Abstract
Sulfate- and sulfite-reducing prokaryotes (SSRP) communities play a key role in both sulfur and carbon cycles. In estuarine ecosystems, sulfate concentrations change with tides and could be limited in tidal freshwater reach or deep sediments. In a subtropical estuary of northern Taiwan in December 2007, we examined the compositional changes of SSRP communities. We examined three sites: from the lower estuarine brackish-water reach (site GR and mangrove vegetation site, GM) to the upper estuarine tidal freshwater reach (site HR), as well as from surface to a 50-cm depth. The partial sequence of sulfite reductase (dsrB) genes was used as a molecular marker of SSRP, linked to polymerase chain reaction and denaturing gradient gel electrophoresis (DGGE) techniques. SSRP communities of the DGGE profiles varied with sites according to one-way analyses of similarities (Global R = 0.69, P = 0.001). Using cluster analysis, the DGGE profile was found to show site-specific clusters and a distinct depth zonation (five, six, and two SSRP communities at the GM, GR, and HR sites, respectively). SSRP composition was highly correlated to the combination of salinity, reduced sulfur, and total organic carbon contents (BIO-ENV analysis, r s = 0.56). After analyzing a total of 35 dsrB sequences in the DGGE gel, six groups with 15 phylotypes were found, which were closely related to marine-freshwater gradient. Moreover, sequences neighboring sulfite-reducing prokaryotes were observed, in addition to those affiliated to sulfate-reducing prokaryotes. Four phylotypes harvested in HR resembled the genus Desulfitobacterium, a sulfite-reducing prokaryote, which failed to use sulfate as an electron acceptor and were active in freshwater and sulfate-limited habitat. The other five phylotypes in the HR reach belonged to the sulfate-reducing prokaryotes of the genera Desulfatiferula, Desulfosarcina, Desulfovibrio, and Desulfotomaculum, which appeared to tolerate low salinity and low sulfate supply. SSRP phylotypes at the mangrove-vegetated GM site (five phylotypes in two groups) were phylogenetically less diverse, when compared with those at the non-mangrove-vegetated GR site (three phylotypes in three groups) and the tidally influenced freshwater HR site (nine phylotypes in five groups). Phylotypes found at GR and GM were all affiliated to marine sulfate-reducing prokaryote strains of the genera Desulfofaba, Desulfobotulus, Desulfatiferula, Desulfosarcina, and Desulfotomaculum. Notably, a phylotype recorded in the surface sediment at GR resembled the genus Desulfobulbus, which was recorded from freshwater environment consisting of the freshwater input at GR during ebb tides.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The dissimilatory reduction of sulfate is a key process in global sulfur cycle and organic matter mineralization in marine and freshwater ecosystems [4, 26, 43]. Sulfate reduction is mainly mediated by a phylogenetically and physiologically diverse group of microorganisms, called sulfate-reducing prokaryotes (SRP), which use sulfate as terminal electron acceptor in oxidizing various types of carbon sources [55]. SRP are a group of obligate anaerobic prokaryotes, taxonomically belonging to five bacterial and two archaeal phyla [5, 18]. Besides the extremely anoxic environments, SRP are also discovered in various environments exposed to temporarily oxic condition including biofilms [53], microbial mats [35], and wastewater treatment systems [33].
Estuarine surface sediment exhibits temporarily oxic, hypoxic, or anoxic conditions with tidal inundation and in which sulfate supplied from the sea will be diluted by freshwater from upstream [42, 66]. Organic matter contributed from human activities besides the river and riparian vegetation also influences the carbon supply to the estuary [28, 66]. Several studies, however, demonstrated that at least certain SRP tolerate a high concentration of oxygen while some SRP, possessing metabolic versatility, can survive in places with a shortage of sulfate [33, 35, 55]. Understanding the composition of SRP communities and which abiotic factors influence their distribution is crucial as natural functions of degraded estuaries need to be restored. Limited data are available on SRP distributional patterns in estuarine wetlands where sulfate concentrations and sediment oxygenation vary constantly with tidal rhythm [48] and with increasing anthropogenic pollution threats.
During the last decades, by using 16S rRNA or functional genes as molecular markers, many new SRP taxa have been recorded [9, 20, 52]. Highly conserved functional genes, such as dsrAB and aprBA, were also applied for SRP, sulfite-reducing prokaryotes (SiRP), or sulfide-oxidizing prokaryotes (SOP) [25, 37, 46, 65]. Dissimilatory sulfite reductase, encoded by dsrAB gene, is a key enzyme which catalyzes the conversion of sulfite to sulfide [37, 65]. Its corresponding sequence of dsrAB genes provides a basis for culture-independent molecular diversity studies of natural assemblages with sulfite-reducing ability, mainly possessed by SRP and SiRP, while using polymerase chain reaction (PCR) primers broadly specific for a large fragment of all known dsrAB genes [16, 36, 37, 49, 65]. In addition, the phylogeny of dsrAB has shown that SOP having oxidative-operating sulfite reductase are monophyletic and distantly related to the SRP or SiRP which use a reductive type of enzyme [46].
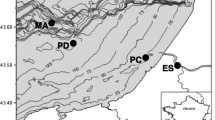
Sulfate and organic matter are essential compartments for dissimilatory SRP growth, and therefore, are important factors controlling their distribution [55]. When the sulfate concentration is low, such as in freshwater environments, SRP is also capable of fermentation and anaerobic oxidation of organic compounds [39, 51]. In the Danshuei River estuary, northern Taiwan (Fig. 1), sea water provides high levels of sulfate to the lower estuary. Toward its upstream reaches, sulfate levels gradually decrease and eventually reach almost zero in tidally influenced freshwater area [29, 44]. Nevertheless, compared with that in the lower estuary, the sulfate-reducing rate in the upper freshwater area was markedly 200-fold higher [11]. Apparently, some unique SRP may be thriving in the sulfate-depleted upper reach. Decaying vegetative debris is a source of organic matter in the mangrove-vegetated habitat in the lower part of this estuary [27, 28]. Previous studies have also demonstrated the importance of redox conditions in affecting the distribution of SRP assemblages [7]. In addition to the supply of organic matter, mangrove is known to influence sediment redox condition due to oxygenation activity surrounding their root tips [41]. Moreover, pore-water sulfate level decreased with depth in the sediment of the lower reach estuary of the Danshuei River [21], while sediment redox recorded in intertidal mud flats also decreased as depth increased [56]. In the present study, we hypothesized that in the Danshuei River estuary, northern Taiwan, (1) SRP communities and gene types differ between tidal freshwater site and lower estuarine sites; (2) SRP communities and gene types differ between mangrove-vegetated and non-vegetated habitats in the lower estuarine area; and (3) their distributions exhibit depth zonation in sediments. We also examined the presence of other dsrAB gene evolving prokaryotes, i.e., SiRP and SOP, in the estuarine system. In this study, molecular fingerprints based on dsrB-targeted denaturing gradient gel electrophoresis (DGGE) were used as a proxy to describe the diversity of SRP. In addition, environmental factors were also measured simultaneously. Finally, we discussed the possible factors controlling the presence of SRP in an estuary along the gradient from brackish-water to freshwater.
