Abstract
The diversity of microbial communities inhabiting two terrestrial volcanic glasses of contrasting mineralogy and age was characterised. Basaltic glass from a <0.8 Ma hyaloclastite deposit (Valafell) harboured a more diverse Bacteria community than the younger rhyolitic glass from ∼150-300 AD (Dόmadalshraun lava flow). Actinobacteria dominated 16S rRNA gene clone libraries from both sites, however, Proteobacteria, Acidobacteria and Cyanobacteria were also numerically abundant in each. A significant proportion (15-34%) of the sequenced clones displayed <85% sequence similarities with current database sequences, thus suggesting the presence of novel microbial diversity in each volcanic glass. The majority of clone sequences shared the greatest similarity to uncultured organisms, mainly from soil environments, among these clones from Antarctic environments and Hawaiian and Andean volcanic deposits. Additionally, a large number of clones within the Cyanobacteria and Proteobacteria were more similar to sequences from other lithic environments, included among these Icelandic clones from crystalline basalt and rhyolite, however, no similarities to sequences reported from marine volcanic glasses were observed. PhyloChip analysis detected substantially greater numbers of phylotypes at both sites than the corresponding clone libraries, but nonetheless also identified the basaltic glass community as the richer, containing approximately 29% unique phylotypes compared to rhyolitic glass.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
During volcanic eruptions, lava that cools quickly, for example by coming into contact with ice or water, quenches rapidly to form glass. Depending on the silica content, the material may form basaltic glass or in the case of silica-rich material, rhyolitic glass or obsidian. In the ocean environment alone, the rate of crustal production may be well over a billion tonnes a year [2] and much of this is in the form of basaltic glass. Volcanic glasses are also common in terrestrial volcanic environments [28].
Investigating the alteration or weathering of volcanic glasses has importance for a number of key areas in Earth and planetary sciences including: (1) the chemical weathering of rocks is well established [19, 37, 45], but less is known about the biological contribution. As rock weathering contributes to the carbonate-silicate cycle and long-term climate by consuming CO2 in rock weathering reactions [7, 16, 17], it is important to elucidate the diversity of organisms that might take part in these reactions. An important first step is to understand the microbial diversity of volcanic materials in relation to their geochemistry; (2) the weathering of volcanic rocks, particularly glasses, contributes nutrients into the biosphere, providing highly fertile agricultural soil and playing an important role in soil formation at the so-called ‘critical zone’ [14]. Investigating the diversity of volcanic glasses will contribute both to understanding the diversity of organisms that can be sustained by weathered volcanic glass and the microbial taxa that might take part in weathering the material during soil neogenesis.
The most intensive efforts to understand the microbial diversity of volcanic glasses have focused on oceanic basaltic glass. Templeton et al. [50] showed the presence of Mn-oxidising bacteria in basalts from Loihi Seamount, although autotrophic Mn-oxidisers were not recovered, suggesting Mn oxidation in heterotrophs is a secondary process [49]. Neutrophilic Fe-oxidising bacteria were isolated from oceanic basaltic glass [18]. In the study of the microbial community of the basaltic glasses of the Knipovich Ridge, Arctic, Thorseth et al. [52] identified heterotrophs and some chemolithotrophs including phylotypes belonging to the ε-Proteobacteria and closely matching with sulphur oxidisers. Stalk-like deposits similar to those produced by Gallionella spp. were observed, but Gallionella spp. were not identified by molecular methods. Iron-reducing organisms were cultured from Arctic Ridge seafloor basaltic glasses by Lysnes et al. [32], suggesting the possibility of an iron cycle within seafloor basalts. They determined the presence of a diversity of other organisms belonging to the Proteobacteria, Chloroflexi, Firmicutes, Actinobacteria and Crenarchaeota of unknown physiology. Phylogenetic analysis of the microbial inhabitants of seafloor basaltic glasses around Hawaii show high diversity [38, 40] which is thought to be linked to the chemolithotrophic use of basaltic glass alteration products [40].
Studies of the microbial communities associated with terrestrial volcanic glasses are in comparison to marine studies relatively scarce. In a study of Icelandic basaltic glass, Cockell et al. [10, 11] showed that most of the organisms corresponded to heterotrophic taxa and there were no sequences obtained from chemolithotrophs, suggesting that they are very rare, if they are present. The dominant phyla were Actinobacteria, Proteobacteria and Bacteroidetes. They attributed these results to the very different environmental conditions in terrestrial basaltic glasses compared to the deep ocean, with the weathered terrestrial basaltic glasses being more similar to soils. Organisms were pervasive in the basaltic glass and microbial abundance was on the order of ∼107 organisms/g.
These results contrast to results by Herrera et al. [24, 25] who studied Icelandic obsidian. Using FISH they showed that the organisms were highly localised and were predominantly associated with phenocrysts of iron and pyroxenes. Microbial enumerations were difficult on account of the localised communities, but Actinobacteria, Acidobacteria and Proteobacteria were prevalent. They attributed this localization to the high silica content of the rock and the likelihood that bioessential cations were difficult for the biota to access.
In the present study, our motivation was to further our understanding of the composition of microbial communities within terrestrial volcanic rocks. We use 16S rRNA gene clone libraries, denaturing gradient gel electrophoresis (DGGE) and microarray analyses to characterise the diversity and richness of Bacteria inhabiting terrestrial Icelandic obsidian and basaltic glasses.
