evaluated in future studies.
Linearity of 3H-Leucine Incorporation
As found by Findlay et al. [29], we observed constant leucine incorporation and no leucine degradation in incubations of up to 60 min (Fig. 5). Also, higher incubation periods could increase leucine degradation and tritium respiration. The nonlinear incorporation would indicate a change in bacterial composition [26] and in its metabolism. On the other hand, short incubations may not stimulate all bacterial assemblages [7]. In addition, a longer period is recommended to observe the effect of metabolic inhibitors. It is difficult to do short field incubations [65].
Environmental factors such as temperature and the location of sampling stations are also important to determine the time of incubation. Tornblom and Sondegaard [91] used distinct incubations times for Z. marina leafs, depending on local temperature, but always between 30 and 60 min. The abundance and composition of bacterial communities can change spatially [97], and we observed the highest rates in Sargassum sampled in the main upwelling-influenced area of the studied sites in the Cabo Frio embayment (data not shown). Unfortunately, it was not possible to measure temperature during sampling, but temperature was the same during the incubations for all compared treatments.
Extraction Efficiency
We observed equivalent results using both acid and alkaline extraction methods but with high variability between replicates. Tornblom and Sondegaard [91] used TCA, sonication, and filtration as an efficient extraction method with low associated error in leafs of Z. marina. In Sargassum blades, we observed that the microcentrifuge procedure was more time-consuming than the filtration procedure but decreased the associated errors and was much more economic. In our study, sonication was more efficient than the vortex procedure, and it increased the dispersion of organic and inorganic aggregates with bacteria [76]. Also, the acid extraction, by stopping the reaction with 5 % TCA, decreases the consumption of chemical compounds. The use of 20 % acetic acid did not change the extraction efficiency in blades of Sargassum, but it may be important in other habitats such as turf algae since carbonates, silt, and sand facilitate adsorption of tritium and amino acids. In this case, it is better to incubate slurries of detritus than the algal matrix [65].
Suggested Technical Protocol
Our protocol is an adaptation of the protocol suggested by Miranda et al. [65] for freshwater periphyton with some modifications. Following Fig. 7, we recommend the incubation of 1 cm2 of the intact blade in 1.5 mL of 0.22-μm filtered local water in microcentrifuge tubes (2.0 mL), with the addition of 40–50 nM 3H-leucine for 1-h incubations. Addition of TCA to a 5 % final concentration stops the reaction and precipitates proteins. If not extracted right away, samples can be frozen for up to 15 days without noticeable leucine degradation by radiolysis. After thawing, the protein extraction procedure follows centrifugation purification until the addition of 1-mL scintillation cocktail. Counting samples the next day reduces chemiluminescence and ensures better mixing between pellet and cocktail. Complete chemiluminescence self-extinction takes up to 4 days, during which sample and blank DPM values change proportionally, leaving the calculated incorporation rates unaffected.
Light Quality During Sample Incubation
The quality of light (natural × fluorescent lamps) and the incubation conditions influenced 3H-leucine incorporation in the studied matrix. Usually, in situ experiments do not yield the same results as laboratory ones [15, 35, 36], and our results showed higher 3H-leucine incorporation in the field incubations. Light quality (natural × fluorescent) includes distinctions in wavelength and ultraviolet radiation. Häder et al. [43] reported the effect of ultraviolet light in bacteria associated with biofilms of Z. marina leaves, and Törnblom and Sondergaard [91] observed that there was an association between auto- and heterotrophic processes. In phototrophic biofilms, dissolved organic nitrogen, as the amino acid leucine, can also be incorporated by phototrophic organisms [32, 81]. Considering that Sargassum biofilm is hetero- and phototrophic, as observed in SEM images, the influence of light on our results is probably associated with the coupling between auto- and heterotrophic processes. Light quality and sample manipulation also affect the functionality of biofilm defense mechanisms [10].
Specific Metabolic Inhibitors
The use of a photosynthesis inhibitor decreased 3H-leucine incorporation more intensely in experiments done in situ, more so under dark conditions. Diuron acts on photosystem II blocking the transport of electrons through the photosynthesis chain reaction [67, 96] and also reduces bacterial survival [58].