Materials and Methods
Environmental Samples
Spatial distributions of SRP were studied, and abiotic factors in the ambient were measured simultaneously by taking pair-sets of sediment cores from three sites in the Danshuei River estuary, northern Taiwan (Fig. 1). In the riverside, along a decreasing salinity and sulfate gradient from the lower toward upper estuary, two sites were selected, one in the lower estuarine brackish-water locality Guandu, GR, with salinity ranging 5.0–22.0 [29] and the other in the upper tidally influenced freshwater locality Hwajiang, HR, with nearly no salinity. The biogeochemical characteristics revealed that reduced sulfur compound in Guandu sediment were two- to fourfold higher than those in Hwajiang [21]. Inversely, sulfate-reducing rates were 200-fold lower (0.0654 M day-1 in Guandu vs. 13.1 M day-1 in Hwajiang, 11). The third site was located also in the lower estuarine Guandu reach but within a mangrove-vegetated area, GM. This vegetation site was chosen in determining whether the existence of vegetation alters the microbial communities.
Each of the three pair-sets of the sediment samples was collected using PVC corer of 10 cm in diameter. During the low tide in December 2007, corers were pushed down approximately 1 m, sealed immediately after collection, and brought to the laboratory within an hour. Sediment cores were cut every 10 up to 50 cm in depth to collect six 1–2-cm thick subsamples. Subsamples for abiotic measurements were done in an argon-filled anti-oxidant bag to prevent the reaction of sulfur compounds with oxygen. These subsamples were then sealed in 50-ml centrifuge test tubes and refrigerated at 4°C until further treatment. Subsamples for microbial community analysis were sealed in 1.5-ml microtubes and stored at −80°C until the environmental DNA were extracted within 2 days.
Measurements of Abiotic Factors
Sediment samples were centrifuged at 4,000 rpm for 15 min. The supernatant was filtered through a 0.45-μm Nuclepore filter to obtain pore-water. Salinity was determined using a refractometer (S/Mill-E, ATAGO, Tokyo, Japan). Sulfate was collected by precipitating dissolved sulfate with barium chloride (BaCl2) in a solution with pH lower than 2. Sulfide was measured using the methylene blue method [15]. Total organic carbon (TOC) content was determined with a CHN analyzer (Perkin Elmer EA 2400 Series II; Wellesley, MA) after sediment samples had been freeze-dried and acid-fumed. Reduced sulfur (acid-volatile sulfur (AVS) and pyrite-S combined) was extracted and determined using diffusion method [30].
Extraction and Purification of DNA
Approximately 1.0 g wet weight of sediment was taken for DNA extraction and purification using the UltraClean Soil DNA kit (MoBio, Solana Beach, CA) according to the manufacturer's instructions. Two microliters of nucleic acid extracts were mixed with 5× bromophenol blue loading buffer (50% [v/v] glycerol, 50 mM Tris/HCl pH 7.5, 5 mM EDTA, 0.05% [w/v] bromophenol blue), electrophoresed on 1% [w/v] agarose (Invitrogen, Carlsbad, CA, USA) containing 0.5 μg ml−1 ethidium bromide (Sigma, Saint Louis, MO), with 1.0× Tris–borate EDTA buffer [57]. Ethidium bromide stained bands were digitally recorded with an ImageQuant 300 Imager system (Amersham Bioscience, Buckinghamshire, UK). Quantification of the extracted DNA was performed by comparing to 100 bp DNA Ladder quantification standard (Geneaid, Taipei County, Taiwan). The analysis of the digital images was carried out using 1D gel analysis in the software package of ImageQuant TL (Amersham Bioscience, Buckinghamshire, UK).
PCR Amplification of Partial dsrB Gene
PCR amplification of ~350-bps dsrB fragment was performed using the primers DSRp2060F and DSR4R [24, 65, 69], to which a 40-bps GC-clamp [50] was added at the 5’-end of the forward primer. Positive controls, as pretested sample from hypoxic sediment, and negative controls with blank were always included in PCR amplification experiments. The reaction was carried in a PxE Thermal Cycler (Thermo Electron Corp., Milford, MA). The 50-μl reaction mixture contained 100 ng of soil DNA, 1× Optimized DyNAzymeTM EXT Buffer, 0.5-μM concentrations of each primer, 200-μM concentrations of each deoxynucleotide, and 1 U of DyNAzyme EXT DNA Polymerase mix. Thermal cycling was followed as described in Geets et al. [24]. The presence and size of the amplification products were determined by agarose (1.5% [w/v]) gel electrophoresis.
DGGE of dsrB Genes
Community compositions, expressed as band patterns, based on dsrB genes were generated using dsrB-based DGGE. A 1-mm-thick 8% polyacrylamide gel with a denaturing gradient consisting of 40% to 70% urea–formamide was used for DGGE. Gels were ran at 60°C for 16 h at 80 V in 1× Tris–acetate–EDTA (TAE) buffer using a Bio-Rad Dcode universal mutation detection system (Bio-Rad Laboratories, Mississauga, ON, Canada). Following electrophoresis, the gels were incubated for 20 min in 1× TAE buffer containing 1× SYBR Gold nucleic acid gel stain (Molecular Probes Europe BV, The Netherlands) and photographed under UV light using an ImageQuant 300 Imager system with ImageQuant TL software (Amersham Bioscience, Buckinghamshire, UK).
Statistical Analysis of DGGE Profile
High-resolution digital DGGE images were analyzed using ImageQuant TL software. For data analysis, the images were normalized using the AlphaEase FC software, version 6.0 (Alpha Innotech, San Leandro, CA); the bands were detected using the band-searching algorithm of the software. After normalization of the gels, only the bands with a peak height intensity exceeding 5.0% of the strongest band in each lane were included in further analyses. Analysis of band patterns was performed with the Dice coefficient using 1% position tolerances and 0.5% optimization for the band migration distance. Clustering of patterns was performed by UPGMA dendrogram with a similarity matrix of Pearson coefficient. The stability of the dendrograms was evaluated by randomizing the sample order 100 times and recalculating the dendrograms with 95% background noise. The percentage of similarity between band patterns is used only to address the relatedness among the DGGE patterns and is not an indication of quantitatively genetic relatedness among respective samples.
The differences of DGGE profile patterns among sampling settings were examined using one-way analyses of similarities (ANOSIM) based on the similarity matrices [13]. Four settings were examined: (1) riverside locality at Guandu and Hwajiang, (2) vegetated and non-vegetated habitats at lower estuarine Guandu, (3) three sites, GM, GR and HR, and (4) six depths.
