Abstract
Sheath blight infection of rice by Rhizoctonia solani Kühn AG1-IA often results in serious yield losses in intensive rice cultivation. Biological control agents (BCAs) have previously been isolated but poor efficiency is often observed when applied under field conditions. This study compares a traditional dual-culture plate assay and a new water-surface microcosm assay for isolation of antagonistic soil bacteria. In the water-surface microcosm assay, floating pathogen mycelium is used as a source for isolation of hyphae-colonizing soil bacteria (HCSB), which are subsequently screened for antagonism. Ten antagonistic soil bacteria (ASB) isolated from a variety of Vietnamese rice soils using dual-culture plates were found to be affiliated with Bacillus based on 16S rRNA gene sequencing. However, all the ASB isolates grew poorly and showed no antagonism in the water-surface microcosm assay. In contrast, 11 (out of 13) HCSB isolates affiliated with Burkholderia sp. all grew well by colonizing the hyphae in the microcosms. Two of the Burkholderia sp. isolates, assigned to B. vietnamiensis based on recA gene sequencing, strongly inhibited fungal growth in both the dual-culture and water-surface microcosm assays; HCSB isolates affiliated to other species or species groups showed limited or no inhibition of R. solani in the microcosms. Our results suggest that HCSB obtained from floating pathogen hyphae can be a new source for isolation of efficient BCAs against R. solani, as the isolation assay mimics the natural habitat for fungal-bacterial interaction in the fields.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sheath blight of rice (Qryzae sativa) caused by Rhizoctonia solani Kühn AG1-IA is a destructive soil borne disease [1], causing high losses in rice production, especially under intensive cultivation with high fertilizer input and several yearly crops [2–4]. Between crops, the pathogen survives mainly as sclerotia in the soil or as mycelium in rice stubble. Infection potential of sclerotia has been found to be three times higher than that of mycelium in plant debris, making sclerotia the main source of infection [5]. Primary plant infection occurs near the water line where sclerotia float and accumulate around the rice plant [1]. Under favorable microclimatic conditions, infection spreads rapidly by means of runner hyphae to upper plant parts and onto adjacent plants causing secondary infections [6]. Despite intensive screening of the rice germplasm accessions, no effective source of resistance towards sheath blight has so far been identified. Hence, sheath blight control through host resistance is not possible at present, due to the low inherent level of resistance in rice [3]. Soil and crop management can lower the disease incidence and severity of sheath blight mainly due to changes in the canopy structure [7]. However, this has not proven adequate to suppress sheath blight incidences to an acceptable low level in intensified cropping systems. As a result, fungicides are widely used for control of the pathogen, although their excessive use might cause severe environmental and health problems. Furthermore, the growing cost of pesticides and consumers demand for pesticide-free food has led to a search for alternatives [2, 8].
An alternative to chemical control of sheath blight is biological control using antagonistic soil microorganisms (for recent reviews see [9, 10]). The success of biological control of fungal diseases is dependent on optimal performance of the biological control agents (BCAs) in the environment, i.e., to live and establish itself in the environment and tolerate different stresses associated with particular ecological niches. Hence, it has been suggested to use isolation techniques that specifically select BCAs for effectiveness under environmental conditions similar to those present in the fields [11]. Despite this, the most common approach to obtain BCAs has been to select antagonistic microorganisms from in vitro dual-culture screening on agar plates. Such screenings of soil bacteria have revealed several isolates showing antagonism against R. solani AG1-IA, e.g., Pseudomonas sp. and Bacillus sp. [12], and specifically Pseudomonas fluorescens [13] and Bacillus pumilus [14]; however, practical application of BCA often fails in the field [15, 16]. As the pathogen is protected against direct attack from the BCA when inside the plant the BCA should preferably interact with the pathogen before plant infection occurs. Since primary infections in paddy rice of the sheath blight disease initiate from floating sclerotia, this study hypothesize that a suitable place to look for ecologically adapted antagonists against R. solani would be the water surface of the irrigation water. This study reports a new method for isolating BCAs from floating R. solani mycelium mimicking the conditions in the rice field at early stages of crop growth, when the effect of the plant on the environment is limited. The new method was compared to the traditional dual-culture plate assay previously used for isolation of BCAs targeting R. solani AG1-IA.
Methods
Soil Types and Fungal Isolates
Soil sampling was conducted in paddy fields of five major provinces of the Cuu Long (Mekong) delta: An Giang, Can Tho, Dong Thap, Tien Giang, and Vinh Long (Table 1). Fields with mature rice plants were sampled. In each field a 20 × 20-cm-wide and 10-cm-deep soil sample was taken. Fifty grams of each soil sample was transferred to a sterile box, stored at 5°C and transported to Denmark for further studies. The pH was measured with a glass electrode (B-212 Twin compact pH meter Horiba, Japan) in a 1:1 (w/v) ratio of soil and water. Electric conductivity (EC) was measured with a conductivity meter (B-173 Twin compact Horiba, Japan), also in a 1:1 (w/v) soil-to-water ratio.
