Abstract
This study reports on the factors involved in regulating the composition and structure of bacterial communities epiphytic on intertidal macroalgae, exploring their temporal variability and the role of copper pollution. Culture-independent, molecular approaches were chosen for this purpose and three host species were used as models: the ephemeral Ulva spp. (Chlorophyceae) and Scytosiphon lomentaria (Phaeophyceae) and the long-living Lessonia nigrescens (Phaeophyceae). The algae were collected from two coastal areas in Northern Chile, where the main contrast was the concentration of copper in the seawater column resulting from copper-mine waste disposals. We found a clear and strong effect in the structure of the bacterial communities associated with the algal species serving as host. The structure of the bacterial communities also varied through time. The effect of copper on the structure of the epiphytic bacterial communities was significant in Ulva spp., but not on L. nigrescens. The use of 16S rRNA gene library analysis to compare bacterial communities in Ulva revealed that they were composed of five phyla and six classes, with approximately 35 bacterial species, dominated by members of Bacteroidetes (Cytophaga-Flavobacteria-Bacteroides) and α-Proteobacteria, in both non-polluted and polluted sites. Less common groups, such as the Verrucomicrobiae, were exclusively found in polluted sites. This work shows that the structure of bacterial communities epiphytic on macroalgae is hierarchically determined by algal species > temporal changes > copper levels.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Marine macroalgae provide a sizable portion of submerged surfaces in intertidal environments; these surfaces constitute suitable substrata for bacterial colonization, including settlement and development, and several types of bacterial–algal interactions have been described. Bacteria are required for completion of different stages of algal development, including the promotion of spore settlement [34], spore germination, and correct morphogenesis in adults [12, 22, 24, 34]. Further, epiphytic bacteria produce inhibitory compounds that protect algae from colonization by other bacteria, invertebrates, or from the effects of herbivory [11, 13, 14]. The relationships described above have been mainly studied using cultured, isolated bacteria. In nature, however, diverse assemblages of bacteria colonize algae [9, 11, 17]. In this context, fewer studies have followed the effects of natural changes, like those induced by latitude or seasonality, on the structure of the macroalgal epiphytic bacterial communities [38]. For instance, spatial variations were observed in bacterial communities associated to Ulva australis, and higher variability in bacterial composition was observed in warmer (Summer and Spring) than in colder seasons (Autumn and Winter) [35]. Seasonal variations in planktonic bacterial communities do occur in marine waters and sediments, and they are associated with changes in physicochemical factors such as temperature, salinity, or pH [28]. Similarly, strong seasonal effects were reported in epibiotic bacterial communities living on corals, suggesting that the magnitude of changes is determined by the nature and composition of host exudates [15].
Conversely, the effect of stressful environmental conditions, such as metal pollution, on the structure of macroalgal epiphytic bacterial communities has received almost no attention. Metals like Cu, Cd, Pb, and Zn, are toxic to most microorganisms when present at high concentrations [10], and they induce changes in the structure of the microbial community by favoring growth of tolerant bacterial groups [23].
An important anthropogenic source of metals is mining, and copper mining waste disposals, particularly those from operations in countries with limited environmental regulations, represent a potential problem for terrestrial and marine ecosystems. The intertidal and subtidal zones in Chañaral Bay, Northern Chile, is a good example of a marine environment where large-scale copper-mine waste disposals have been poured on the coast for several decades with major negative impacts on the biota [4, 5]. Dissolved copper in these coastal waters is one or two orders of magnitude higher than in pristine areas [1]. Several studies have focused on the changes experienced by the community structure of macro-organisms conformed by assemblages of algae and invertebrates ([25] and references therein), dependent of physicochemical features of the water column, including temperature, pH, and metal speciation [1, 18, 30]. Available information on the microbial assemblages in this area is limited, and a pioneering study on the culturable fraction of the epiphytic bacteria from Ulva compressa in this area found no differences in taxa composition between polluted and non-polluted sites [29]. Recent studies, based on analysis of variability of environmental 16S rRNA gene sequences, demonstrated that bacterial community structure is mainly determined by the environment i.e. water column, sediment, rock, or algae [26].