Methods
Field Site and Sampling
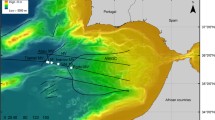
Sampling locations (Fig. 1a) and methods have been described previously [24]. Briefly, obsidian samples were collected at obsidian outcrops formed during the ∼150-300 AD formation of the Dómadalshraun lava flow (64º2.01′N, 19º7.75′W). Naturally weathered obsidian samples were taken from a ∼1 m3 block of rock using a rock hammer (Fig. 1b). Weathered hyaloclastite/basaltic glass produced during a subglacial volcanic eruption in the Pleistocene [13] was collected some 20.8 km distant, at Valafell (location 64º4.83′N, 19º32.53′ W), near Mt. Hekla in southern Iceland. Basaltic glass weathers to the clay-like material palagonite [46], which imparts a brown colouration to the otherwise black basaltic glass. The material examined here is therefore a mixture of unaltered basaltic glass and palagonite with its associated organisms and is hereinafter referred to as basaltic glass/palagonite. Samples were removed from blocks that lay on the ground (Fig. 1c).
Location of Valafell (star) and Dómadalshraun (circle) sampling sites in Southern Iceland (a). Obsidian samples were retrieved from the Dómadalshraun lava flow (64º1.76′N, 19º6.37′W), formed∼AD 150-300 (b), while basaltic glass/palagonite samples were taken from hyaloclastite deposited at Valafell (64º4.83′N, 19º32.53′W) <0.8 Myr (c)
Electron Microprobe Analysis (EMPA)
The major elemental composition of the rocks was measured by electron microprobe analysis on carbon-coated thin sections (obsidian) or carbon-coated resin-blocks (palagonite) using a Cameca SX100 electron microprobe (Department of Earth Sciences, Open University, Milton Keynes, UK), as previously described [11, 25].
Porosity and Surface Area Measurements
Obsidian and basaltic glass/plagonite surface area was determined by Brunauer-Emmett-Teller (BET) analysis (five point; Imperial College, London, UK) [6] using ∼3 mm3 fragments of material. Porosity was determined by mercury intrusion porosimetry (MCA Services, Meldreth, Cambridge, UK). For both BET and mercury intrusion porosimetry, fragments of rock pooled from six different samples were examined.
Extraction of Total DNA from Rock Samples
DNA was extracted from four (obsidian) or five (basaltic glass/palagonite) samples of each glass type. For each sample, 10 g of rock was crushed to a powder in a metal cyclinder [23]. The total DNA was directly extracted from the crushed rock using the PowerMax Soil DNA Isolation Kit (MoBio Laboratories, Cambridge, UK) according to the manufacturer’s instructions.
Denaturing Gradient Gel Electrophoresis
Microbial communities in individual samples, in addition to a composite sample for each glass type (see below), was analysed by DGGE analysis of the 16S rRNA genes using a nested PCR protocol. The first round of amplification was performed with primers pA [5] and com2 [43], targeting the V1-V9 hypervariable regions, while the second round of amplification was performed with the GC-clamped forward primer 338 [34] in combination with com2, targeting V3-V9. Amplification of the pA-com2 region was performed in 50 µl reactions with 0.2 µM each primer, 200 µM each dNTP (New England Biolabs), 2.5 U Taq DNA polymerase, 1.5 mM MgCl2, 1× PCR buffer (200 mM Tris-HCl (pH 8.4), 500 mM KCl; Invitrogen Corporation, Paisley, UK) and 5 ng of template DNA. One microlitre of unpurified product was used as template in the second round of amplification with 0.4 µM each primer, 160 µM each dNTP, 2.5 mM MgCl2, 2 U Taq and 1× PCR buffer (as described above). For each primer set, 25 amplification cycles were performed in a G-Storm GS1 thermal cycler (GRI Ltd, Essex, UK) as follows; 94 °C for 1 min, annealing at 55 °C and extension at 72 °C, each for 40 s (pA-com2) or 30 s (338GC-com2). Initial denaturations and final extensions (at 94 °C and 72 °C respectively) were performed for 5 min. Each sample was amplified in duplicate with 338GC-com2 and products combined and purified (Illustra™ GFX™, GE Healthcare, Buckinghamshire, UK). Three hundred and fifty nanograms of each purified PCR product was resolved on a 6% polyacrylamide gel containing a 30-70% denaturant gradient, where 100% is defined as 7 M urea and 40% (v/v) formamide. The composite basaltic glass/palagonite product was included in multiple lanes as a reference. Gels were run using a D-Code Universal Mutation Detection System (Bio-Rad, Hercules, CA, USA) at 75 V for 18 h in 1× TAE at 60 °C. Bands were visualised with SYBR® Green I (Invitrogen Corporation, Paisley, UK) and fingerprints analysed with GelComparII® (Applied Maths, Sint-Martens-Latem, Belgium) based on presence-absence of bands in each sample. A UPGMA dendrogram was constructed using the Jaccard binary coefficient and 1,000 bootstrap replicates. The DGGE presence-absence matrix was used to generate a triangular similarity matrix based on the Bray Curtis index [31] using PRIMER5 software [9]. The analysis of similarity (ANOSIM) routine was used to examine the statistical significance of differences between the DGGE profiles (significance data reported as p values). Spearman correlation tests were performed to test for a correlation between bacterial community structure and mineralogy.