The relationships between six abiotic factors (salinity, and concentration of sulfate, sulfide, and reduced sulfur, as well as TOC content and sampling depth) and DGGE profile patterns were determined using BIO-ENV routine. BIO-ENV uses generalized Mantel test to examine associations between biotic and abiotic data expressed as Spearman rank correlation coefficients [13]. The matrices of DGGE profile patterns were directly come from the aforementioned cluster analysis, and the abiotic-factor matrices were based on Euclidean distance [14]. ANOSIM and BIO-ENV analyses were performed using PRIMER version 5.23 software package [14].
Sequence of dsrB DNA Fragments
A total of 35 DGGE bands including nine bands from site GR, 17 bands from site HR, and nine bands from site GM were excised and sequenced. Bands excised from the DGGE fingerprints were dissolved in 20 μl of deionized H2O and incubated overnight at 4°C in order to elute the DNA from the gel. Five microliters of the solution was used in a PCR reaction with the corresponding GC-clamped primers. These amplicons were purified with a Clean Up-M kit (Viogene) as specified by the manufacturer. Using dye terminator chemistry, DNA sequences were determined by direct sequencing with a DNA sequencer model 3730 (Applied Biosystems, Foster City, CA). The primer used for sequencing was DSR4R. The sequences obtained were compared with public database of amino acid sequences using BLAST to determine their potential taxonomic affiliation [2]. These sequences have been deposited in the NCBI nucleotide sequence databases under accession numbers HQ413099–HQ413133.
Phylogenetic Affiliation Analysis
The dsrB gene sequences were aligned as amino acid sequences before being aligned against their closest relatives. Forty-seven sequences were chosen from the database [63], including 14 SRP, 3 SiRP, 9 SOP, and 21 environmental sequences. Phylogenetic analyses were performed using MEGA version 5.0 [60, 61]. Only unambiguously aligned positions were used. The dsrB matrices were analyzed by distance-matrix of neighbor-joining (NJ) [58], maximum parsimony (MP) [23], and maximum likelihood (ML) [22]. Here, 1,000 bootstraps were performed for the NJ, MP, and ML algorithms to analyze the relatedness of the dsrB genes. Multifurcations were created at the appropriate basal node when branching patterns were only supported in greater than 70% of the bootstrap resamplings. In this phylogenetic tree, the lowest determined taxon whose sequences were grouped within a concordant branch is termed a “phylotype” hereafter to indicate the taxonomic identification of our dsrB sequences.
Results
Abiotic Characteristics
All the abiotic factors measured exhibited site differences, while the differences were less for depth zonation in the Danshuei River estuary (Fig. 2). Pore-water salinity decreased from the lower estuarine brackish-water reach to the upper tidal freshwater reach in the range of 0 at HR to a high of 19 at GR. Sedimentary salinity in the two brackish-water sites, particularly in GR, increased with depth. The salinities at the GM site were lower than those at GR, and at GM, a minimum of 6.0 appeared at the top 10-cm layer (Fig. 2). Pore-water sulfate concentration also decreased from the brackish-water reach to the tidal freshwater reach, ranging from 0 to 6.7 mM in the lower estuary to a constant 0 mM in the upper estuary. At both brackish-water GR and GM sites, the sulfate concentrations peaked at the depth of 20 cm below the surface, with values of 6.7 and 5.2 mM, respectively, and then sharply decreased to zero in further deep layers (Fig. 2). Comparatively, the pore-water sulfide concentrations at HR were higher than those at GR and GM, while GM had the lowest levels (2.1–6.0 μM at HR vs. 0–3.0 μM at GR and 0–1.0 μM at GM, Fig. 2). TOC contents at GM ranged from 1.2% to 1.8% (weight-to-weight) and were higher than those at GR and HR with TOC contents generally <1.0% (Fig. 2). Reduced sulfur contents decreased from the brackish-water reach to the tidal freshwater reach, with ranges from 4.6 to 15.2 and 0.8 to 2.3 mg g-1, respectively (Fig. 2).
Composition of SRP
The band patterns of the DGGE profiles of the dsrB gene revealed that the composition of SRP varied between the sites in the Danshuei River estuary (Fig. 3). The DGGE profile patterns were well separated in two riverside localities, as well as mangrove and non-mangrove-vegetated habitats (Table 1). Eight major bands (labeled as A, B, C, D, E, F, G, and H) were chosen to express the distribution of the dominant groups of SRP. Position A was mainly distributed in the GR and GM. Positions B and F appeared at sites GM and HR. Positions C and E occurred at both the riverside sites GR and HR. Positions D and H were recorded from all the three sites, whereas position G was only recognized at site HR. Although the band patterns did not significantly differ with depth as the three sites were pooled together (Table 1), the bands within each site seemed to exhibit variations of depth zonation (Fig. 3).
DGGE profiles of dsrB gene at site GR, GM, and HR. Letter labels on the left column refer to dominant bands, while those on the right column refer to sites where dominant bands were observed. On the DGGE profiles, the bands marked in rectangle with dash line were chosen for further sequencing. In summary, band D and H were found at all three sites. Band A was found at sites GR and GM; band B and F were found at sites GM and HR; band C and E were found at sites GR and HR; and band G was only found at site HR. Each of the six depths sampled is shown on top
DGGE profiles were separated into three distinct clusters using cluster analysis (Fig. 4). With the exception of surface sample at site GR (labeled as GR00), the community compositions of dsrB gene were well classified by site. The first cluster included five communities from all the subsurface layers at site GR (10–50 cm, labeled as GR10, GR20, GR30, GR40, and GR50; Fig. 4), with similarity values varying from 0.55 to 0.89. Two sub-clusters of middle (GR10, GR20, and GR30) and deep (GR40, GR50) zones were further separated along the depth. The second cluster consisted of the entire dsrB gene communities from site HR, with two distinct communities and similarity values ranging from 0.67 to 1.00. Here, the compositions of dsrB gene showed little zonal variation in the depth below the surface, where the composition differed greatly from that in the surface layer (HR00, Fig. 4). The last cluster consisted of all the dsrB gene communities from the site GM as well as the surface layer of site GR, with similarity values ranging from 0.17 to 0.93. Five communities were vertically separated at site GM, and the most similar community was observed in the deep layers of 40–50 cm (GM40 and GM50).
The DGGE profile patterns mostly correlated with a combination of salinity, TOC content, and reduced sulfur concentration according to BIO-ENV analysis (matching correlation, r s = 0.56, Table 2). Furthermore, the DGGE profile patterns also exhibited high correlations with a combination of TOC content and salinity (r s = 0.52) as well as and single salinity factor (r s = 0.51; Table 2). In addition, the profile patterns were also affected by reduced sulfur concentration, although to a lesser extent (r s = 0.45; Table 2).