Biofilms are known for their ability to decrease antibiotic action, and such are a challenge in medical and deontological areas [49]. This is associated with the biofilm width and with chemical characteristics of extracellular polymeric matrixes [20, 22]. Wunder et al. [100] concluded that antibiotic speciation and molecular size are important factors affecting the interactions between antibiotics and biofilms. It may explain in part the variable inhibition by antibiotics used in field and laboratory incubations described herein (Fig. 6). Streptomycin caused a higher inhibition of 3H-leucine incorporation than penicillin. Following Jawetz et al. [45], streptomycin inhibits bacterial protein synthesis and acts mainly in mycobacterium with a lipid-rich cellular wall. On the other hand, penicillin acts mainly on Gram-positive bacteria since it inhibits cell wall synthesis and induces bacterial lysis. However, the permeability of cell walls differs between bacteria since protein composition varies in type and concentration. Chloramphenicol is a synthetic bacteriostatic with a wide action that directly inhibits protein synthesis. Chloramphenicol-dependent inhibition of bacterial protein biosynthesis is mainly due to prevention of peptide chain elongation [83]. Nair et al. [69] observed a better efficiency of chloramphenicol on marine bacteria than streptomycin. On the other hand, penicillin was more efficient against pigmented bacteria while chloramphenicol affected nonpigmented bacteria.
Sulfate-reducing bacteria are important in the sulfur cycle in most marine environments, but the presence of sodium molybdate did not change 3H-leucine incorporation rates in incubations done in the laboratory. Molybdate enters cells via a sulfate transport system and interferes with the formation of adenosine phosphosulfate, leading to deprivation of reduced sulfur compounds for growth, forming adenosine phosphomolybdate in the cell [71].
Sharma et al. [85] observed that fungi were more active in older Sargassum-associated detritus and that detritus protein content increased while the concentration of amino acid decreased. In our experiments, the addition of cycloheximide (eukaryotic inhibitor) decreased mean 3H-leucine incorporation by almost 50 %, suggesting a eukaryote–prokaryote coupling. Cycloheximide is a protein synthesis inhibitor which blocks the translocation step in elongation [5]. Besides fungus, diatoms and Sargassum are eukaryotes that should have a strict association with heterotrophic bacteria.
References
Almeida A, Cunha A, Fernandes S, Sobral P, Alcantara F (2007) Copper effects on bacterial activity of estuarine silty sediments. Estuar Coast Shelf Sci 73:743–752
Ashen JB, Goff LJ (2000) Molecular and ecological evidence for species specificity and coevolution in a group of marine algal-bacterial symbioses. Appl Environ Microbiol 66:3024–3030
Atlas RM, Bartha R (1993) Microbial ecology—fundamentals and applications. Benjamin/Cummings Publishing Company, Redwood
Baath E, Pettersson M, Soderberg KH (2001) Adaptation of a rapid and economical microcentrifugation method to measure thymidine and leucine incorporation by soil bacteria. Soil Biol Biochem 33:1571–1574
Baliga BS, Pronczuk AW, Munro HN (1969) Mechanism of cycloheximide inhibition of protein synthesis in a cell-free system prepared from rat liver. J Biol Chem 244:4480–4489
Bell RT (1986) Further investigation of the isotope dilution approach for estimating the degree of participation of [3H]thymidine in DNA synthesis in studies of aquatic bacterial production. Appl Environ Microbiol 52:1212–1214
Buesing N, Gessner MO (2003) Incorporation of radiolabeled leucine into protein to estimate bacterial production in plant litter, sediment, epiphytic biofilms, and water samples. Microb Ecol 45:291–301
Buesing N, Marxsen J (2005) Theoretical and empirical conversion factors for determining bacterial production in freshwater sediments via leucine incorporation. Limnol Oceanogr-Met 3:101–107