PCR Amplification, Cloning and Sequencing of Bacterial 16S rRNA Genes
For both obsidian and basaltic glass/palagonite, equal amounts of DNA from four individual samples were pooled to yield composite samples (samples Pal1, Pal2, Pal3 and Pal5 for basaltic glass/palagonite, and all four obsidian samples). These composites were used to construct 16S rRNA gene clone libraries with primers pA and com2. Conditions were similar to those described above, exceptions being an increase in MgCl2 (2 mM), cycle number (30) and an increase in the final extension time to 10 min. Products were resolved on 1% agarose, gel purified (Illustra™ GFX™, GE Healthcare, Buckinghamshire, UK) and cloned into pCR®4-TOPO® vector before transforming into chemically competent One Shot® TOP10 Escherichia coli (Invitrogen Corporation, Paisley, UK). Clones were selected from plates at random and vector inserts were sequenced with primer pA (MCLAB, California, USA). Putative chimaeras were identified in the clone libraries by submitting sequences to the CHECK CHIMERA programme of the RDP and to the Bellerophon server (Huber et al., 2004). Phylogenetic searches were performed with SeqMatch (RDP) and the Basic Local Alignment Search Tool [1]. Diversity and richness estimators were computed with DOTUR [41]. Coverage estimates were calculated manually [8, 21]. Clone libraries were compared at species (97%) and genus (95%) sequence similarity levels using the Libshuff [44] function available in MOTHUR [42]. Neighbour-joining phylogenetic trees were constructed in MEGA4 [47] using the Jukes-Cantor [29] nucleotide substitution model and 1,000 bootstrap replicates.
GenBank Accession Numbers
16S rRNA gene clone sequences have been deposited in GenBank under accession numbers GU219531 to GU219829.
PhyloChip Analysis
The bacterial community present in obsidian and basalt glass/palagonite was investigated by PhyloChip analysis of the composite samples. The 16S rRNA genes (V1-V9 regions) were amplified using an 8-temperature gradient PCR and universal primers 27f and 1492r [30]. Amplification was performed at each temperature, using 3 µl of template, 300 nM each primer, 1× Ex Taq buffer, 2 mM MgCl2, 200 µM each dNTP, 25 µg bovine serum albumin (Roche Applied Science, Indianapolis, IN, USA) and 0.625 U Ex Taq (TaKaRa Bio, Fisher Scientific, Pittsburg, PA, USA). The following thermocycling conditions were used in an iCycler (Bio-Rad, Hercules, CA, USA): 95 °C for 3 min, 30 cycles of 95 °C for 30 s, 48-58 °C for 30 s and 72 °C for 2 min, with a final extension for 10 min at 72 °C. Products from each annealing temperature were combined and concentrated to 40 µl or less (Microcon YM-100 filters, Millipore, Billerica, MA, USA) before DNA quantification on a 2% agarose E-gel using the Low Range Quantitative DNA Ladder (Invitrogen Carlsbad, CA, USA).
PhyloChip (second generation, G2) details are presented elsewhere [3, 4, 15]. Five hundred nanograms of purified PCR product was spiked with a mix of amplicons of known concentrations, fragmented with DNAse I (Invitrogen, Carlsbad, CA, USA), and biotin-labelled according to the manufacturer’s instructions for Affymetrix Prokaryotic arrays. Labelled products were hybridised overnight at 48 °C and 60 rpm. Arrays were washed, stained and scanned as previously described. Probe selection, scoring, data acquisition and analysis have been described elsewhere [3, 4, 15]. Data obtained from the cel files (produced from the GeneChip Microarray Analysis Suite, version 5.1) were normalised using the method described by Ivanov et al. [26], and log10 transformed to compensate for slight variations in probe responses on different chips. A positive fraction (pf) cutoff value of 0.9 was used to select operational taxonomic units (OTUs) for further investigation. This value means that 90% of the probe pairs in a probe set had to be scored positive in order for the probe set intensity value (a trimmed mean) to be used further in data analyses. A probe pair was scored positive when the perfect-match probe intensity minus the mis-match probe intensity was at least 130 times greater than the squared noise value and when the perfect-match intensity was 1.3 times the mis-match intensity.
Results
Mineral Analysis
The quantitative mineral composition of the rock samples was determined by EMPA (Table 1). Results support the basaltic nature of samples from Valafell, with high levels of CaO and MgO, and basaltic SiO2 and K2O concentrations. The mean concentrations of these minerals in samples from the Dómadalshraun lava flow indicate a rhyolitic nature, with high K2O and SiO2 (>72%), and low MgO and CaO.
Porosity and Surface Area
The porosity of the basaltic glass/palagonite and obsidian material was 25.8 and 11.1% respectively, while the surfaces areas were approximately 109 and <0.3 m2/g, respectively.
DGGE Comparison of Bacterial Communities
Three hundred and fifty nanograms of purified PCR product was loaded for both composite samples and for each of the individual samples, to facilitate direct comparisons of bacterial community structure among glass types. Cluster analysis performed on the presence-absence of bands clearly separated the samples on the basis of mineralogy (Fig. 2). Obsidian samples shared at least 58% similarity, while the basaltic glass/palagonite samples shared a minimum similarity of ∼54%. Only one sample, Pal5 did not cluster with its counterparts. ANOSIM performed on the data also supported the separation of obsidian and basaltic glass/palagonite samples, where Pal5 groups most closely with other basaltic glass/palagonite samples (Supplemental Information Figure 1).