Phylogenetic Affiliations of the Studied Partial Sequences of dsrB Gene
The phylogenic tree of partial sequences of dsrB gene is shown in Fig. 5. Most of the sequences were affiliated to those of δ-Proteobacteria of sulfate reducers, while some sequences were related to those of the spore-forming sulfate reducers. One sequence, HR3F, resembled that of SiRP; however, no sequence showed affinity to that of SOP. Six clades with 15 gene phylotypes were identified according to the phylogenic tree. The clade groups 1, 2, 4, 5, and 6 comprised more than two gene phylotypes, while only the clade group 3 had one phylotype (Fig. 5). In addition, sequences of groups 1 and 2 were found from all the three sites, whereas the sequence of group 3 was found at site GR only. In contrast, sequences of groups 4, 5, and 6 were primarily recorded from site HR, which comprised a freshwater environment.
Phylogenetic tree of dsrB gene based on DGGE bands using neighbor-joining method (NJ). Bootstrap values (>70%) of NJ as well as maximum parsimony and maximum likelihood methods are shown in the aforementioned order on the left of the relevant nodes. The first two letters refer to sites and the third numeral refers to depths. The last letter refers to major bands, A–H, which are shown in Fig. 3. SRP: sulfate-reducing prokaryotes, SiRP: sulfite-reducing prokaryotes
Sequences of the first clade, group 1, were affiliated to the family Desulfobacteraceae. The first clade consisted of six phylotypes. The 1a phylotype, comprising GM5B sequence, was close to the dsrB sequence of Desulfofaba fastidiosa strain (AY268892, 87% similarity of protein sequences). The 1b phylotype, consisting of HR3E sequence, was closely related to the dsrB sequence of Desulfatiferula olefinivorans (DQ826725, 90% similarity). The 1c phylotype, possessing HR0F sequence, was closely related to the dsrB sequence of Desulfosarcina variabilis (AF191907, 77% similarity). The 1d phylotype, comprising six sequences of GM2D, GR0H, GR1D, GR1H, GR2H, and GR4D, was similar to samples from Estuarine (FJ748824) and polluted harbor sediment (DQ112199), as well as landfill leachate aquifer (EF065086; 84–98%, 85–94%, and 84–99% similarity, respectively). The 1e phylotype, possessing four sequences of GM3A, GM4A, GM4D, and GM5A, was clustered with environmental clone from salt marsh (AY741561), coastal bay (AM408826), and anoxic basin sediments (AM236161; 90–93%, 91–92%, and 93–96% similarity, respectively). The 1f phylotype, consisting of only two sequences of GM0H and GM1H, was not found in both the identified strain and environmental clones in the current databases.
The third clade, group 3, was related to the family Desulfobulbaceae. The GR0D sequence was similar to the dsrB sequence of uncultured Desulfobulbus sp. (FJ544736, 81% similarity) and an environmental clone from landfill leachate aquifer (EF065035, 78% similarity).
The forth clade, group 4, was related to the family Desulfovibrionaceae. Two phylotypes were observed in this clade (Fig. 5). The 4a phylotype, possessing HR0B sequence, was clustered with the dsrB sequence of Desulfovibrio salexigens (CP001349, 97% similarity) and Desulfovibrio butyratiphilus (AB490775, 63% similarity), as well as an environmental clone from sulfidogenic sludge biofilter (EU350982, 92% similarity). The 4b phylotype, consisting of HR2C sequence, was similar to the dsrB sequence of Desulfovibrio oxyclinae (AY626034), Desulfovibrio longus (AB061540), and Desulfovibrio aminophilus (AY626029; 86%, 80%, and 77% similarity, respectively).
The second clades, group 2, were related to the spore-forming SRP of the family Peptococcaceae. Two phylotypes were classified in this clade. The 2a phylotype, comprising GM5F, was neighbored to clusters associated with dsrB sequences of the genus Desulfotomaculum. The 2b phylotype, comprising GR2C, GR3A, GR3F, and HR5E sequences, was closely related to the dsrB sequences of Desulfotomaculum thermobenzonicum (AJ310432), Desulfotomaculum kuznetsovii (AF273031) and Desulfotomaculum acetoxidan (AY015580; 64–71%, 65–71%, and 68–78% similarity, respectively), the environmental clones from fjord subsurface water (78–82% similar to AY865325), landfill leachate aquifer (84–94% similarity to EF065066), marshland sediment (78–81% similarity to AM901630), and estuarine sediment (81–96% similarity to AY953404).
The fifth and sixth clades, groups 5 and 6, were mainly related to freshwater environmental sequences, and some of the members appeared to be affiliated to the identified SiRP of the family Peptococcaceae. Two phylotypes were observed in the fifth clade (Fig. 5). The 5a phylotype consisted of four sequences of HR1H, HR2D, HR3H, and HR5G, and was closely related to the environmental clones reported in freshwater-influenced estuarine sediments (76–96% similarity to AY953407, AY953408, and FJ748823), low-sulfate acidic fen (69–79% similarity to AY167469), and low-sulfate landfill leachate aquifer (75–79% similarity to EF065023). The 5b phylotype, possessing HR1D, HR1E, and HR4D sequences, was not found in either the identified strain or environmental clones in the current databases.
The sixth clade consisted of two phylotypes. The 6a phylotype, having four sequences of HR2G, HR2H, HR4G and HR5F, was similar to a partial sequence from biological filter reactor (82–100% similarity to AB507433) and low-sulfate landfill leachate aquifer (71% similarity to EF065042). Nevertheless, the 6b phylotype, with HR3F sequence, did not match with either the identified strain or environmental clones in the current databases.
Discussion
In the estuary of the Danshuei River, SSRP communities varied between the lower estuarine brackish-water reach and the upper tidal freshwater reach and also differed between mangrove-vegetated and non-vegetated habitats. These discrepancies may be attributed to sulfate availability, freshwater input, oxygen stress, and the presence or absence of mangrove vegetation.
Effects of Salinity, Reduced Sulfur, and Organic Matters on SSRP Communities
The SSRP community structures in the Danshuei River are found to be highly associated with a combination of salinity, reduced sulfur concentration, and TOC content (Table 2), reflecting the effects of spatial gradient in salinity, deposition of reduced sulfur, and vegetation, respectively. When compared with these three factors, sulfate concentration appears to be less related to the SSRP community structures, because the correlation value decreased from 0.56 to 0.47 when the sulfate concentration was considered as an additional abiotic factor. Nevertheless, sulfate concentration may play a role indirectly in influencing the SSRP community distribution. Two processes, physical and biological, seem to be involved. First, the decrease in the sulfate level is a companion event with changing salinity along a marine-freshwater gradient. Second, this diminish is a metabolic consumption of sulfate by SRP [7, 8]. The sulfate concentration was positively correlated with water salinity, while negatively correlated with the deposition of sulfate-reducing products, such as reduced sulfur compound, AVS, and pyrite combined, particularly in a close system including deep sediments in lower estuarine area [7].
Previous studies have reported that organic matter is one of the important factors controlling SRP distribution [55]. Our results also show that the TOC content is greater at the mangrove-vegetated site GM than at non-vegetated site GR, suggesting that the level of organic matter derived from mangrove litter differs between these two sites, and in turn, is likely to be involved in the differentiation of SRP communities. In addition, certain SRP communities have been observed in habitats where fermentation is prevalent [39, 51]. Based on these data, we believe that the accumulation of organic matter can benefit growth of dissimilar SRP communities. Nevertheless, the pathway that becomes dominant depends on the levels of sulfate.