Burke C, Thomas T, Lewis M, Steinberg P, Kjelleberg S (2011) Composition, uniqueness and variability of the green algae Ulva australis. ISME J 5:590–600
Burmolle M, Webb JS, Rao D, Hansen LH, Sorensen SJ, Kjelleberg S (2006) Enhanced biofilm formation and increased resistance to antimicrobial agents and bacterial invasion are caused by synergistic interactions in multispecies biofilms. Appl Environ Microbiol 72:3916–3923
Cho BC, Azam F (1990) Biogeochemical significance of bacterial biomass in the oceans euphotic zone. Mar Ecol Prog Ser 63:253–259
Chrzanowski TH (1988) Consequences of accounting for isotopic dilution in thymidine incorporation assays. Appl Environ Microbiol 54:1868–1870
Christofoletti R, Almeida TVV, Ciotti AM (2011) Environmental and grazing influence on spatial variability of intertidal biofilm on subtropical rocky shores. Mar Ecol Prog Ser 424:15–23
Coelho-Souza SA (2009) Atividade de bactérias planctônicas e epifíticas do sistema de ressurgência do Cabo Frio, RJ. PhD thesis, UFRJ, 166p
Coelho-Souza SA, Guimaraes JRD, Mauro JBN, Miranda MR, Azevedo S (2006) Mercury methylation and bacterial activity associated to tropical phytoplankton. Sci Tot Environ 364:188–199
Coelho-Souza SA, Guimaraes JRD, Miranda MR, Poirier H, Mauro JBN, Lucotte M, Mergler D (2011) Mercury and flooding cycles in the Tapajós river basin: the role of periphyton of a floating macrophyte (Paspalum repens). Sci Tot Environ 409:2746–2753
Coelho-Souza SA, Lopez MS, Guimaraes JRD, Coutinho R, Candella R (2012) Biophysical interactions in the Cabo Frio upwelling system, Southeastern Brazil. Braz J Ocanogr. in press
Cole JJ (1982) Interactions between bacteria and algae in aquatic ecosystems. An Rev Ecol Syst 13:291–314
Cooksey KE, Cooksey B (1995) Adhesion of bacteria and diatoms to surfaces in the sea—a review. Aquat Microb Ecol 9:87–96
Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappinscott HM (1995) Microbial biofilms. An Rev Microbiol 49:711–745
Cury JC, Araujo FA, Coelho-Souza SA, Peixoto RS, Oliveira JAL, Santos HF, Davila AMR, Rosado AS (2011) Microbial diversity of a Brazilian coastal region influenced by an upwelling system and anthropogenic activity. PLoS One 6:e16553. doi:10.1371/journal.pone.0016553
Davey ME, O’Toole GA (2000) Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev 64:847–867
Dixon JL, Turley CM (2001) Measuring bacterial production in deep-sea sediments using 3H-thymidine incorporation: ecological significance. Microb Ecol 42:549–561
Eardly DF, Carton MW, Gallagher JM, Patching JW (2001) Bacterial abundance and activity in deep-sea sediments from the eastern North Atlantic. Prog Oceanogr 50:245–259
Fenchel T (1988) Marine plankton food chains. An Rev Ecol Syst 19:19–38
Ferguson RLE, Buckley EN, Palumbo AV (1984) Response of marine bacterioplankton to differential filtration and confinement. Appl Environ Microbiol 47:49–55
Ferreira CEL, Goncalves JEA, Coutinho R (2001) Community structure of fishes and habitat complexity on a tropical rocky shore. Environ Biol Fishes 61:353–369
Ferreira CEL, Goncalves JEA, Coutinho R, Peret AC (1998) Herbivory by the dusky damselfish Stegastes fuscus (Cuvier, 1830) in a tropical rocky shore: effects on the benthic community. J Exp Mar Biol Ecol 229:241–264
Findlay SG, Meyer JL, Edwards RT (1984) Measuring bacterial production via rate of incorporation of [3H]thymidine into DNA. J Microbiol Met 2:57–72
Fischer H, Pusch M (1999) Use of the [14C]leucine incorporation technique to measure bacterial production in river sediments and the epiphyton. Appl Environ Microbiol 65:4411–4418
Fleurence J (1999) Seaweed proteins: biochemical, nutritional aspects and potential uses. Trends Food Sci Technol 10:25–28
Flores E, Herrero A (2005) Nitrogen assimilation and nitrogen control in cyanobacteria. Biochem Soc Trans 33(Pt 1):164–167
Furtado ALD, Casper P (2000) Different methods for extracting bacteria from freshwater sediment and a simple method to measure bacterial production in sediment samples. J Microbiol Met 41:249–257
Gao K (1990) Seasonal variation of photosynthetic capacity in Sargassum horneri. Japan J Phycol 38:25–33