R. solani isolate TG-2 was obtained from sheath blight lesions of rice plants collected from the Tien Giang province. In short, small pieces of plant material were cut from the junction between diseased and healthy tissues using a sterilized scalpel. The samples were surface-sterilized in 1% sodium hypochlorite for 15 s and then washed three times in sterilized distilled water. The samples were subsequently placed on a sterilized filter paper to remove excess of moisture, before transfer to water agar plates and incubation at 28°C. To obtain a pure pathogen isolate, single-hyphae tips were transferred multiple times to new water agar plates. After isolation, the pathogen was grown routinely on potato dextrose agar (PDA) plates unless stated otherwise. TG-2 was subsequently tested to verify its pathogenic potential towards rice.
Enumeration of the Bacterial Population
For each soil sample, 1 g was suspended in 9 ml of sterile 0.9% NaCl solution (Merck, Germany), vortexed for 3 min and used to prepare a series of tenfold dilutions in 0.9% NaCl. Colony forming units (CFUs) were determined on R2A agar (Merck, Germany) after incubation at 28°C for 3 days. CFUs were expressed per g of dry soil; dry weight was determined after desiccation at 105°C for 24 h.
Bacterial Isolates
Isolation of Antagonistic Soil Bacteria (ASB Isolates)Using Traditional Dual-Culture Plate Assay
Enumeration and isolation of bacteria was conducted according to Quan et al. [17] with some modifications. After spreading diluted soil suspensions onto R2A plates, the center of each plate was inoculated with an 7-mm-diameter agar plug excised from a 4-day-old culture of the R. solani (see below). After 3 days of incubation at 28°C, total bacterial colonies were counted. Bacterial colonies showing inhibition zones against the pathogen were counted as antagonistic and subsequently isolated by repeated streaking onto R2A plates. Bacterial isolates were grown in Luria-Bertani (LB) broth (28°C) over night, mixed 1:1 with 85% glycerol and stored in cryotubes at −80°C for further use.
Isolation of Hyphae-Colonizing Soil Bacteria Using Water-Surface Microcosm Assay
R. solani TG-2 was grown on half-strength PDA plates (Difco Laboratories, USA) for 5 days. After this time, the whole plate was covered by mycelium and the fungus was assumed to have depleted the agar for nutrients, as significant sclerotia formation had started. Agar plugs were cut 1–2 cm from the edge of the mycelium with a 1-cm-diameter flamed cork borer and transferred (hyphae up) to the center of 9-cm-diameter Petri dishes. Immediately, 20 ml mineral nutrients (2 g l−1 of Yeast Nitrogen Source without Amino Acids, YNS-AA, Difco Laboratoires, USA) was poured into the microcosms and inoculated with 100 μl of bacterial soil suspension reaching 1,000 CFU per microcosm. This addition of mineral nutrients was necessary in order for R. solani to spread over the water surface, as it did not spread on the surface of pure water. The bacterial soil suspension was prepared by diluting soil in sterile 0.9% NaCl. The microcosms were incubated at 28°C for 7 days before isolation of hyphae-colonizing soil bacteria (HCSB).
To isolate HCSB the mycelia in the microcosms were transferred with sterile forceps to sterilized test tubes containing 1.5 ml of 0.9% NaCl and 1.5-g sterilized, 2-mm-diameter glass beads. To remove the bacteria from the hyphae, vortexing was performed for 5 min before the resulting suspension was filtered through sterilized glass wool to remove the hyphae. The bacterial suspensions obtained were then diluted 1:1,000 in 0.9% NaCl before 100 μl of the diluted suspension was spread onto R2A plates and incubated at 28°C for 3 days. HCSB were isolated and stored as described for the ASB isolates from the dual-culture plate assay.
Antagonism Towards R. solani
Antagonism by the isolates was evaluated using both the dual-culture plate assay and the water-surface microcosm assay. Ten ASB isolates and 13 HCSB isolates were selected to represent all colony morphologies observed among the isolates.
Dual-Culture Plate Assay
Bacteria from 1-day-old colonies were streaked in two lines (2.5 cm for each) on 9-cm R2A plates from the margin towards the center of the plate. A 7-mm-diameter agar plug of R. solani was placed at the center. Plates without bacterial streaks served as controls. Four replicates were made for each bacterial isolate and plates were incubated at 28°C for 72 h. After incubation, images of the plates were recorded digitally by a scanner and the areas colonized by the fungus were measured as for the water-surface microcosm assay below. Antagonism was evaluated as inhibition ratio, similar to percent inhibition of radial growth [18], using the equation
where A 1 is mycelium area on control plates without bacteria and A 2 is mycelium area on plates with bacteria.