Cluster analysis of glass bacterial communities based on DGGE (30-70% denaturant) community fingerprints. The Jaccard UPGMA dendrogram was generated from the presence-absence matrix in GelComparII®. Values at nodes represent percentage similarities. Basaltic glass/palagonite samples from Valafell and rhyolitic glass/obsidian samples from Dόmadalshraun are represented by Pal and Obs, respectively. Samples ending in C are the composites for the given site
16S rRNA Gene Clone Library Analyses
One hundred and twenty-four and 175 chimaera-free clones were sequenced from the obsidian and basaltic glass/palagonite 16S rRNA gene libraries, respectively. One of the obsidian clones (deposited as GU219604) was determined to be of mitochondrial origin. Rarefaction analysis at both 97% (species) and 95% (genus) sequence similarity levels indicated that libraries were not sampled to saturation (Fig. 2). Coverage estimates for the obsidian library (0.66 and 0.73) were higher than for basaltic glass/palagonite (0.48 and 0.66) at both levels of similarity. Diversity and richness estimates for both libraries suggest a richer, more diverse community within the basaltic glass/palagonite, with Chao1 richness estimates 147 and 111 in obsidian compared to 216 and 131 in basaltic glass at 97% and 95% sequence similarities, respectively. The corresponding Shannon diversity indices were 3.93 and 3.80 for obsidian and 4.28 and 4.08 for basaltic glass, derived using 120 sequences for each library type aligned over 620 nucleotides. Non-parametric Shannon indices were also calculated at 97% and 95% sequence identities (Fig. 3). Values were higher than for Shannon indices, but again indicated higher diversity at the Valafell site at both taxonomic levels (not shown).
The distribution of clones within their respective phyla is represented in Fig. 4. Actinobacteria dominated the basaltic glass/palagonite library (43% of clones sequenced), followed by Proteobacteria, Acidobacteria, Cyanobacteria and Bacteroidetes. The phylum Actinobacteria was also the largest phylum in the obsidian library (25.6%), followed by Cyanobacteria, Acidobacteria and Proteobacteria. The comparison of libraries by Libshuff analysis (10,000 randomizations) demonstrated that they are significantly different (P < 0.0001). Libshuff analysis revealed a combined 116 OTUs at 97% sequence identities among libraries. Of these, 46 were unique to the obsidian library, 57 unique to the basaltic glass/palagonite library, and the remaining 13 OTUs common to both. The phylogenetic relationship of these OTUs is demonstrated in Fig. 5, where each OTU is represented by one clone. All the phyla identified in our libraries contained clones most similar to sequences from soil environments. Indeed, many of the phyla were dominated by sequences most similar to soil 16S rRNA gene clone libraries sequences deposited in public databases. The Actinobacteria, which are represented by two main clusters (Fig. 5b) displayed the greatest sequence similarities to prairie, grassland, Antarctic, Hawaiian and Socompa volcano soil sequences. Trembling aspen rhizosphere sequences also displayed great similarity to some of our Actinobacteria and to the Gemmatimonadetes and Planctomycetes clones in our libraries. Sequences from the Socompa Volcano study [12] and Hawaiian and Antarctic soils also feature among our Acidobacteria, Bacteroidetes, Cyanobacteria and Proteobacteria phyla. Sequences from prairie and polychlorinated biphenyl-contaminated soil also display great similarities to many of our Acidobacteria. A large proportion of clones within the Cyanobacteria and Proteobacteria phyla are most closely related to those retrieved from lithic environments elsewhere.
Neighbour-joining phylogenetic tree of 16S rRNA gene clone sequences from basaltic glass/palagonite and obsidian libraries. The tree was generated using nucleotide positions 112-770 (E. coli numbering). a full phylogenetic tree, b Actinobacteria phylum as represented by wedge in a. Clones are represented at the OTU-level (defined at 97% sequence similarities) by one sequence from each Libshuff-identified OTU. OTU designations are followed in parenthesis by the number of clones represented by that OTU in basaltic glass/palagonite and obsidian libraries, respectively. OTUs are highlighted in bold, and database sequences from other from Icelandic clone libraries are in bold italics. Bootstrap values (1,000 replicates) are shown where they exceed 60%. The scale bar represents 2% estimated sequence divergence. Aquifex sp. was used as an outgroup
PhyloChip Analysis and Comparison with Clone Libraries
Bacterial communities from both glass types were compared using a high-density oligonucleotide microarray, the G2 PhyloChip. Thirty-six and 41 phyla were detected in obsidian and basaltic glass/palagonite, respectively, representing 295 and 337 subfamilies within 1323 and 1710 OTUs. A comparison at the OTU-level revealed that the basaltic glass/palagonite contained a significantly higher proportion of unique phylotypes, more than twice that of the obsidian (Table 2). For a more direct comparison with PhyloChip results, clone library sequences were classified using the G2 chip taxonomy classifer provided through greengenes (http://greengenes.lbl.gov/cgi-bin/nph-classify_G2.cgi). In so doing, 85 and 60 OTUs were identified in basaltic glass/palagonite and obsidian libraries, respectively (11 phyla in each). Within these OTUs 10.3% (obsidian) and 17.6% (basaltic glass/palagonite) of subfamilies were unique to the clone libraries alone, being undetected by microarray. Conversely, approximately 88% of subfamilies detected by PhyloChip in each rock type were not detected by clone library analysis.
Discussion
Igneous volcanic glasses are a major contributor to the global CO2 cycle through silicate weathering reactions, and to understand the potential role of microbes in this rock weathering process requires knowledge of the microbial diversity contained therein. While the microbial communities inhabiting marine glasses have been comparatively well documented, information regarding the terrestrial glasses is scarce. In the present study, we sought to build upon earlier work on terrestrial crystalline basalt and rhyolite [23] and characterise the bacterial communities in their glassy counterparts.
Bacterial Diversity in Terrestrial Icelandic Volcanic Glasses
Analyses of the bacterial communities inhabiting basaltic and rhyolitic/obsidian glasses not only revealed substantial bacterial diversity and abundant novel 16S rRNA gene sequences, but furthermore that the bacterial communities within each glass type differed quite substantially. Obsidian was characterised by a lower diversity and richness, as observed by 16S rRNA gene microarray and clone library analyses. Kelly et al. (in review) also observed lower diversity and richness in crystalline rhyolite compared to crystalline basalt, thus suggesting a possible link between mineralogy and community structure. DGGE analysis of bacterial communities also clustered samples on the basis of glass mineralogy, with the exception of one basaltic glass/palagonite sample (Pal5).