Gao K, Umezaki I (1989) Comparative studies of photosynthesis in different parts of Sargassum thunbergii. Jap J Phycol 37:7–16
Gao K, Umezaki I (1989) Studies on diurnal photosynthetic performance of Sargassum thunbergii. I. Changes in photosyntesis under natural sunlight. Jap J Phycol 37:89–98
Gasol JM, Doval MD, Pinhassi J, Calderón-Paz JI, Guixa-Boixareu N, Vaqué D, Pedrós-Alió C (1998) Diel variations in bacterial heterotrophic production in the Nortwestern Mediterranean Sea. Mar Ecol Prog Ser 164:125–133
Godoy EAS, Coutinho R (2002) Can artificial beds of plastic mimics compensate for seasonal absence of natural beds of Sargassum furcatum? ICES J Mar Sci 59:S111–S115
Golberg K, Eltzov E, Schnit-Orland M, Marks RS, Kushmaro A (2011) Characterization of quorum sensing signals in coral-associated bacteria. Microb Ecol 61:783–792
Guenther M, Gonzalez-Rodriguez E, Carvalho WF, Rezende CE, Mugrabe G, Valentin JL (2008) Plankton trophic structure and particulate organic carbon production during a coastal downwelling-upwelling cycle. Mar Ecol Prog Ser 363:109–119
Guimaraens MA, Goncalves JEA, Lourenco SO, Coutinho R (2008) Sensitivity analyses of population biomass dynamics for Ulva spp. and Sargassum furcatum at the Cabo Frio upwelling region of Brazil. J Biol Syst 16:579–596
Guimaraes JRD, Mauro JBN, Meili M, Sundbom M, Haglund AL, Coelho-Souza SA, Hylander LD (2006) Simultaneous radioassays of bacterial production and mercury methylation in the periphyton of a tropical and a temperate wetland. J Environ Manag 81:95–100
Hader DP, Kumar HD, Smith RC, Worrest RC (1998) Effects on aquatic ecosystems. J Photochem Photobiol 46:53–68
Hietanen S, Tuominen L, Kuparinen J (1999) Benthic bacterial production in the northern Baltic Sea measured using a modified C-14 leucine incorporation method. Aquat Microb Ecol 20:13–20
Jawetz E, Melnick JL, Adelberg EA (1984) Microbiologia Médica. Lange Medical Publications, Rio de Janeiro/San Diego
Jellett JF, Li WKW, Dickie PM, Boraie A, Kepkay PE (1996) Metabolic activity of bacterioplankton communities assessed by flow cytometry and single carbon substrate utilization. Mar Ecol Prog Ser 136:213–225
Kanagasabhapathy M, Sasaki H, Haldar S, Yamasaki S, Nagata S (2006) Antibacterial activities of marine epibiotic bacteria isolated from brown algae of Japan. An Microbiol 56:167–173
Keil RG, Kirchman DL (1992) Bacterial hydrolysis of protein and methylated protein and its implications for studies of protein-degradation in aquatic systems. App Environ Microbiol 58:1374–1375
Kierek-Pearscon K, Karatan E (2005) Biofilm development in bacteria. Adv Appl Microbiol 57:79–111
King GM, Garey MA (1999) Ferric iron reduction by bacteria associated with the roots of freshwater and marine macrophytes. App Environ Microbiol 65:4393–4398
Kirchman D (2001) Measuring bacterial biomass production and growth rates from leucine incorporation in natural aquatic environments. Met Microbiol 30:227–237
Kirchman D, Knees E, Hodson R (1985) Leucine incorporation and its potential as a measure of protein synthesis by bacteria in natural aquatic systems. Appl Environ Microbiol 49:599–607
Koczan JM, Lenneman BR, McGrath MJ, Sundin GW (2011) Cell surface attachment structures contribute to biofilm formation and xylem colonization by Erwinia amylovora. Appl Environ Microbiol 77:7031–7039
Lachnit T, Blumel M, Imhoff JF, Wahl M (2009) Specific epibacterial communities on macroalgae: phylogeny matters more than habitat. Aquat Biol 5:181–186
Levipan HA, Quinones RA, Urrutia H (2007) A time series of prokaryote secondary production in the oxygen minimum zone of the Humboldt current system, off central Chile. Prog Oceanogr 75:531–549
Long RA, Azam F (2001) Antagonistic interactions among marine pelagic bacteria. App Environ Microbiol 67:4975–4983
Long RA, Azam F (2001) Microscale patchiness of bacterioplankton assemblage richness in seawater. Aquat Microb Ecol 26:103–113
López-Doval JC, Ricart M, Guasch H, Romani AM, Sabater S, Munoz I (2010) Does grazing pressure modify diuron toxicity in a biofilm community? Arch Environ Contaminat Toxicol 58:955–962