Water-Surface Microcosm Assay
R. solani was grown as described for the isolation of HCSB and an agar plug with the fungus was placed in the center of 100 × 20 mm (diameter × height) Nunclon™ cell culture dishes (Thermo scientific, USA). Finally, 5 ml of bacterial suspension containing approximately 104 cells ml−1 in YNS-AN (2 g l−1) from bacterial colonies grown for 24 h on R2A was added to the dishes. The hydrophilic surface Nunclon™ cell culture dishes reduced the meniscus of the water surface making it possible to use only 5 ml of bacterial suspension in the microcosm dishes. The low volume was necessary to enable quantitative measurements of fungal growth by scanning. Five replicate dishes for each bacterial isolate were incubated at 28°C for 4 days.
At day 4 samples for bacterial CFU determination were taken from each microcosm, between the agar plug and the edge of the dish; furthermore, fungal growth was determined in each microcosm using a standard office flatbed scanner (CanoScan 9950 F). The scanner was operated in a dark room with the scanner lid open to create a dark-field type image. Using the contrast enhancement histogram function of the scanner program (CnoScan Toolbox 4.9) single hyphae growing on the water surface could be visualized. The extent of the fungal growth was marked digitally on the image, and the area of the dish colonized by the fungus was measured using the scientific image analysis freeware ImageJ 1.32 made available by NHS (http://rsbweb.nih.gov/ij). Antagonism was evaluated as described for the dual-culture assay.
Identification and Genes of Antagonism
Cell Lysis
Individual bacterial colonies were picked from agar plates using a sterile loop and resuspended in 5 ml LB broth (10 g Bacto™ Tryptone, Difco Laboratories, USA; 5 g Bacto™ Yeast Extract, Difco Laboratories, USA; 10 g NaCl, Merck, Germany; pH 7). The cell suspension was incubated on a shaker at 28°C for 24 h. One milliliter of bacterial suspension was transferred to a 1.5-ml Eppendorf tube and centrifuged at 10,000×g for 5 min. The supernatant was discarded and the pellet was washed twice in PBS by centrifuging and resuspending. Finally, the pellet was resuspended in 100 μl Milli-Q water. The bacterial suspension was boiled for 10 min at 99°C in a heating block and subsequently transferred immediately to ice. The cell lysate was stored at −20°C.
PCR and Phylogenetic Identification
For initial phylogenetic identification of both ASB and HCSB isolates, PCR was carried out using primers 341-F [19] and 1492-R [20] which amplify partial 16S rRNA genes from most known Eubacteria. The PCR reaction was carried out in a 50-μl setup containing 1× HF reaction buffer (provided with the polymerase), 1.5 mM MgCl2, 200 μM of each dNTP, 0.5 μM of each primer, 1 U of Phuison DNA polymerase (Finnzymes, Finland) and 2 μl of cell lysate. The PCR program consisted of an initial heating step at 98°C for 30 s, followed by 30 cycles of initial denaturation at 98°C for 10 s, primer annealing at 57°C for 20 s, elongation at 72°C for 1 min and a final elongation step for 5 min. Amplicons were checked for quality and quantity by electrophoresis on a 0.8% agarose gel, followed by ethidium bromide staining for 5 min before visualization using the Gel Doc system (Biorad, USA).
The HCSB were further characterized by PCR using primers BCR1 and BCR2 targeting the recombinase A (recA) gene [21]. PCR was performed as described above with the following modifications to the reaction mixture: 250 μM of each dNTP, 0.8 μM of each primer and 5 μl of cell lysate were used. The PCR annealing temperature was here raised to 58°C. PCR amplicons were purified using QIAquick PCR purification kit (Qiagen, USA) following the manufacturer's instructions, and subsequently sequenced at the Uppsala Genome Center, Uppsala, Sweden. For sequencing, the internal primers BCR3 and BCR4 were used [21]. Sequences obtained were trimmed using the CLCbio software (CLCbio, Denmark). Subsequently, a BLAST search (http://blast.ncbi.nlm.nih.gov/Blast.cgi) was completed in order to identify the isolates based on similarity to known database sequences. A selection of reference sequences were downloaded from GenBank and included in the alignment for phylogenetic analysis of the isolates. As the two sequencing products were not overlapping, the missing sequence was trimmed out of the reference strains to obtain 639 unambiguously aligned nucleotides; aligned sequences were subject to phylogenetic analysis using the default settings in Sequence Viewer version 6.3 (CLCbio, Denmark). All sequences were deposited in GenBank with the following accession numbers: JF429974-JF430005.