The close spatial proximity of microorganisms to biotic surfaces generates specific intercellular interactions and creates complex and highly differentiated communities [13]. Marine microbial communities are probably sensing signal networks mediated by chemical substances produced by the algal host, which probably modulate the level of host specificity as demonstrated in bacterial communities associated to different co-occurring eukaryotic host [21] in corals [37] and sponges [17]. Some evidence indicates the presence of strikingly similar microbial communities associated to the same host but quite different between different species of hosts. As macroalgae are heterogeneous organisms in the chemical nature of their exudates, it is possible to propose that the structure of macroalgal epiphytic bacterial communities should present a species-specific relationship strongly depending on the algal species serving as host. Furthermore, and along with other factors, copper levels could modify the structure of these bacterial communities.
To test this hypothesis we compared the community structure of bacterial assemblages from different macroalgal species, and assessed the change in community structure induced by time and by copper pollution. Epiphytic bacterial communities living on algal surfaces were studied using the culture-independent terminal-restriction fragment length polymorphism (T-RFLP) and clonal analysis approaches. Three algal species from five study sites were included in the study.
Methods
Study Area
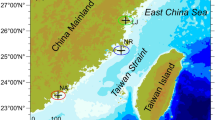
The study was carried out along 85 km of shoreline around Chañaral Bay (26° 15′ S; 69° 34′ W), Northern Chile (Fig. 1). The coast around Chañaral is located in a desert area characterized by stable environmental conditions during the year and phenomena like rain, flooding and large urban settlements and their associated wastes disposals, all of which could mask the effect of copper enrichment, are absent. Therefore, it constitutes a unique opportunity to study in isolation the effects of copper enrichment, as the principal disrupting factor, on bacterial communities in marine environments. Samples were collected in January, May, July, and October 2005, and in January, May, and October 2006 in Pan de Azúcar (26°08.2′S; 70°39.3′W), Canal Palito (discharge point), Palito 200 (26°15.8′S; 70°40.6′W), Achurra, (26°18.4′ S, 70°39.8′W) and Zenteno (26°54.1′S; 70°48.5′W). Pan de Azúcar and Zenteno are non-polluted sites with Cu+2 levels of 0.4 and 0.1 nM, respectively, considered normal for seawater [1, 30]. In contrast, Canal Palito, Palito 200, and Achurra are copper-enriched sites, with Cu+2 levels of 5.1, 4.9, and 2.5 nM, respectively [1, 30]. Parameters such as temperature, salinity, pH, dissolved oxygen, and other metals (Cd, Zn, Hg, Fe, and As) were similar for all the study sites [1, 18, 30].
Algal Species, Sampling Collection and DNA Extraction
Three algal species were selected for this study: Scytosiphon lomentaria and Lessonia nigrescens (Phaeophyceae) and Ulva spp (U. compressa + U. intestinalis, Chlorophyta). A total of 63 triplicate samples (n = 189) were obtained from three seaweed species, in seven sampling periods (within a 2-year period), and in five sampling sites. All samples included several individuals each; for Ulva and Scytosiphon 10-g wet weight, and for Lessonia three fronds, were used in each sample.
Ulva is a cosmopolitan genus abundant in a wide spectrum of coastal habitats, including the copper-enriched rocky beaches around Chañaral [25]. S. lomentaria was present only in impacted sites and L. nigrescens, which occurs naturally in control sites only [25], was also present in Palito 200 after transplanted to the site 1 year before the beginning of this study [8]. The three algal species were used to characterize and compare their epiphytic bacterial community structures during 2005. For assessing temporal variations and changes in the epiphytic communities associated with copper levels, samples of L. nigrescens and Ulva spp. were collected in 2005 and 2006. S. lomentaria was excluded from this comparative analysis because it was present only in the polluted sites and was not observed in all sampling periods. DNA extraction from the epiphytic bacterial communities was done using an UltraClean Soil DNA isolation kit (MoBio Laboratories, Solana Beach, CA, USA) based on a previous report [26]. Genomic DNA concentrations were determined by electrophoresis and comparison against a standard DNA ladder (Gene ruler™ 100-bp DNA Ladder Plus, Fermentas Life Sciences) in 1% agarose gels.