SSRP Communities in the Tidal Freshwater Reach
Composition of SSRP communities, varying among the three study sites, was found to be highly related to the marine-freshwater gradient, reflecting positive correlations with the changes in the sulfate concentration and salinity. The importance of this gradient was also reported in studies on the distribution of denitrifiers in the same Danshuei River estuary [21] and the diversity of dsrAB gene sequences from mudflats in France [42].
SRP are normally distributed in sulfate-enriched but oxygen-depleted habitat, particularly in marine environment. Notably, our results show that both SRP and SiRP, for instance, members of groups 4, 5, and 6, occur in the tidal freshwater reach, which lack salinity and sulfate (Figs. 2 and 5). However, when examining sulfide concentration, it was higher in the tidal freshwater reach (site HR) than in the brackish-water reach (sites GR and GM). In addition, previous reports also showed that sulfate reduction was still rather active in this tidal freshwater reach (sulfate reducing rate at GR vs. HR—0.0654 vs. 13.1 mole m-2 day-1) [11]. We ruled out the possibility that sulfate is derived from salt intrusion with tides because no evidence showed the availability of sulfate at this site [62] (this study). Alternatively, leaking of some sulfurous compounds from a deeply buried ore or degradation and transformation from sulfur-containing pollutants may occur because the HR river bank has been a garbage dump site for decades [31]. Consequently, some unique SRP or SiRP strains may have been selected for this kind of low-sulfate and low-salinity setting. We found that band G series groups (HR2G, HR4G, and HR5G) detected from the HR area were phylogenetically affiliated to those recorded from low-sulfate acidic fen or landfill leachate aquifer of freshwater environment (Fig. 5 of this study) [55, 67]. In freshwater environments with limited sulfate supply, many Desulfobulbus, Desulfovibrio, and Desulfomicrobium species grow by fermenting pyruvate to form acetate, carbon dioxide, and hydrogen, in which acetogenic growth of SRP becomes dominant [6, 10, 39, 51].
Deposition of Reduced Sulfur Compounds and Vertical Distribution of SSRP Communities
On the basis of band patterns on DGGE profile, the SSRP communities in the brackish-water reach at Guandu exhibited more diverse compositions in deep than in shallow zones, and the delineation seemed to appear between 30- and 40-cm depth at site GR and between surface and 10 cm at site GM (Fig. 4). This discrepancy on the compositions between deep and shallow zones suggests that the SSRP in the present study sites are capable of utilizing different ecological setting, including saline and freshwater habitats, sulfate shortage, and deposition of reduced sulfur compound (Table 2) as shown in other studies [8, 26]. At the GR site, a sharp decrease in the pore-water sulfate and increase in reduced sulfur occurred at 20- and 40-cm depth, respectively (sulfate concentration at 20- vs. at 30-cm depth, 6.7 vs. 0.8 mM; reduced sulfur concentration at 30- vs. at 40-cm depth, 6.2 vs. 15.2 mg g-1; Fig. 2). At the GM site, sulfate depletion also occurred at around 20-cm depth, whereas reduced sulfur compounds began to accumulate 10 cm below the surface, a depth much shallower than that at GR. These data reflect that there exists a transitional zone, below which the sediments become steadily anoxic and reductive, thus favoring SRP growth because SRP is known to be obligate anaerobic prokaryotes [5]. In contrast, the SSRP band patterns at the HR site exhibited little depth variation. In particular, their compositions were rather similar through all depths below the 10-cm layer surface (Fig. 4). This vertically invariant compositional pattern may again be attributed to a constant anoxic condition but also to the scant occurrence of sulfate and reduced sulfur compounds through all depth layers.
Oxygen Stress on SSRP Communities in the Surface Sediments and Vegetated Habitat
The DGGE profiles of the dsrB gene at the three study sites revealed that the SSRP communities in the surface of 0–1 cm were less diverse than those in the deep layers of below 10-cm in depth (Fig. 4). This zonation pattern is likely to be related to oxygen stress. Oxygen in waterlog sediment penetrates only several millimeters of the top sediment [41], and the oxygenated depth is often deprived of SRP [7]. Nevertheless, certain species of SRP, such as Desulfovibrio and Desulfobulbus spp., can still survive in micro-aerobic condition or even tolerate high concentrations of oxygen [19, 35]. Based on these data, the SSRP communities found in the surface sediments in the present study may consist of oxygen-tolerable species that differ taxonomically from those dwelling in deep, anoxic environments.
At the site GM, the dissimilarity of the SRP assemblages increased as the depth decreased, and the SRP taxonomic richness (number of bands) became lower as they were distributed in shallower areas (Fig. 3). These trends suggest that the presence of vegetation may cause the disturbance of surface sediment, thus influencing the composition of the SRP communities. Holmer and Storkholm [26] proposed that macrophytes could affect sulfate reduction by altering the redox conditions of the sediments. Bioturbation, caused by emergent macrophytes or by various activities of macrobenthos, have been found to have negative effects on SRP distribution [41, 47]. It has been recorded that macroinfauna, such as polychaetes and crabs, are abundant in the Danshuei GM area, when compared with those in non-vegetated area [32]. The macrobenthos’ burrowing, foraging, or irrigating activities can rework and oxygenate sediments up to a depth of 5–10 cm by polychaetes [54] and even deeper by crabs. We presume that oxygenation by mangrove roots and aeration by macroinfauna may act together to decrease the SRP richness at the GM site.
Phylogenic Relationships of dsrB Gene Sequences in the Tidal Freshwater Reach
The sequences of dsrB gene exhibited phylogenetically distinct SSRP communities among the three studied sites and the underlined processes were closely related to the marine-freshwater gradient (Fig. 5). The SSRP communities in the tidal freshwater habitat (HR) exhibited relatively high diversity. They were not only affiliated to prokaryotes living in freshwater habitats but also to those from marine environments. SSRP communities in the HR shared only one phylotype, 2b, with those at the other two sites in the lower reach, indicating the uniqueness of the dsrB genes in the HR. The phylotypes distributed in brackish lower estuarine GR and GM areas all showed affinity to marine SRP clades, whereas the phylotypes clustered to groups 5 and 6 were all from the freshwater habitat HR and resembled SiRP, particularly the phylotype 6b. Moreover, these freshwater-related phylotypes at HR were phylogenetically close to the genus Desulfitobacterium. Prokaryotes of this genus fail to use sulfate as an electron acceptor and are not active in typical marine environment [12, 64]. The other neighboring phylotypes in groups 5 and 6 were also reported from an environmental setting characterized as freshwater area of an estuary [42], low sulfate area in coastal zone [38, 67], low-sulfate acid fen [45], or biological filter reactor [68].