Screening for Genes of Antagonism
The HCSB isolates were finally subjected to PCR targeting genes coding for common antifungal metabolites in Gram-negative BCAs: pyrrolnitrin, pyoluteorin, 2,4-DAPG, phenazine [22], and hydrogen cyanide [23]. The PCR was set up in 20-μl reactions containing: 1× Phusion HF buffer, 200 μM of each dNTP, 0.5 μM of each primer, and 0.4 U Phusion DNA polymerase and 1 μl of cell lysate as template. Cycling conditions were: initial heating at 98°C for 30 s, followed by 30 cycles of 98°C for 15 s, 63°C (67°C for hydrogencyanide detection) for 30 s and 72°C for 30 s, followed by a final step at 72°C for 10 min. P. fluorescens Pf5 and P. fluorescens 2–79 were used as positive controls in PCR.
Results
All soils were slightly acidic (pH 4.3 to 5.8), and EC values fell in the range of 0.1–1 mS/cm, well in correspondence with records of these alluvial type soils [24].
Antagonistic Soil Bacteria from Dual-Culture Plate Assays
The bacterial antagonists towards R. solani TG-2 in the soil samples, as determined by plate counts, represented a fraction of 2–20% of the total CFUs; An Giang province soil showed a lower proportion (2–5%) than the other soils (10–20%) but total CFU counts were also higher at An Giang (1.2 × 108 g dry soil−1) compared to the other soils (approximately 2–7 × 107 g dry soil−1). Hence, all soils had a density of culturable bacterial antagonists of approximately 5 × 105 g dry soil−1. It should be noted that the total CFU counts are underestimations of the true number of bacteria in the fields, as some bacteria might be inhibited by the fungus.
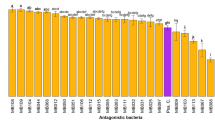
Using the dual-culture assay, a total of 180 ASB were isolated from the rice paddy soils. Only three different colony morphologies were observed for the 180 isolates and ten isolates representing the three morphologies, were selected for further studies. Phylogenetic identification as based on the 16S rRNA gene sequencing revealed that they were all Bacillus sp., most closely resembling B. pumilus and Bacillus thuringiensis (Table 2). When tested for antagonism in the dual-culture assay, Bacillus sp. isolates C-30.26, A-10.2, and A-7.15 revealed inhibition ratios of 0.39, 0.44, and 0.25 when tested against R. solani TG-2, respectively (Table 2). All other Bacillus sp. isolates showed inhibition ratios of only approximately 0.1. When the Bacillus sp. isolates were subsequently tested for antagonism in the water-surface microcosm assay, only two strains (A-7.24 and V-6.10) were found to be slightly antagonistic to R. solani (Table 2). Further, all Bacillus sp. isolates grew poorly or not at all in the water-surface microcosm, i.e., neither decreased nor increased their cell density (CFU per milliliter) more than within one order of magnitude, after 4 days of incubation (Table 2).
Antagonistic Hypha-Colonizing Soil Bacteria (HCSB Isolates) from Water-Surface Microcosms
Microscopic observations confirmed the ability of the bacteria to colonize hyphae in the microcosm. After 24 h, no firm attachment of bacteria was observed, whereas observations after 48 h showed heavy colonization of the hyphae (Fig. 1). Thirteen colonies representing different morphologies were selected. Eleven were identified as Burkholderia sp., primarily from the Burkholderia cepacia species complex, based on 16S rRNA gene sequencing. Isolates belonging to the B. cepacia species complex (BCC) were further characterized based on the gene sequence of recA (Table 2), a housekeeping gene involved in recombination and DNA repair [25, 26]. Phylogenetic analysis of the recA sequences revealed that most isolates formed a distinct group related to Burkholderia diffusa while two isolates (A-7.3 and V-16.5) were closely related to Burkholderia vietnamiensis (Fig. 2). The two B. vietnamiensis isolates were both strong antagonists against R. solani TG-2 in the dual-culture assay, showing inhibition ratios of 0.59 and 0.71, respectively (Table 2). In contrast, all isolates clustering with B. diffusa showed limited or no inhibition. Intriguingly, B. vietnamiensis A-7.3 and V-16.5 were also the only isolates strongly inhibiting R. solani in the water-surface microcosm assay (Table 2, Fig. 3). None of the B. diffusa isolates showed notable antagonism and three of them, all from the Can Tho province (C-19.2, C-20.3, and C-30.1), actually stimulated mycelium growth. Screenings for the presence of selected genes known from Pseudomonas sp. and Burkholderia sp. to be involved in production of antifungal metabolites were negative for all the Burkholderia sp. isolates. All Burkholderia sp. isolates grew well in the microcosms and densities (CFU per milliliters) generally increased by three to four orders of magnitude after 4 days of incubation, in stark contrast to the Bacillus sp. isolates inability to grow in the same assay (Table 2).