While all analysis methods point to differences in bacterial communities existing between rock types, variability exists between the methods used and their resolving power. DGGE analysis cannot fully determine richness or indeed diversity [27, 36]. As observed in the current study, the number of bands present, averaging 34 for basaltic glass and 30 for obsidian, are substantially below any richness estimates derived by other analyses. PhyloChip analysis detected up to a ninefold higher abundance of taxa (obsidian) than the corresponding predicted clone library richness estimate, contrary to results obtained elsewhere, where good agreement between the methods was generally observed [54], although agreement between microarray and clone libraries is not always been observed [39]. Potential reasons for this discrepancy have been discussed by Kelly et al. (in review) and include differences in DNA amplification protocols, the use of substantially larger quantities of DNA for microarrays compared to clone library construction and the use of a shorter segment of the 16S rRNA gene for clone library construction, all likely contributing to the underestimation of bacterial diversity by the clone libraries, factors also relevant in the study of Rastogi et. al. [39]. PhyloChip analysis also has methodological limitations which it is prudent to consider. Firstly, the choice of an appropriate cut-off or pf value (positive fraction) for OTU detection. This has been examined elsewhere and our choice of 0.90 as a cut-off has been rigorously tested [3, 15]. Kelly et al. (in review) examined the effects of an increased pf cutoff on the abundance of different phyla and found only minor differences. The second and perhaps most important consideration is the reduced capacity of the PhyloChip to detect novel taxa, as previously discussed [15, 54]. In the present study, 15% of obsidian and 34% of basaltic glass/palagonite 16S rRNA gene clone sequences displayed <85% similarity to sequences currently deposited within the RDP database. Additionally, as many of our clones display the greatest similarities to more recent uncultured database sequences, is it likely that the PhyloChip too has severely underestimated the true phylogenetic diversity within these glasses. This supposition is also supported by the presence in the clone libraries of up to 17.6% subfamilies unique to clone libraries.
The above considerations aside, PhyloChip and 16S clone libraries largely agree in the presence-absence of the major bacterial phyla (Table 2). In the present study, phyla representing at least 1% of the detected OTUs by PhyloChip were generally detected by clone libraries, as also observed by Kelly et al. (in review) in the comparison of bacterial communities from crystalline volcanic rocks. Notable exceptions are the γ- and ε-Proteobacteria, and the Spirochaetes, each of which were undetected in libraries. As clone libraries were not sampled to saturation, failure to detect low abundance (<3%) phyla may be explained in terms of incomplete sampling, however the failure to detect γ-Proteobacteria constituting >10% of microarray OTUs is more difficult to account for and may be related to the failure of the clone library primers to amplify these sequences. Kelly et al. (in review) detected γ-Proteobacteria in crystalline rocks by PhyloChip and 16S rRNA gene clone libraries, but used a different primer set for constructing the libraries. We have examined a subset of the database sequences corresponding to the γ-Proteobacteria OTUs which were detected by the PhyloChip in the basaltic glass/palagonite sample and found suitable binding sites for both pA and com2 primers, thus suggesting that a bias against the γ-Proteobacteria occurred during the clone library PCR, or that either probe mis-hybridization occurred or an inappropriate pf cut-off was chosen (or a combination of these) during PhyloChip analysis.
In addition to the differences in diversity and richness, the obvious difference in the relative percentages or indeed phyla ranking observed between microarray and clone libraries is noteworthy. This observation has also been made previously by Kelly et al. (in review) and by Rastogi et al. [39] in their comparison of contaminated and uncontaminated soil. Although the glass clone libraries were revealed to be significantly different by Libshuff analysis and varied in the abundance of each phylum, microarrays indicated a very similar phylum distribution among glass types. At both OTU- and sf-level, however, it is apparent that the basaltic glass/palagonite contained a significant fraction (19% and 29.2%, respectively) of unique phylotypes. We suggest however that given the likelihood that many additional novel phylotypes went undetected by the PhyloChip, as discussed above, these figures may not be representative of the true difference in glass bacterial communities
The vast majority of terrestrial Icelandic glass clones share the greatest sequence homologies with uncultured organisms, principally from soil environments, consistent with the observation that they represent very different organisms than those inhabiting deep ocean basaltic glasses [10]. The levels of diversity and richness reported here in volcanic glasses is comparable to those reported in soil environments and higher than richness reported in oceanic basalt [40]. The richness of basaltic glass bacteria is also higher than that reported for Icelandic crystalline basalt (Kelly et al., in review).
In the present study, sequences from volcanic soil habitats such as the Socompa volcano in the Andes [12] and Hawaiian volcanic deposits [20] are abundant among the Actinobacteria¸ Acidobacteria, Cyanobacteria, Bacteroidetes and to a lesser extent, Proteobacteria. Sequences from these Andean and Hawaiian studies also displayed homologies to the crystalline basalt and rhyolitic clones within Actinobacteria, Bacteroidetes, Acidobacteria, Proteobacteria and Verrucomicrobia phyla identified by Kelly et al. (in review) and may represent ecotypes adapted to volcanic environments.