López MS, Coutinho R (2010) Positive interaction between the native macroalgae Sargassum sp. and the exotic bivalve Isognomon bicolor? Braz J Oceanogr 58:69–72
Lourenço SO, Barbarino E, De-Paula JC, Pereira LOS, Marquez UML (2002) Amino acid composition, protein content and calculation of nitrogen-to-protein conversion factors for 19 tropical seaweeds. Physiol Res 50:233–241
Marinho-Soriano E, Fonseca PC, Carneiro MAA, Moreira WSC (2006) Seasonal variation in the chemical composition of two tropical seaweeds. Biores Technol 97:2402–2406
Marxsen J (1996) Measurement of bacterial production in stream-bed sediments via leucine incorporation. FEMS Microbiol Ecol 21:313–325
Matsuo Y, Suzuki M, Kasai H, Shizuri Y, Harayama S (2003) Isolation and phylogenetic characterization of bacteria capable of inducing differentiation in the green alga Monostroma oxyspermum. Environ Microbiol 5:25–35
McArthur JV (2006) Microbial ecology—an evolutionary approach. Elsevier, San Diego
Miranda MR, Guimaraes JRD, Coelho-Souza SA (2007) [3H]Leucine incorporation method as a tool to measure secondary production by periphytic bacteria associated to the roots of floating aquatic macrophyte. J Microbiol Met 71:23–31
Mouget JL, Dakhama A, Lavoie MC, Delanoue J (1995) Algal growth enhancement by bacteria—is consumption of photosynthetic oxygen involved? FEMS Microb Ecol 18:35–43
Murray RE, Cooksey KE, Priscu JC (1986) Stimulation of bacterial-DNA synthesis by algal exudates in attached algal-bacterial consortia. App Environ Microbiol 52:1177–1182
Nadell CD, Xavier JB, Foster KR (2009) The sociobiology of biofilms. FEMS Microbiol Rev 33:206–224
Nair S, Chandramohan D, Bharathi PAL (1992) Differential sensitivity of pigmented and non-pigmented marine bacteria to metals and antibiotics. Wat Res 26:431–434
Olson JB, Kellogg CA (2010) Microbial ecology of corals, sponges, and algae in mesophotic coral environments. FEMS Microbiol Ecol 73:17–30
Patidar SK, Vinod T (2005) Effect of molybdate on methanogenic and sulfidogenic activity of biomass. Bioresour Technol 96:1215–1222
Pérez MT, Hortnagl P, Sommaruga R (2010) Contrasting ability to take up leucine and thymidine among freshwater bacterial groups: implications for bacterial production measurements. Environ Microbiol 12:74–82
Petit M, Alves GP, Lavandier P (1999) Phytoplanktonic exudation, bacterial reassimilation and production for three diel cycles in different trophic conditions. Archiv Fur Hydrobiol 146:285–309
Pollard PC, Moriarty DJW (1984) Validity of the tritiated thymidine method for estimating bacterial growth rates: measurement of isotope dilution during DNA synthesis. Appl Environ Microbiol 48:1076–1083
Ramalho L (2001) Distribuição espaço-temporal da epifauna séssil de Sargassum furcatum KUETZING (Pheophyta: Fucales) e sua mímica, na Ilha de Cabo Frio, Arraial do Cabo, RJ. phD thesis, UFPR, 92 p
Ramsay AJ (1984) Extraction of bacteria from soil: efficiency of shaking or ultrasonication as indicated by direct counts and autoradiography. Soil Biol Biochem 16:475–481
Rao D, Webb JS, Kjelleberg S (2005) Competitive interactions in mixed-species biofilms containing the marine bacterium Pseudoalteromonas tunicata. Appl Environ Microbiol 71:1729–1736
Rao D, Webb JS, Kjelleberg S (2006) Microbial colonization and competition on the marine alga Ulva australis. Appl Environ Microbiol 72:5547–5555
Rao D, Webb JS, Holmstrom C, Case R, Low A, Steinberg P, Kjelleberg S (2007) Low densities of epiphytic bacteria from the marine alga Ulva australis inhibit settlement of fouling organisms. Appl Environ Microbiol 73:7844–7852
Ras M, Lefebure D, Derlon N, Paul E, Girbal-Neuhauser E (2011) Extracellular polymeric substances diversity of biofilms grown under contrasted environmental conditions. Wat Res 45:1529–1538
Roeselers G, van Loosdrecht MCM, Muyzer G (2008) Phototrophic biofilms and their potential applications. J App Phycol 20:227–235