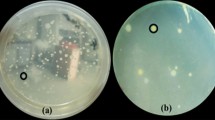
Digitally contrast enhanced scanning images of Rhizoctonia solani growth in the air–water interface in the presence of the two antagonistic HCSB V-16.5 and A-7.3. Bacteria and fungus were co-inoculated at the start of the experiment. Each microcosm was inoculated with 5 × 104 bacterial cells. The half moon-shaped appearances around the fungal discs are light reflections from the disks caused by the scanning procedure
Discussion
The majority of potential BCA isolates selected by their antagonism in traditional dual-culture assays (ASB) showed no antagonism towards R. solani in the microcosm assay. Furthermore, none of them were able to proliferate in the microcosm assay. The only two ASB isolates able to significantly reduce growth of the fungus did not grow in the microcosm, and thus seemed unable to depend only on the carbon/energy from live hyphae. Interestingly, all ASB isolates could be assigned to Bacillus sp., which are commonly found in screenings for potential BCAs in soil [9, 27–29]. The reason for the Bacillus sp. isolates failure to control fungal growth in the water-surface microcosm assay could be that their production of antifungal compounds or general survival in the habitat was suppressed by environmental factors and/or lack of nutrients, supported by the fact that Firmicutes, including Bacillus sp., are not normally part of the bacterioneuston community inhabiting the surface water of aquatic habitats [30]. In the microcosm assay, lack of a suitable carbon/energy source is more reasonable than lack of nutrients, since there was ample supply of mineral nutrients.
Contrary, all HCSB isolates were considered hyphotrophic, i.e., utilizing hyphae for growth, as they were able to proliferate in the microcosm with living R. solani hyphae as their only available carbon/energy source. Truly HCSB have so far only been described and studied in relation to their potential positive interaction with mycorrhiza [31–35], but to our knowledge, neither in relation to the irrigation water and sheath blight disease specifically, or the neuston habitat in general. Based on 16S rRNA sequencing the majority of HCSB isolates was identified as Burkholderia sp., especially within the BCC. Members of Burkholderia sp. are previously found in rice soil and rhizosphere habitats [36] but have also been identified as an important part of the bacterioneuston of lakes [37] resembling the air–water interface of irrigated rice fields. Traits of Burkholderia sp. strains may encompass antifungal genes and the group is known to produce a wide range of secondary metabolites active against R. solani, e.g., pyrrolnitrin, phenazine [38–40], cepaciamide A [41], and some unknown compounds [17, 42]. Furthermore, chitinolytic activity has been reported for Burkholderia and Pseudomonas strains [43, 44]. In this study, a phenotypic diversity in respect to interaction with R. solani was demonstrated within the Burkholderia genus. Some isolates showed strong antagonism against the fungus while others stimulated the fungal growth. To further elucidate the phylogenetic relationship of the BCC strains, a second round of identification was performed. As the recA gene has a higher polymorphism than the 16S rRNA gene it has previously been used to identify strains within the BCC complex [45]. The analysis revealed that the two highly antagonistic strains could be assigned specifically to B. vietnamiensis, a species within the BCC originally isolated from Vietnamese rice paddy soil [46]. The remaining isolates of the BCC formed a distinct clade (bootstrap value of 98) showing affiliation to B. diffusa. Since there were no obvious signs of cell wall degradation seen by microscopy, this study did not go further into a search for potential chitinolytic activity of the strains, and it was anticipated that the antagonistic potential was due to production of antagonistic compounds. However, no genes coding for common antifungal compounds in Burkholderia and Pseudomonas spp. [22] were detected in any of the strains, indicating a production of other antifungal compounds in the two exceedingly antagonistic B. vietnamiensis strains. Failure to detect genes for known antagonistic compounds have previously been noted for the strain B. vietnamienses LMG10929 isolated from rice rhizosphere in Vietnam despite its antagonistic potential towards R. solani in a dual-culture assay [47].
Burkholderia sp. seems to have a large variety of life styles in natural environments and isolates belonging to the BCC have been used as plant disease antagonists, plant growth promoters, and degraders of toxic substances (reviewed by [48]). B. diffusa has previously been isolated from a variety of habitats [49], but there are no previous reports on its presence within the rice paddy system or neuston communities in general. Our isolates T-22.3 and T-17.3 could be affiliated with Burkholderia kururiensis strains KP23 and ATSB13 isolated from rhizosphere soil and aquifer samples, respectively [50]. In line with our findings, strains belonging to this species have previously been found in rice field soils and have been tested for antagonism towards R. solani, although none was observed [51]. Isolate V-6.1 was affiliated with Staphylococcus sp. of which several strains have been isolated from rice seeds, and a subgroup has proven to be antagonistic to R. solani [52]. In contrast, V-6.4 was affiliated with Dyella sp., which has to our knowledge never been isolated from rice soils.