In contrast to the Actinobacteria, Proteobacteria clones from glass 16S rRNA gene libraries display little overlap in sequence homologies with sequences from volcanic environments, being dominated by 15 α-Proteobacteria clones most similar to a cryptoendolithic sandstone clone (EU751317). None of the Proteobacteria clones from the crystalline Icelandic rocks of Kelly et al. (in review) share appreciable similarities with those from the present glass libraries, suggesting that crystalline and glassy rock types harbour different Proteobacteria ribotypes. Many of the other glass phyla however do contain sequences similar to crystalline basalt and rhyolite clone sequences from Iceland (Kelly et al. in review), although the rhyolite clones are not included in Fig. 5 due to their comparatively shorter lengths.
The Cyanobacteria cluster of Fig. 5 similarly contains abundant endolith-type sequences, similar to those from sandstone (EF522285), quartz (FJ790633) and an underground tomb (FN298014) suggesting that the cryptoendolithic way of life may be an important determinant of cyanobacterial community composition in terrestrial volcanic glass. Furthermore, >50% of clone sequences within the Cyanobacteria cluster are more similar to algae. The presence of algae in Icelandic volcanic glass has been observed by us previously by microscopy (unpublished). The presence in both glass types of significant proportions of phototroph-derived sequences (∼12-17%) suggests a potential role of algae and cyanobacteria in supporting the diverse heterotrophic bacterial communities in these low organic carbon habitats. It should be mentioned here that some of the 16S rRNA gene clone sequences from basaltic glass and obsidian displayed great similarities to those retrieved by Herrera et al. [25] and Cockell et al. [10] in libraries generated from a single sample of each of the present glass types, from the same sites as the present study. Those clone libraries, however, were generated from smaller 16S fragments and as such, to enhance the phylogenetic resolutions in the present study were not included in the phylogenetic trees here. Furthermore, the use of longer sequences and their generation from a composite rather than a single sample resulted in differences in the relative abundances of phyla observed between libraries from the same rock type.
Possible Factors Influencing Microbial Community Composition in Volcanic Glasses
The greater diversity observed in the older, weathered basaltic glass cannot be attributed to any one cause, however, we suggest that apart from differences in age and location (the latter undoubtedly resulting in differences in environmental conditions), mineralogical differences and porosity may have contributed to the elevated richness of basaltic glass compared to obsidian. Basaltic glass weathers to palagonite [46, 51], a soft secondary weathering material whose cations may be more amenable to microbial access compared to the solid rock. Individual samples may well have different quantities of palagonite, which could influence the microbial communities within the rocks. This could be one factor that might account for why Pal5 clustered differently from the other basaltic glass/palagonite samples by DGGE analysis. Two additional key differences between the glasses relating to mineralogy are silica and bioessential cation contents. Wolff-Boenisch et al. [53] showed that at 25 °C and pH 4, basaltic glass weathers approximately an order of magnitude faster than silica-rich glass. As the cations are bound within the glass silica matrix, the high silica concentrations in obsidian would be expected to limit the availability of bioessential cation, which is reflected in the calculated release rates of elements from glass based on silica content [53]. Basaltic glass contains a higher percentage of the bioessential cations, Mg, Ca and Fe than obsidian, but lower concentrations of Na and K, which might influence the types of microorganisms that thrive by selecting for those with nutrient requirements that can be met within each rock type. As discussed by Kelly et al. (in review), rock mineralogy also influences albedo, thereby influencing temperatures experienced within the rock [22]. Obsidian is darker than the weathered basaltic glass and small differences in temperatures experienced on the rock surface and within the rock might well influence the communities. An additional, indirect effect of mineralogy is the difference in rock porosity. At more than double the porosity of obsidian and >300-fold larger surface area, basaltic glass offers a substantially larger area for microbial attachment and colonisation. Porosity will also affect the uptake and retention of water, thus influencing the microbial communities within.
Another confounding factor in the present study is the contrasting age of the parent rocks. Substrate age is known to have an influence on microbial communities. Mason et al. [33] analysed the microbial communities within marine basalts of varying age and locations from the East Pacific Rise. T-RFLP analysis revealed that the old samples (ranging in age from a few thousand to approximately 3 Ma) clustered together, while the younger basalts formed a separate cluster. Elsewhere, Lysnes et al. [32] examined marine basalts from the Arctic spreading ridges, also of varying ages. The authors found that Actinobacteria were absent in the youngest samples but demonstrated increasing abundance with sample age, being the most abundant group in the oldest basalt (1 Ma). Even on shorter timescales, an increase in soil Chao 1 diversity indices from 111 to 378 was observed at 99% sequence similarities [48], while Nemergut et al. [35] observed an increase of approximately 60% in the phylogenetic diversity of soils from <1 year to 1-4 year, levelling off in 20-year soils. In the current study, the older age of the basaltic glass may have resulted in increased diversity on two levels that are intrinsically linked to glass composition, namely succession and weathering rates. Studies such as those of Nemergut et al. [35] and Tarlera et al. [48] attribute the observed increases in soil microbial diversity over time to successional changes in the microbial communities. Succession may indeed contribute to the increased diversity in the present study, however the older age of the basaltic glass means that it has been subjected to weathering for a longer period, thus facilitating an increase in the surface area for microbial attachment and nutrient leaching, thereby providing a more clement microbial environment.
In conclusion, this study shows that terrestrial volcanic glasses host remarkably diverse bacterial communities. Rock composition and thus weathering rate are likely to play an important role in influencing this diversity. However, this study highlights the potentially complex involvement of other factors such as age and porosity in determining the endolithic communities in volcanic terrains.