Samuelsson MO, Kirchman DL (1990) Degradation of adsorbed protein by attached bacteria in relationship to surface hydrophobicity. App Environ Microbiol 56:3643–3648
Schlünzen F, Zarivach R, Harms J, Bashan A, Tocilj A, Albrecht R, Yonath A, Franceschi F (2001) Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature 413:814–821
Seymour JR, Mitchell JG, Pearson L, Waters RL (2000) Heterogeneity in bacterioplankton abundance from 4.5 millimetre resolution sampling. Aquat Micrb Ecol 22:143–153
Sharma S, Raghukumar C, Raghukumar S, Sathepathak V, Chandramohan D (1994) Thraustochytrid and fungal component of marine detritus. II. Laboratory studies on decomposition of the brown alga Sargassum cinereum J. Ag. J Exp Mar Biol Ecol 175:227–242
Sieburth JM, Tootle JL (1981) Seasonality of microbial fouling on Ascophyllum nodosum (L.) Lejosl., Fucus vesiculosus L., Polysiphonia lanosa (L.), and Chondrus crispus Stackh. J Phycol 17:57–64
Smith DC, Azam F (1992) A simple, economical method for measuring bacterial protein synthesis rates in seawater using 3H-leucine. Mar Microb Food Webs 6:107–114
Smith JE, Shaw M, Edwards RA, Obura D, Pantos O, Sala E et al (2006) Indirect effects of algae on coral: algae-mediated, microbe-induced coral mortality. Ecol Lett 9:835–845
Staufenberger T, Thiel V, Wiese J, Imhoff JF (2008) Phylogenetic analysis of bacteria associated with Laminaria saccharina. FEMS Microb Ecol 64:65–77
Tibbles BJ, Davis CL, Harris JM, Lucas MI (1992) Estimates of bacterial productivity in marine-sediments and water from a temperate salt-marsh lagoon. Microb Ecol 23:195–209
Tornblom E, Sondergaard M (1999) Seasonal dynamics of bacterial biomass and production on eelgrass Zostera marina leaves. Mar Ecol Prog Ser 179:231–240
Tujula NA, Crocetti GR, Burke C, Thomas T, Holmstrom C, Kjelleberg S (2010) Variability and abundance of the epiphytic bacterial community associated with a green marine Ulvacean alga. ISME J 4:301–311
Underwood AJ (1997) Experiments in ecology. Cambridge University Press, Cambridge, p 504
Valentin JL (2001) The Cabo Frio upwelling system, Brazil. In: Seeliger U, Kjerfve B (eds) Coastal marine ecosystems of Latin America. Springer, Berlin, pp 97–105
Van Wambeke F, Christaki U, Bianchi M, Psarra S, Tselepides A (2000) Heterotrophic bacterial production in the Cretan Sea (NE Mediterranean). Prog Oceanogr 46:205–216
Velthuys BR (1981) Electron-dependent competition between plastoquinone and inhibitors for binding to photosystem II. FEBS Let 126:277–281
Wahl M (2008) Ecological level and interface ecology: epibiosis modulates the interactions between host and environment. Biofouling 24:427–438
Williams CJ, Boyer JN, Jochem FJ (2009) Microbial activity and carbon, nitrogen, and phosphorus content in a subtropical seagrass estuary (Florida Bay): evidence for limited bacterial use of seagrass production. Mar Biol 156:341–353
Wotton RS (2011) EPS (Extracellular Polymeric Substances), silk, and chitin: vitally important exudates in aquatic ecosystems. JN Am Benthol Soc 30:762–769
Wunder DB, Bosscher VA, Cok RC, Hozalski RM (2011) Sorption of antibiotics to biofilm. Wat Res 45:2270–2280
Zar JH (1999) Biostatistical analysis. Prentice-Hall, New Jersey, p 663
Acknowledgments
We thank the IEAPM logistic support, especially to the Grupo de Quimica, the Department of Oceanography, Division of Biotechnology Lab team, and LEG 10 team. This study would not have been concluded without the help of Maria Helena Baeta-Neves, Antônio Casarin, Dagles Viana dos Reis, William Romão, and Carlos Eduardo Leite Ferreira. We also thank Dr. Stuart Jenkins and the anonymous referees who contributed to the final format of this manuscript. This work was supported by CAPES, CNPq, and FAPERJ.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Coelho-Souza, S.A., Miranda, M.R., Salgado, L.T. et al. Adaptation of the 3H-Leucine Incorporation Technique to Measure Heterotrophic Activity Associated with Biofilm on the Blades of the Seaweed Sargassum spp. . Microb Ecol 65, 424–436 (2013). https://doi.org/10.1007/s00248-012-0116-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-012-0116-9