In conclusion, we have obtained two promising BCAs effective against R. solani causing sheath blight in rice. Furthermore, we have provided new insight into the importance of incorporating habitat simulation when selecting potential biocontrol agents. The targeting HCSB using a novel water-surface microcosm assay is a new approach, and seems more successful than conventional isolation of antagonistic bacteria from bulk soil or the rhizosphere based on dual-culture assays, in its potential for obtaining antagonistic bacteria able to control R. solani under field conditions. In essence, the new approach first selects bacteria able to grow in the hyphal environment and secondly screens for antibiosis of these selected isolates in a similar environment. The approach presented in this study gains theoretical support by a recent work [53], where an ideal BCA was suggested to be a BCA with a low growth rate (or low dying rate) in the target habitat, but a high growth rate on diseased tissue (i.e., the fungus). Thus, in natural rice field water with fierce bacterial competition, a BCA being especially adapted to grow on, or induce the leakage of, hyphae-derived carbon/energy from the pathogen is expected to profit considerably while at the same time inhibiting the pathogen. Further studies will reveal the mechanism of the antagonistic behavior as well as a validation of the biocontrol ability under field conditions.
References
Hashiba T, Kobayashi T (1996) Rice diseases incited by Rhizoctonia species. In: Sneh B, Jabaji-Hare S, Neate S, Dijst G (eds) Rhizoctonia species: taxomony, molecular biology, ecology, pathology and disease control. Kluwer Academic Publishers, Dordrecht, pp 331–340
Mew TW, Cottyn B, Pamplona R, Barrios H, Xiangmin L, Zhiyi C, Fan L, Nilpanit N, Arunyanart P, Kim PV, Du PV (2004) Applying rice seed-associated antagonistic bacteria to manage rice sheath blight in developing countries. Plant Dis 88:557–564
Mew TW, Leung H, Savary S, Cruz CMV, Leach JE (2004) Looking ahead in rice disease research and management. Crit Rev Plant Sci 23:103–127
Savary S, Castilla NP, Elazegui FA, McLaren CG, Ynalvez MA, Teng PS (1995) Direct and indirect effects of nitrogen supply and disease source structure on rice sheath blight spread. Phytopathology 85:959–965
Kobayashi T, Mew TW, Hashiba T (1997) Relationship between incidence of rice sheath blight and primary inoculum in the Philippines: mycelia in plant debris and sclerotia. Ann Phytopathol Soc Jpn 63:324–327
Lee FN, Rush MC (1983) Rice sheath blight: a major rice disease. Plant Dis 67:829–832
Willocquet L, Fernandez L, Savary S (2000) Effect of various crop establishment methods practised by Asian farmers on epidemics of rice sheath blight caused by Rhizoctonia solani. Plant Pathol 49:346–354
Gerhardson B (2002) Biological substitutes for pesticides. Trends Biotechnol 20:338–343
Whipps JM (2001) Microbial interactions and biocontrol in the rhizosphere. J Exp Bot 52:487–511
Compant S, Duffy B, Nowak J, Clément C, Barka EA (2005) Use of plant growth-promoting bacteria for biocontrol of plant diseases: principles, mechanisms of action, and future prospects. AEM 71:4951–4959
Kloepper JW (1996) Host specificity in microbe-microbe interactions. Bioscience 46:406–409
Kanjanamaneesathian M, Kusonwiriyawong C, Pengnoo A, Nilratana L (1998) Screening of potential bacterial antagonists for control of sheath blight in rice and development of suitable bacterial formulations for effective application. Australas Plant Pathol 27:198–206
Kazempour MN (2004) Biological control of Rhizoctoni solani, the causal agent of rice sheath blight by antagonistics bacteria in greenhouse and field conditions. Plant Pathol J 3:88–96
Luo JY, Xie GL, Li B, Luo YC, Zhao LH, Wang X, Liu B, Li W (2005) Gram-positive bacteria associated with rice in China and their antagonists against the pathogens of sheath blight and bakanae disease in rice. Rice Sci 12:213–218
Haas D, Défago G (2005) Biological control of soil-borne pathogens by fluorescent Pseudomonads. Nat Rev Microbiol 3:307–319
Alabouvette C, Olivain C, Steinberg C (2006) Biological control of plant diseases: the European situation. Eur J Plant Pathol 114:329–341