References
Altschul SF, Madden TL, Schäffer AA, Zhang Z, Zhang Z, Miller W, Lipman D (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database serach programs. Nucl Acids Res 25:3389–3402
Bach W, Edwards KJ (2003) Iron and sulfide oxidation within the basaltic ocean crust: implications for chemolithoautotrophic microbial biomass production. Geochim Cosmochim Acta 67:3871–3887
Brodie EL, DeSantis TZ, Joyner DC, Baek SM, Larsen JT, Andersen GL, Hazen TC, Richardson PM, Herman DJ, Tokunaga TK et al (2006) Application of a high-density oligonucleotide microarray approach to study bacterial population dynamics during uranium reduction and reoxidation. Appl Environ Microbiol 72:6288–6298
Brodie EL, DeSantis TZ, Parker JPM, Zubietta IX, Piceno YM, Andersen GL (2007) Urban aerosols harbor diverse and dynamic bacterial populations. Proc Natl Acad Sci 104:299–304
Bruce KD, Hiorns WD, Hobman JL, Osborn AM, Strike P, Ritchie DA (1992) Amplification of DNA from native populations of soil bacteria by using the polymerase chain reaction. Appl Environ Microbiol 58:3413–3416
Brunauer S, Emmett P, Teller E (1938) Adsorption of gases in multimolar layers. J Am Chem Soc 60:309–319
Caldeira K (1995) Long-term control of atmospheric carbon dioxide; low-temperature seafloor alteration or terrestrial silicate-rock weathering? Am J Sci 295:1077–1114
Chao A, Yang MCK (1993) Stopping rules and estimation for recapture debugging with unequal failure rates. Biometrika 80:193–201
Clarke KR (1993) Non-parametric multivariate analyses of changes in community structure. Aust J Ecol 18:117–143
Cockell CS, Olsson-Francis K, Herrera A, Kelly L, Thorsteinsson T, Marteinsson V (2009) Bacteria in weathered basaltic glass, Iceland. Geomicrobiol J 26:491–507
Cockell CS, Olsson-Francis K, Herrera A, Meunier A (2009) Alteration textures in terrestrial volcanic glass and the associated bacterial community. Geobiol 7:50–65
Costello EK, Halloy SRP, Reed SC, Sowell P, Schmidt SK (2009) Fumarole-supported islands of biodiversity within a hyperarid, high-elevation landscape on Socompa Volcano, Puna de Atacama, Andes. Appl Environ Microbiol 75:735–747
Crovisier J-L, Advocat T, Dussossoy J-L (2003) Nature and role of natural alteration gels formed on the surface of ancient volcanic glasses (natural analogs of waste containment glasses). J Nucl Mater 321:91–109
Dahlgren R, Shoji S, Nanzyo M (1993) Mineralogical characteristics of volcanic ash soils. In: Shoji S, Nanzyo M (eds) Volcanic ash soils genesis, properties and utilization. Elsevier, pp 101-143
DeSantis T, Brodie E, Moberg J, Zubieta I, Piceno Y, Andersen G (2007) High-density universal 16S rRNA microarray analysis reveals broader diversity than typical clone library when sampling the environment. Microbial Ecol 53:371–383
Dessert C, Dupré B, François LM, Schott J, Gaillardet J, Chakrapani G, Bajpai S (2001) Erosion of Deccan Traps determined by river geochemistry: impact on the global climate and the 87Sr/86Sr ratio of seawater. Earth Planet Sc Lett 188:459–474
Dessert C, Dupré B, Gaillardet J, Francois LM, Allegre CJ (2003) Basalt weathering laws and the impact of basalt weathering on the global carbon cycle. Chem Geol 202:257–273
Edwards KJ, Rogers DR, Wirsen CO, McCollom TM (2003) Isolation and charcterization of novel psychrophilic, neutrophilic, Fe-oxidizing, chemolithotrophic à- and g-Proteobacteria from the deep sea. Appl Environ Microbiol 69:2906–2913
Gislason SR, Eugster HP (1987) Meteoric water-basalt interactions. II: a field study in N.E. Iceland. Geochim Cosmochim Acta 51:2841–2855
Gomez-Alvarez V, King GM, Nüsslein K (2007) Comparative bacterial diversity in recent Hawaiian volcanic deposits of different ages. FEMS Microbiol Ecol 60:60–73
Good IJ (1953) The population frequencies of species and the estimation of population parameters. Biometrika 40:237–264
Hall K, Lindgren SB, Jackson P (2005) Rock albedo and monitoring of thermal conditions in respect of weathering: some expected and some unexpected results. Earth Surf Processes and Landf 30:801–811
Herrera A, Cockell CS (2007) Exploring microbial diversity in volcanic environments: a review of methods in DNA extraction. J Microbio Meth 70:1–12
Herrera A, Cockell CS, Self S, Blaxter M, Reitner J (2009) A cryptoendolithic community in volcanic glass. Astrobiol 9:369–382
Herrera A, Cockell CS, Self S, Blaxter M, Reitner J, Arp G, Dröse W, Thorsteinsson T, Tindle AG (2008) Bacterial colonization and weathering of terrestrial obsidian in Iceland. Geomicrobiol J 25:25–37
Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV et al (2009) Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139:485–498
Jackson CRR, Eric E, Churchill PF (2000) Denaturing gradient gel electrophoresis can fail to separate 16S rDNA fragments with multiple base differences. Mol Biol Today 1:49–51
Jacobsson SP, Gudmundson MT (2008) Subglacial and intraglacial volcanic formations in Iceland. Jokull 58:179–196
Jukes TH, Cantor CR, Munro HN (1969) Evolution of protein molecules. In: (eds) Mammalian protein metabolism. Academic Press, pp 21-132
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt EG, M. (eds) Nucleic acid techniques in bacterial systematics. pp 115-148