Epiphytic Bacterial Community Structure Analysis
16S rRNA gene sequences for T-RFLP analyses were amplified by PCR using the primer pairs specific for Bacteria 8F (5′ AGAGTTTGATCCTGGCTCAG 3′) and 1392R (5′ ACGGGCGGTGTGTAC 3′), using 10 ng of DNA as template in each amplification reaction. T-RFLP analyses were carried as previously described [26]; briefly, separate T-RFLP profiles were generated for each endonuclease (MspI and HhaI), which were used to estimate community indexes as richness (R′), evenness (E), and Shannon diversity (H′). All T-RFLP profiles from one endonuclease digestion/DNA sample corresponded to a data set, and each different T-RF was considered as an OTU (operational taxonomic unit). ANOVAs were used to compare these indexes, followed by post hoc Tukey’s tests. The T-RF data were analyzed with the multivariate statistical software Primer 5 (Primer-E Ltd, Plymouth, UK), and non-metric multidimensional scaling (NMDS) analyses were used to group data according to their similarity as previously described [6, 7]. One-way or two-way crossed analyses of similarity (ANOSIM) were used to examine the statistical significance of the grouping [6] and ranking of factor’s effects. A test for similarity of percentages (SIMPER) was used to calculate the contribution of individual OTUs to the similarity and dissimilarity among samples [7].
Cloning, Sequencing and Phylogenetic Analysis
Clone libraries were prepared from bacterial 16S rRNA gene amplicons obtained from epiphytic communities from Ulva, as described above. Amplicons (fragment lengths ∼1,300 bp) were ligated into the TOPO-TA vector (Invitrogen Life Technologies, Carlsbad, CA), and E. coli DH5α was transformed with the recombinant plasmids according to the manufacturer’s instructions. A total of 79 clones were analyzed and those with the correct fragment size (42) were sequenced with the 1392R primer, using the ABI PRISM Big Dye Terminator Cycle Sequencing Kit and ABI3100 Genetic Analyzer, according to the manufacturer’s protocol (Applied Biosystems Inc. Foster City, CA, USA). Partial sequences of 16S rRNA gene were used for phylogenetic analyses. To determine their approximate phylogenetic affiliation these sequences (∼795 to 914 bp) were aligned with those available in GenBank using CLUSTAL X software. Phylogenetic analyses with PAUP* software used a neighbor-joining algorithm (distance analyses), and insertions and deletions (gaps) were treated as sites of new stage of character. For tree construction, the genera and their accession numbers were: Aquimarina (AM990846.1), Flexibacter (AB078070.1), Lewinella (EU371935.1), Erytrobacter (AY739662.1), Sulfitobacter (EF202614.1), Granulosicoccus (EF495228.1), Saccharophagus (AF055269.1), Rhodopirellula (EF589354.1), Rubritalea (AB297806.1), Verrucomicrobiae (AM25983.1), Stanieria (AB039009.1), and (as outgroup) Sulfolobus metallicus (EU419200.1). Bootstrap re-sampling was applied to assess support for individual nodes using 1,000 bootstrap replicates. The 16S rRNA gene sequences reported in this study have been deposited in the GenBank database under the accession numbers GQ247326 to GQ247367.
Results
The Structure of the Epiphytic Bacterial Community is Strongly Host-Dependent
No significant changes in total richness, diversity, and evenness indexes were observed in bacterial communities from the same algal host and among bacterial communities from different algal species, independently of sampling sites and sampling times, with the exception of Scytosiphon in Palito (Electronic Supplementary Material, Table S1, Fig. S1). Nevertheless, clearly distinguishable T-RFLP profiles, showing the epiphytic bacterial community structure, were observed for the three algal species in digestions obtained with the endonucleases MspI and HhaI (representative T-RFLP profiles are shown in Fig. 2).