Quan CS, Zheng W, Liu Q, Ohta Y, Fan SD (2006) Isolation and characterization of a novel Burkholderia cepacia with strong antifungal activity against Rhizoctonia solani. Appl Microbiol Biotechnol 72:1276–1284
Whipps JM (1987) Effect of media on growth and interactions between a range of soil borne glasshouse pathogens and antagonistic fungi. New Phytol 107:127–142
Amann RI, Binder BJ, Olson RJ, Chisholm SW, Devereux R, Stahl DA (1990) Combination of 16S rRNA-targeted oligonucleotide probes with flow-cytometry for analyzing mixed microbial-populations. Appl Environ Microbiol 56:1919–1925
Lane DJ (1991) 16 S/23 S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. John Wiley and Sons, New York, pp 115–175
Mahenthiralingam E, Bischof J, Byrne SK, Radomski C, Davies JE, Av-Gay Y, Vandamme P (2000) DNA-based diagnostic approaches for identification of Burkholderia cepacia complex, Burkholderia vietnamiensis, Burkholderia multivorans, Burkholderia stabilis, and Burkholderia cepacia genomovars I and III. J Clin Microbiol 38:3165–3173
de Souza JT, Raaijmakers JM (2003) Polymorphisms within the prnD and pltC genes from pyrrolnitrin and pyoluteorin-producing Pseudomonas and Burkholderia spp. FEMS Microbiol Ecol 43:21–34
Svercel M, Duffy B, Défago G (2007) PCR amplification of hydrogen cyanide biosynthetic locus hcnAB in Pseudomonas spp. J Microbiol Meth 70:209–213
Hoa NM, Singh U, Samonte HP (1998) Potassium supplying capacity of some lowland rice soils in the Mekong delta. Better Crops Int 12:11–15
Karlin S, Weinstock GM, Brendel V (1995) Bacterial classifications derived from RecA protein-sequence comparisons. J Bacteriol 177:6881–6893
Eisen JA (1995) The RecA protein as a model molecule for molecular systematic studies of bacteria: comparison of trees of RecAs and 16 S rRNAs from the same species. J Mol Evol 41:1105–1123
Kim HS, Park J, Choi SW, Choi KH, Lee GP, Ban SJ, Lee CH, Kim CS (2003) Isolation and characterization of Bacillus strains for biological control. J Microbiol 41:196–201
Cazorla FM, Romero D, Pérez-García A, Lugtenberg BJJ, de Vicente A, Bloemberg G (2007) Isolation and characterization of antagonistic Bacillus subtilis strains from the avocado rhizoplane displaying biocontrol activity. J Appl Microbiol 103:1950–1959
Hu QP, Xu JG, Song P, Song JN, Chen WL (2008) Isolation and identification of a potential biocontrol agent Bacillus subtilis QM3 from Qinghai yak dung in China. World J Microbiol Biotechnol 24:2451–2458
Hervas A, Casamayor EO (2009) High similarity between bacterioneuston and airborne bacterial community compositions in a high mountain lake area. FEMS Microbiol Ecol 67:219–228
Timonen S, Jørgensen KS, Haahtela K, Sen R (1998) Bacterial community structure at defined locations of Pinus sylvestris - Suillus bovinus and Pinus sylvestris—Paxillus involutus mycorrhizospheres in dry pine forest humus and nursery peat. Can J Microbiol 44:499–513
Mansfeld-Giese K, Larsen J, Bødker L (2002) Bacterial populations associated with mycelium of the arbuscular mycorrhizal fungus Glomus intraradices. FEMS Microbiol Ecol 41:133–140
Aspray TJ, Jones EE, Whipps JM, Bending GD (2006) Importance of mycorrhization helper bacteria cell density and metabolite localization for the Pinus sylvestris-Lactarius rufus symbiosis. FEMS Microbiol Ecol 56:25–33
Toljander JF, Artursson V, Paul LR, Jansson JK, Finlay RD (2006) Attachment of different soil bacteria to arbuscular mycorrhizal fungal extraradical hyphae is determined by hyphal vitality and fungal species. FEMS Microbiol Lett 254:34–40
Scheublin TR, Sanders IR, Keel C, van der Meer JR (2010) Characterisation of microbial communities colonising the hyphal surfaces of arbuscular mycorrhizal fungi. ISME J 4:752–763
Jha B, Thakur MC, Gontia I, Albrecht V, Stoffels M, Schmid M, Hartmann A (2009) Isolation, partial identification and application of diazotrophic rhizobacteria from traditional Indian rice cultivars. Eur J Soil Biol 45:62–72
Kalwaslińska A, Kesy J, Donderski W (2008) Biodegradation of carbendazim by epiphytic and neustonic bacteria of eutrophic Chełmzyńskie. Pol J Microbiol 57:221–230
Cartwright DK, Chilton WS, Benson DM (1995) Pyrrolnitrin and phenazine production by Pseudomonas cepacia, strain 5.5B, a biocontrol agent of Rhizoctonia solani. Appl Microbiol Biotechnol 43:211–216