Legendre P, Legendre L (1998) Numerical ecology. Elsevier Science, Amsterdam
Lysnes K, Thorseth IH, Steinsbu BO, Øvreås L, Torsvik T, Pedersen RB (2004) Microbial community diversity in seafloor basalt from the Arctic spreading ridges. FEMS Microbiol Ecol 50:213–230
Mason OU, Di Meo-Savoie CA, Van Nostrand JD, Zhou J, Fisk MR, Giovannoni SJ (2008) Prokaryotic diversity, distribution, and insights into their role in biogeochemical cycling in marine basalts. ISME J 3:231–242
Muyzer G, DeWaal EC, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59:695–700
Nemergut D, Anderson S, Cleveland C, Martin A, Miller A, Seimon A, Schmidt S (2007) Microbial community succession in an unvegetated, recently deglaciated soil. Microbial Ecol 53:110–122
Nikolausz M, Sipos R, Révész S, Székely A, Márialigeti K (2005) Observation of bias associated with re-amplification of DNA isolated from denaturing gradient gels. FEMS Microbiol Lett 244:385–390
Oelkers EH, Gislason SR (2001) The mechanism, rates and consequences of basaltic glass dissolution: I. An experimental study of the dissolution rates of basaltic glass as a function of aqueous Al, Si and oxalic acid concentration at 25 °C and pH = 3 and 11. Geochim Cosmochim Acta 65:3671–3681
Orcutt B, Bailey B, Staudigel H, Tebo BM, Edwards KJ (2009) An interlaboratory comparison of 16S rRNA gene-based terminal restriction fragment length polymorphism and sequencing methods for assessing microbial diversity of seafloor basalts. Environ Microbiol 11:1728–1735
Rastogi G, Osman S, Vaishampayan P, Andersen G, Stetler L, Sani R (2010) microbial diversity in uranium mining-impacted soils as revealed by high-density 16S microarray and clone library. Microbial Ecol 59:94–108
Santelli CM, Orcutt BN, Banning E, Bach W, Moyer CL, Sogin ML, Staudigel H, Edwards KJ (2008) Abundance and diversity of microbial life in ocean crust. Nature 453:653–656
Schloss PD, Handelsman J (2005) Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol 71:1501–1506
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ et al (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541
Schwieger F, Tebbe CC (1998) A new approach to utilize PCR single-strand-conformation polymorphism for 16S rRNA gene-based microbial community analysis. Appl Environ Microbiol 64:4870–4876
Singleton DR, Furlong MA, Rathbun SL, Whitman WB (2001) Quantitative comparisons of 16S rRNA gene sequence libraries from environmental samples. Appl Environ Microbiol 67:4374–4376
Stefansson A, Gislason SR (2001) Chemical weathering of basalts, Southwest Iceland: effect of rock crystallinity and secondary minerals on chemical fluxes to the ocean. Am J Sci 301:513–556
Stroncik N, Schmincke H-U (2002) Palagonite—a review. Int J Earth Sci 91:680–697
Tamura KD, J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Tarlera S, Jangid K, Ivester AH, Whitman WB, Williams MA (2008) Microbial community succession and bacterial diversity in soils during 77,000 years of ecosystem development. FEMS Microbiol Ecol 64:129–140
Tebo BM, Johnson HA, McCarthy JK, Templeton AS (2005) Geomicrobiology of manganese(II) oxidation. Trends Microbiol 13:421–428
Templeton A, Staudigel H, Tebo B (2005) Diverse Mn(II)-oxidizing bacteria isolated from submarine basalts at Loihi Seamount. Geomicrobiol J 22:127–139
Thorseth IH, Furnes H, Tumyr O (1991) A textural and chemical study of Icelandic palagonite of varied composition and its bearing on the mechanism of the glass-palagonite transformation. Geochim Cosmochim Acta 55:731–749
Thorseth IH, Torsvik T, Torsvik V, Daae FL, Pedersen KSP (2001) Diversity of life in ocean floor basalt. Earth Planet Sc Lett 194:31–37
Wolff-Boenisch D, Gislason SR, Oelkers EH, Putnis CV (2004) The dissolution rates of natural glasses as a function of their composition at pH 4 and 10.6, and temperatures from 25 to 74 °C. Geochim Cosmochim Acta 68:4843–4858
Yergeau E, Schoondermark-Stolk SA, Brodie EL, Dejean S, DeSantis TZ, Goncalves O, Piceno YM, Andersen GL, Kowalchuk GA (2008) Environmental microarray analyses of Antarctic soil microbial communities. ISME J 3:340–351
Acknowledgements
This work was made possible and supported by the Leverhulme Trust (project number F/00 269/N). We thank Andy Tindle for the provision of the microprobe facilities (Department of Earth Science, Open University, UK). The authors are also grateful to Steve Blake and Steve Self (Earth and Environmental Sciences, Open University, UK) for helpful discussions and advice, and to Steve Summers for performing the analysis of similarity (Planetary and Space Sciences Research Institute, Open University, UK).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental Information Figure 1
ANOSIM analyis of DGGE profiles. Pal basaltic glass/palagonite samples from Valafell, Obs rhyolitic glass/obsidian samples from Dόmadalshraun. Samples ending in C are composites for the given glass type (GIF 77 kb)
Rights and permissions
About this article
Cite this article
Kelly, L.C., Cockell, C.S., Piceno, Y.M. et al. Bacterial Diversity of Weathered Terrestrial Icelandic Volcanic Glasses. Microb Ecol 60, 740–752 (2010). https://doi.org/10.1007/s00248-010-9684-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-010-9684-8