The marine-related phylotypes found at HR are affiliated to the genera Desulfatiferula (family Desulfobacteraceae), Desulfosarcina, Desulfovibrio (family Desulfovibrionaceae), and Desulfotomaculum (family Peptococcaceae). All the genera are capable to adapt to or tolerate low salinity or low sulfate supply [38, 42, 67]. In addition, they are found from a wide range of various environmental settings, such as from freshwater area of estuary [42] or coastal low sulfate area [38, 67]. Among them, D. aminophilus, which possesses the ability to break down amino acid anaerobically, was not only found in estuarine and marine environments, but also in freshwater area [3]. In a biofilter where the process of sulfate reduction was inhibited, Desulfovibrio-related fragments were also detected [59].
Moreover, the aforementioned prokaryotes and the others that the phylotypes from the site HR resembled are known to be capable of degrading pollutants [20, 31, 40]. For instance, the SiRP Desulfitohacterium dehalogenans and Desulfitobacterium hafniense are capable of performing dechlorination of chlorinated aromatic compounds [12, 64]. The SRP D. thermobenzonicum and D. variabilis can use benzoate or similar aromatic compounds as a carbon source [40, 62]. The SRP D. olefinivorans has the ability to scavenge aliphatic hydrocarbons of crude oil. In addition, some of these prokaryotes have been isolated from brackish sediment of a wastewater decantation facility of an oil refinery [17]. We believe that the sediments in the HR area may have been exposed to anthropogenic pollution, because the adjacent river bank is used for garbage dumping and landfill [31] (the corresponding author’s personal observation). Therefore, the group of SRP and SiRP that have ability to break down pollutants can colonize the HR area.
Phylogenetic Relationships of dsrB Gene Sequences in the Brackish-water Reach
In the present study, the prokaryotes found in the brackish-water reach (GR and GM) all belong to SRP clades. When compared with those observed in GR and HR, the SRP phylotypes in the GM habitat were less diverse and dominated by group 1 sequences (i.e., 1a, 1d, 1e, and 1f, Fig. 5). Such homogeneous phylotype constituent may be attributed to a constant supply of sulfate and organic substrate (TOC content was the highest among the three sites, Fig. 2). Besides, one unique phylotype, group 3, was found at saline GR. Prokaryotes that exhibited close affinity to group 3 were reported from freshwater habitats, reflecting their physiological versatility [6, 39, 51].
Most of the phylotypes determined from the GR and GM sediments were affiliated to certain typical marine SRP such as the members of the genera Desulfofaba (family Desulfobacteraceae), Desulfobotulus (family Desulfobacteraceae), Desulfatiferula (family Desulfobacteraceae), Desulfosarcina (family Desulfobacteraceae), and Desulfotomaculum (family Peptococcaceae). The accumulated data demonstrate that these prokaryotes are detected from marine systems (Fig. 5), including salt marsh [4], lower estuary close to sea [34], coast with high sulfate [67], polluted harbor [69], and anoxic abyssal basin [43]. In addition, the D. fastidiosa-related strain was isolated from a sulfate–methane transition zone [1]. Some SRP, such as Desulfovibrio and Desulfomicrobium species, can oxidize lactate and ethanol to acetate when hydrogen is efficiently removed by hydrogen-consuming methanogens [6]. Sulfate reducers were also the dominant acetogenic prokaryotes in a methanogenic reactor that was used to treat whey [10]. In the present study from 40 to 50-cm depth at the GM site, we found a sequence, GM5B, which was closely related to D. fastidiosa. As sulfates have been very much depleted in this deep zone [11] (Fig. 2), we speculate that this strain may coexist with the methanogens that involve methanogenesis.
The phylotypes found at the GR site were phylogenetically affiliated to an unidentified clone of the genus Desulfobulbus (family Desulfobulbaceae; Fig. 5). Desulfobulbus-related prokaryotes appeared to grow by fermenting in the absence of an external electron acceptor, such as in the freshwater environment that lacks sulfate [42] or landfill leachate from polluted aquifer with low sulfate level (<1 mM) [67]. The GR reach is located in the lower estuary of the Danshuei River and its salinity normally exceeds 22.0 [29]. However, the surface sediment at GR is occasionally or periodically subjected to freshwater [44], in which salinity could be lower than 10 (Fig. 2). Sulfate concentration lower than 2 mM has also been recorded in the deep layers in the vicinity of this site [21]. It is not uncommon that freshwater- and low-sulfate-associated Desulfobulbus SRP can occur in lower estuarine habitat with higher salinity because it has been reported that this strain is distributed in anoxic sediments of freshwater, brackish-water, and seawater [39]. As a result, such distribution pattern suggests that Desulfobulbus-related SRP found in GR surface sediments is capable of surviving in fluctuating salinity and sulfate concentrations.
References
Abildgaard L, Ramsing NB, Finster K (2004) Characterization of the marine propionate-degrading, sulfate-reducing bacterium Desulfofaba fastidiosa sp. nov. and reclassification of Desulfomusa hansenii as Desulfofaba hansenii comb. nov. Int J Syst Evol Microbiol 54:393–399
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Baena S, Fardeau ML, Labat M, Ollivier B, Garcia JL, Patel BK (1998) Desulfovibrio aminophilus sp. nov., a novel amino acid degrading and sulfate reducing bacterium from an anaerobic dairy wastewater lagoon. Syst Appl Microbiol 21(4):498–504
Bahr M, Crump BC, Klepac-Ceraj V, Teske A, Sogin ML, Hobbie JE (2005) Molecular characterization of sulfate-reducing bacteria in a New England salt marsh. Environ Microbiol 7:1175–1185
Barton LL, Fauque GD (2009) Biochemistry, physiology and biotechnology of sulfate-reducing bacteria. Adv Appl Microbiol 68:41–98
Bryant MP, Campbell LL, Reddy CA, Crabill MR (1977) Growth of Desulfovibrio in lactate or ethanol media low in sulfate in association with H2-utilizing methanogenic bacteria. Appl Environ Microbiol 33:1162–1169
Canfield DE, Thamdrup B, Kristensen E (2005) Aquatic geomicrobiology. Adv Mar Biol 48:1–600