Rosales AM, Thomashow L, Cook RJ, Mew TW (1995) Isolation and identification of antifungal metabolites produced by rice-associated antagonistic Pseudomonas spp. Phytopathology 85:1028–1032
EL-Banna N, Winkelmann G (1998) Pyrrolnitrin from Burkholderia cepacia: antibiotic activity against fungi and novel activities against streptomycetes. J Appl Microbiol 85:69–78
Jiao Y, Yoshihara T, Ishikuri S, Uchino H, Ichihara A (1996) Structural identification of cepaciamide A, a novel fungitoxic compound from Pseudomonas cepacia D-202. Tetrahedron Lett 37:1039–1042
Mao S, Lee SJ, Hwangbo H, Kim YW, Park KH, Cha GS, Park RD, Kim KY (2006) Isolation and characterization of antifungal substances from Burkholderia sp. culture broth. Curr Microbiol 53:358–364
Nielsen MN, Sørensen J (1999) Chitinolytic ativity of Pseudomonas fluorescens isolates from barley and sugar beet rhizosphere. FEMS Microbiol Ecol 30:217–227
Ogawa K, Yoshida N, Kariya K, Ohnishi C, Ikeda R (2002) Purification and characterization of a novel chitinase from Burkholderia cepacia strain KH2 isolated from the bed log of Lentinus edodes, Shiitake mushroom. J Gen Appl Microbiol 48:25–33
Payne GW, Vandamme P, Morgan SH, LiPuma JJ, Coenye T, Weightman AJ, Jones TH, Mahenthiralingam E (2005) Development of a recA gene-based identification approach for the entire Burkholderia genus. Appl Environ Microbiol 71:3917–3927
Gillis M, Van TV, Bardin R, Goor M, Hebbar P, Willems A, Segers P, Kersters K, Heulin T, Fernandez MP (1995) Polyphasic taxonomy in the genus Burkholderia leading to an emended description of the genus and proposition of Burkholderia vietnamiensis sp. nov. for N2-fixing isolates from rice in Vietnam. Int J Syst Bacteriol 45:274–289
Schmidt S, Blom JF, Pernthaler J, Berg G, Baldwin A, Mahenthiralingam E, Eberl L (2009) Production of the antifungal compound pyrrolnitrin is quorum sensing-regulated in members of the Burkholderia cepacia complex. Environ Microbiol 11:1422–1437
Chiarini L, Bevivino A, Dalmastri C, Tabacchioni S, Visca P (2006) Burkholderia cepacia complex species: health hazards and biotechnological potential. Trends Microbiol 14:277–286
Vanlaere E, LiPuma JJ, Baldwin A, Henry D, De Brandt E, Mahenthiralingam E, Speert D, Dowson C, Vandamme P (2008) Burkholderia latens sp. nov., Burkholderia diffusa sp. nov., Burkholderia arboris sp. nov., Burkholderia seminalis sp. nov. and Burkholderia metallica sp. nov., novel species within the Burkholderia cepacia complex. Int J Syst Evol Microbiol 58:1580–1590
Anandham R, Indira Gandhi P, Kwon SW, Sa TM, Kim YK, Jee HJ (2009) Mixotrophic metabolism in Burkholderia kururiensis subsp. thiooxydans subsp. nov., a facultative chemolithoautotrophic thiosulfate oxidizing bacterium isolated from rhizosphere soil and proposal for classification of the type strain of Burkholderia kururiensis as Burkholderia kururiensis subsp. kururiensis subsp. nov. Arch Microbiol 191:885–894
Yang JH, Liu HX, Zhu GM, Pan YL, Xu LP, Guo JH (2008) Diversity analysis of antagonists from rice-associated bacteria and their application in biocontrol of rice diseases. J Appl Microbiol 104:91–104
Cottyn B, Debode J, Regalado E, Mew TW, Swings J (2009) Phenotypic and genetic diversity of rice seed-associated bacteria and their role in pathogenicity and biological control. J Appl Microbiol 107:885–897
Jeger MJ, Jeffries P, Elad Y, Xu XM (2009) A generic theoretical model for biological control of foliar plant diseases. J Theor Biol 256:201–214
Acknowledgment
This study was supported by a research grant from the Danish Ministry of Foreign Affairs on “Integrated disease and nutrient management in intensive rice production systems in Vietnam” (104.DAN.8.L.727). We thank Dorthe Ganzhorn and Kirsten Henriksen for excellent laboratory assistance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Cuong, N.D., Nicolaisen, M.H., Sørensen, J. et al. Hyphae-Colonizing Burkholderia sp.—A New Source of Biological Control Agents Against Sheath Blight Disease (Rhizoctonia solani AG1-IA) in Rice. Microb Ecol 62, 425–434 (2011). https://doi.org/10.1007/s00248-011-9823-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-011-9823-x