Representative T-RFLP profiles of epiphytic bacterial communities of three algal species from Palito 200, sampled in October 2005. T-RFLP profiles were obtained with HhaI endonuclease DNA digestion. Fragment sizes found in only one algal species are marked with an asterisk and fragment sizes shared by the three species are in boldface
However, when the three algal species were compared without considering their origin (i.e. polluted or non-polluted sites), the structure of their epiphytic bacterial communities differed significantly. Comparison of the bacterial community structures by NMDS analyses showed, in all cases, moderate to highly significant differences for the three algal species: R MspI/HhaI = 0.62/0.67 in May, R = 0.33/0.37 in July, and R = 0.55/0.56 in October (all with p = 0.001). Data for May and October 2005 (Fig. 3) clearly indicated that epiphytic bacterial communities were more similar within an algal host than between algal hosts. SIMPER analyses used to identify which OTUs made a higher contribution to the differences between these bacterial community structures showed that the average values for dissimilarities between bacterial communities associated with the brown algae Lessonia and Scytosiphon (58.2%) were lower than for Ulva and Lessonia (79.1%) or Ulva and Scytosiphon (86.2%) (data not shown). The main dissimilarities were due to changes in the relative abundances of a few OTUs that are present in all communities, represented by T-RFs of 83, 93, 193, 212, 239, 354, 571, and 583 bp (Fig. 2, in bold). As an example, T-RF of 212 bp represented ∼17% of the relative abundances of the bacterial fragment sizes found in the community from Ulva, but only 0.8% of that in Lessonia and 3.2% in Scytosiphon. Other OTUs were exclusively observed in a particular host (T-RFs marked with an asterisk in Fig. 2), and thus contributing to enhance inter-host differences in bacterial community structure.
3D-NMDS plots of T-RFLP data profiles of bacterial communities associated to three algal species. The plots correspond to samples collected in May 2005 (upper) and October 2005 (bottom). ■ L. nigrescens, ▼Ulva spp, ● S. lomentaria. T-RFLP data correspond to bacterial community DNA digested with HhaI endonuclease. All sampling sites were included in every case
The Structure of These Epiphytic Bacterial Communities is Also Modified by Temporality, But Not Locality
As the algal species was the main factor determining the structure of the bacterial communities, the T-RFLP profiles for bacterial communities from each host species were analyzed separately. The T-RFLP data sets were first compared by ANOSIM using year as an ordering factor. These comparisons showed significant (p = 0.001 in all cases) inter-annual variations with R MspI/HhaI = 0.56/0.47 for Ulva, and R = 0.61/0.51 for Lessonia. S. lomentaria was not included in this part of the study, because it was present only in the polluted sites and was not observed in all sampling periods.
Significant intra-annual variations were also found using month as ordering factor (Fig. 4). For bacterial communities from Ulva spp., ANOSIM showed R MspI/HhaI = 0.67/0.89 (p = 0.001) for samples taken in 2005, and R = 0.63/0.90 (p = 0.001) for 2006 (data not shown). Values for epiphytic bacterial communities on L. nigrescens were R = 0.90/0.94 (p = 0.001) for 2005 and R = 0.76/0.63 (p = 0.001) for 2006 (data not shown). These observations indicate a strong temporal effect on the structure of the bacterial communities living on the surface of both hosts.
2D-NMDS plots for T-RFLP data from bacterial communities from Ulva spp. and L. nigrescens in 2005. Plots were obtained with data from bacterial community DNA digested with MspI endonuclease. Triangle, January; inverted triangle, May; square, July; and diamond, October. All sampling sites were included in every case. R-rank values correspond to R values obtained for all comparisons performed by ANOSIM analyses
Comparisons using locality as ordering factor did not show a significant difference for epiphytic bacterial communities from Ulva (R MspI/HhaI = 0.049/−0.009 in samples taken in 2005, and 0.103/0.059 in 2006, p > 0.05) and Lessonia (R MspI/HhaI = 0.014/0.005 in 2005, and 0.094/0.242 in 2006, p > 0.05).