Capone DG, Kiene RP (1988) Comparison of microbial dynamics in marine and freshwater sediments: contrasts in anaerobic carbon catabolism. Limnol Oceanogr 33:725–749
Castro H, Reddy KR, Ogram A (2002) Composition and function of sulfate-reducing prokaryotes in eutrophic and pristine areas of the Florida Everglades. Appl Environ Microbiol 68:6129–6137
Chartrain M, Zeikus JG (1986) Microbial ecophysiology of whey biomethanation: characterization of bacterial trophic populations and prevalent species in continuous culture. Appl Environ Microbiol 51:188–196
Chen CP, Wu JT, Lin S, Lin, HJ, Hsieh HL, Liu WC, Chen CC, Li LA (2001) A suitable plan for the sediment removal to reduce the oxygen exhaustion in the main stream Tanshui River and its tributary Keelung River. Taiwan Environmental Protection Administration, Taipei, Taiwan. EPA-90-G107-02-104 (in Chinese)
Christiansen N, Ahring BK (1996) Desulfitobacterium hafniense sp. nov. an anaerobic, reductively dechlorinating bacterium. Int J Syst Bacteriol 46:442–448
Clarke KR, Ainsworth M (1993) A method of linking multivariate community structure to environmental variables. Mar Ecol Prog Ser 92:205–219
Clarke KR, Gorley RN (2001) ‘Primer V5: User Manual/Tutorial. Plymouth, Plymouth Marine Laboratory
Cline JD (1969) Spectrophotometric determination of hydrogen sulfide in natural waters. Limnol Oceanogr 14(3):454–458
Cottrell MT, Cary SC (1999) Diversity of dissimilatory sulfite reductases genes of bacteria associated with deep-sea hydrothermal vent polychaete annelid Alvinella pompejana. Appl Environ Microbiol 65:1127–1132
Cravo-Laureau C, Labat C, Joulian C, Matheron R, Hirschler-Réa A (2007) Desulfatiferula olefinivorans gen. nov., sp. nov., a long-chain n-alkene-degrading, sulfate-reducing bacterium. Int J Syst Evol Microbiol 57:2699–2702
Devereux R, Delaney M, Widdle F, Stahl DA (1989) Natural relationships among sulfate-reducing Eubacteria. J Bacteriol 171:6689–6695
Dilling W, Cypionka H (1990) Aerobia respiration in sulfate-reducing bacteria. FEMS Microbiol Lett 71:123–127
Dhillon A, Teske A, Dillon J, Stahl DA, Sogin ML (2003) Molecular characterization of sulfate-reducing bacteria in the Guaymas basin. Appl Environ Microbiol 69:2765–2772
Fan LF, Shieh WY, Wu WF, Chen C-P (2006) Distribution of nitrogenous nutrients and denitrifier strains in estuarine sediment profiles of the Tanshui River, northern Taiwan. Estuar Coast Shelf Sci 69(3–4):543–553
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Fitch WM (1971) Toward defining the course of evolution: minimum change for specified tree topology. Syst Zool 20:406–416
Geets J, Borrernans B, Diels L, Springael D, Vangronsveld J, van der Lelie D, Vanbroekhoven K (2006) DsrB gene-based DGGE for community and diversity surveys of sulfate-reducing bacteria. J Microbiol Meth 66:194–205
Hipp WM, Pott A, Thum-Schmitz N, Faath I, Dahl C, Trüper HG (1997) Towards the phylogeny of APS reductases and sirohaem sulfite reductases in sulfate-reducing and sulfur-oxidizing prokaryotes. Microbiology 143:2891–2902
Holmer M, Storkholm P (2001) Sulphate reduction and sulphur cycling in lake sediments: a review. Freshwater Biol 46:431–451
Hsieh HL (1995) Spatial and temporal patterns of polychaete communities in a subtropical mangrove swamp—influences of sediment and microhabitat. Mar Ecol Prog Ser 127:157–167
Hsieh HL, Chen CP, Chen YG, Yang HH (2002) Diversity of benthic organic matter flows through polychaetes and crabs in a mangrove estuary: 13 C and 34S signals. Mar Ecol Prog Ser 227:145–155
Hsieh H-L, Fan L-F, Chen C-P, Wu J-T, Liu W-C (2010) Effects of semidiurnal tidal circulation on the distribution of holo- and meroplankton in a subtropical estuary. J plankton Res 32(6):829–841
Hsieh YP, Yang CH (1997) Pyrite accumulation and sulfate depletion affected by root distribution in a Juncus (Needle Rush) Salt Marsh. Estuaries 20(3):640–645
Hsu M-S, Fu JC, Lin S-H (2001) The influence of garbage dump on flood in Tanshui River. J Taiwan Conservancy 49(4):1–13 (in Chinese)
Huang SC, Shih SS, Ho YS, Chen CP, Hsieh HL (2011) Restoration of shorebird-roosting mudflats by partial removal of estuarine mangroves in northern Taiwan. Restor Ecol (in press)
Ito T, Okabe S, Satoh H, Watanabe Y (2002) Successional development of sulfate-reducing bacterial populations and their activities in a wastewater biofilm growing under microaerophilic conditions. Appl Environ Microbiol 68:1392–1402
Jiang L, Zheng Y, Peng X, Zhou H, Zhang C, Xiao X, Wang D (2009) Vertical distribution and diversity of sulfate-reducing prokaryotes in the Pearl River estuarine sediments, Southern China. FEMS Microbiol Ecol 70:249–262
Jonkers HM, Koh IO, Behrend P, Muyzer G, de Beer D (2005) Aerobic organic carbon mineralization by sulfate-reducing bacteria in the oxygen-saturated photic zone of a hypersaline microbial mat. Microb Ecol 49:291–300
Karkhoff-Schweizer RR, Huber DPW, Voordouw G (1995) Conservation of the genes for dissimilatory sulfite reductase from Desulfovibrio vulgaris and Archaeoglobus fulgidus allows their detection by PCR. Appl Environ Microbiol 61:290–296
Klein M, Friedrich M, Roger AJ, Hugenholtz P, Fishbain S, Abicht H, Blackall LL, Stahl DA, Wagner M (2001) Multiple lateral transfers of dissimilatory sulfite reductase genes between major lineages of sulfate-reducing prokaryotes. J Bacteriol 183:6028–6035
Kuever J, Rainey F, Widdel F (2005) Desulfovibrio Kluyver and van Niel 1936, 397 AL. In: Brenner DJ, Krieg NR, Garrity GM, Staley JT, Boone DR, Vos PD, Goodfellow M, Rainey FA, Schleifer K-H (eds) Bergey’s Manual® of Systematic Bacteriology, Vol. 2, The Proteobacteria, Part C, The Alpha-, Beta-, Delta-, and Epsilonproteobacteria. Springer, New York, USA, pp 926–938
Kuever J, Rainey F, Widdel F (2005) Desulfosarcina Widdel 1981, 382VP. In: Brenner DJ, Krieg NR, Garrity GM, Staley JT, Boone DR, Vos PD, Goodfellow M, Rainey FA, Schleifer K-H (eds) Bergey’s Manual® of Systematic Bacteriology, Vol. 2, The Proteobacteria, Part C, The Alpha-, Beta-, Delta-, and Epsilonproteobacteria. Springer, New York, USA, pp 981–984
Kuever J, Rainey F, Widdel F (2005) Desulfobulbus Widdel 1981, 382 VP (Effective publication: Widdel 1980, 374). In: Brenner DJ, Krieg NR, Garrity GM, Staley JT, Boone DR, Vos PD, Goodfellow M, Rainey FA, Schleifer K-H (eds) Bergey’s Manual® of Systematic Bacteriology, Vol. 2, The Proteobacteria, Part C, The Alpha-, Beta-, Delta-, and Epsilonproteobacteria. Springer, New York, USA, pp 988–992
Laanbroek HJ (2010) Methane emission from natural wetlands: interplay between emergent macrophytes and soil microbial processes. A mini-review. Ann Bot 105:141–153
Leloup J, Quillet L, Berthe T, Petit F (2006) Diversity of the dsrAB (dissimilatory sulfite reductase) gene sequences retrieved from two contrasting mudflats of the Seine estuary, France. FEMS Microbiol Ecol 55:230–238
Leloup J, Loy A, Knab NJ, Borowski C, Wagner M, Jørgensen BB (2007) Diversity and abundance of sulfate-reducing microorganisms in the sulfate and methane zones of a marine sediment, Black Sea. Environ Microbiol 9:131–142
Liu WC, Hsu MH, Kuo AY (2002) Modeling of hydrodynamics and cohesive sediment transport in Tanshui River estuarine system, Taiwan. Mar Pollu Bull 44:1076–1088
Loy A, Kusel K, Lehner A, Drake H, Wagner M (2004) Microarray and functional gene analyses of sulfate-reducing prokaryotes in low-sulfate, acidic fens reveal cooccurrence of recognized genera and novel lineages. Appl Environ Microbiol 70:6998–7009
Meyer B, Kuever J (2007) Molecular analysis of the distribution and phylogeny of dissimilatory adenosine-59-phosphosulfate reductase-encoding genes (aprBA) among sulfuroxidizing prokaryotes. Microbiology 153:3478–3498
Meysman FJR, Middelburg JJ, Heip CHR (2006) Bioturbation: a fresh look at Darwin’s last idea. Trends Ecol Evol 21(12):688–695
Miletto M, Loy A, Antheunisse AM, Loeb R, Bodelier PLE, Laanbroek HJ (2008) Biogeography of sulfate-reducing prokaryotes in river floodplains. FEMS Microbiol Ecol 64:395–406
Minz D, Flax JL, Green SJ, Muyzer G, Cohen Y, Wagner M, Rittmann BE, Stahl DA (1999) Diversity of sulfate-reducing bacteria in oxic and anoxic regions of a microbial mat characterized by comparative analysis of dissimilatory sulfite reductase genes. Appl Environ Microbiol 65:4666–4671
Muyzer G, De Waal EC, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes encoding for 16S rRNA. Appl Environ Microbiol 59:695–700
Muyzer G, Stams AJM (2008) The ecology and biotechnology of sulphate-reducing bacteria. Nature Rev Microbiol 6:441–454
Nercessian O, Bienvenu N, Moreira D, Prieur D, Jeanthon C (2005) Diversity of functional genes of methanogens, methanotrophs and sulfate-reducers in deep-sea hydrothermal environments. Environ Microbiol 7:118–132
Nocker A, Lepo JE, Martin LL, Snyder RA (2007) Response of estuarine biofilm microbial community development to changes in dissolved oxygen and nutrient concentrations. Microb Ecol 54:532–542
Pearson TH, Rosenberg R (1978) Macrobenthic succession in relation to organic enrichment and pollution of the marine environment. Oceanogr Mar Biol Rev Ann Rev 16:229–311
Rabus R, Hansen T, Widdel F (2006) Dissimilatory sulfate- and sulfur-reducing prokaryotes. In: Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E (eds) The Prokaryotes, Vol. 2. Springer, New York, USA, pp 659–768
Rosenberg R, Nilsson HC, Diaz RJ (2001) Response of benthic fauna and changing sediment redox profiles over a hypoxic gradient. Estuar Coast Shelf Sci 53:343–350
Sambrook J, Russell DW (2001) Molecular Cloning: A Laboratory Manual, 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York
Saitou N, Nei M (1987) The neighbour-joining method: a new method for reconstructing phylogenetic trees. Mo Biol Evol 4:406–425
Schwermer CU, Ferdelman TG, Stief P, Gieseke A, Rezakhani N, van Rijn J, de Beer D, Schramm A (2010) Effect of nitrate on sulfur transformations in sulfidogenic sludge of a marine aquaculture biofilter. FEMS Microbiol Ecol 72:476–484
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, and Kumar S (2011) MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Molecular Biology and Evolution (in press)
Tasaki M, Kamagata Y, Nakamura K, Mikami E (1991) Isolation and characterization of thermophilic benzonation of the thermophilic benzoate-degrading, sulfate-reducing bacterium, Desulfotomaculum thermobenzoicum sp. nov. Arch Microbiol 155:348–352
Thompson DB, Ravussin E, Bennett PH, Bogardus C (1997) Structure and sequence variation at the human leptin receptor gene in lean and obese Pima Indians. Hum Mol Genet 6(5):675–679
Utkin I, Woese C, Wiegel J (1994) Isolation and characterization of Desulfitobucterium dehalogenans gen. nov., sp. nov., an anaerobic bacterium which reductively dechlorinates chlorophenolic compounds. Int J Sys Bacteriol 44:612–619
Wagner M, Roger AJ, Flax JL, Brusseau GA, Stahl DA (1998) Phylogeny of dissimilatory sulfite reductases supports an early origin of sulfate respiration. J Bacteriol 180:2975–2982
Wen LS, Jiann KT, Liu KK (2008) Seasonal variation and flux of dissolved nutrients in the Danshuei Estuary, Taiwan: a hypoxic subtropical mountain river. Estuar Coast Shelf Sci 78:694–704
Wu X-J, Pan J-L, Liu X-L, Li D-T, Yang H (2009) Sulfate-reducing bacteria in leachate-polluted aquifers along the shore of the East China Sea. Can J Microbiol 55:1–11
Yamashita T, Yamamoto-Ikemoto R, Sakurai E, Aikawa K, Kaneko E (2010) Treatment of municipal wastewater using an anaerobic–anoxic–oxic biological filter reactor packed with carbon fibers and aerated with microbubbles. Sustain Environ Res 20(4):205–211
Zhang W, Song L-S, J-S KI, Lau C-K, Li X-D, Qian P-Y (2008) Microbial diversity in polluted harbor sediments II: Sulfate-reducing bacterial community assessment using terminal restriction fragment length polymorphism and clone library of dsrAB gene. Estuar Coast Shelf Sci 76:682–691
Acknowledgments
We sincerely thank Mr. Timothy M. Davidson for reading the manuscript. The authors are very grateful to the anonymous reviewers for their constructive comments. This study was funded through Thematic Research Program by Academia Sinica, Taiwan. This study complies with the current laws of Taiwan in which it was performed.
Author information
Authors and Affiliations
Corresponding author
Additional information
Sen-Lin Tang and Hwey-Lian Hsieh contributed equally.
Rights and permissions
About this article
Cite this article
Fan, LF., Tang, SL., Chen, CP. et al. Diversity and Composition of Sulfate- and Sulfite-Reducing Prokaryotes as Affected by Marine-Freshwater Gradient and Sulfate Availability. Microb Ecol 63, 224–237 (2012). https://doi.org/10.1007/s00248-011-9912-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-011-9912-x