Copper Enrichment Produces Shifts in the Epiphytic Bacterial Communities
In order to analyze the effects of copper on the structure of epiphytic bacterial communities living on Ulva spp. or on L. nigrescens, T-RFLP data sets from each month were analyzed separately, and comparisons using polluted/non-polluted as ordering factor were done. NMDS plots for bacterial communities from Ulva showed significant differences between control and copper-enriched sites (Fig. 5). In contrast, high levels of copper did not modify the structure of the bacterial community associated to L. nigrescens (Fig. 5). SIMPER analyses showed that the main contribution to distinguish the communities from Ulva were OTUs with fragment sizes of 441, 507, and 497 bp, which exhibited different relative abundances depending on whether the host was collected in a pristine or in a copper-enriched site. Other T-RFs with low representation were exclusive for either control sites or for impacted sites, as T-RF of 386 bp and T-RF of 358 bp, respectively. For Lessonia communities, T-RFs of 497, 507, and 84 bp possessed the highest contribution. Nevertheless, they did not show significant differences between their relative abundances in control and impacted sites (data not shown).
3D-NMDS plots for T-RFLP data from epiphytic bacterial communities living on Ulva spp. and on L. nigrescens from control and copper-impacted sites. Symbols represent control (open) and copper-enriched sites (closed). T-RFLP data were obtained from bacterial community DNA digested with MspI endonuclease. R-rank values correspond to ANOSIM analyses for multiple comparisons with MspI and HhaI, from samples obtained in 2005 and 2006
Considering that algal host was the main factor determining the community structure of epiphytic bacteria, the hierarchy among sampling month and copper level was assessed for the two species by two-way crossed ANOSIM tests. The results showed significant differences in the community structure of epiphytic bacteria on Ulva and Lessonia, among months (0.596 ≤ R MspI/HhaI ≤ 0.971, p < 0.001) and copper levels (0.228 ≤ R MspI/HhaI ≤ 0.949, p ≤ 0.005), showing that month effect was higher than copper effect for all analyses (Electronic Supplementary Material Table S2).
The Specific Bacteria Contributing Significantly to the Epiphytic Communities in Ulva Are Different in Copper-Polluted and Non-polluted Environments
To identify the bacterial taxa with higher influence on the structure of the epiphytic community on Ulva and also those responsible for the differences between communities from non-polluted and polluted sites, 16S rRNA gene libraries were constructed. A total of 35 ribotypes were identified from 79 clones analyzed, and a BLAST search was used to determine their closest sequence available in GenBank database. These sequences were affiliated to six taxonomic categories: CFB (Cytophaga-Flavobacteria-Bacteroidetes), α-Proteobacteria, γ-Proteobacteria, Verrucomicrobiae, Planctomycetes, and Cyanobacteria. Members of CFB (16 ribotypes represented by 49.3% of the clones) and α-Proteobacteria (six ribotypes, 15.2% of the clones), followed by Verrucomicrobiae (three ribotypes, 5.1% of the clones), and γ-Proteobacteria (two ribotypes, 8.9% of the clones) dominate bacterial communities. The less-abundant groups were Cyanobacteria and Planctomycetes (both with one ribotype, 1.3% of the clones). Fifteen clones distributed in six ribotypes (19% of total clones) were classified by BLAST as uncultured bacterium, and they were added to a bacterial 16S rRNA gene-sequence tree in order to visualize their relationships (Fig. 6). Based on phylogenetic analyses the uncultured clones were assigned to: CFB (one ribotype, 8.9%, seven clones), α-Proteobacteria (four ribotypes, 8.9%, seven clones), and Planctomycetes (one ribotype, 1.3%, one clone). Of all clones analyzed, 19 ribotypes showed sequences with similarity values higher than 97%, and 16 ribotypes presented sequences with similarities between 89 and 96%. Taxa belonging to Verrucomicrobiae, on the other hand, showed values close to 92%.
Neighbor-joining tree of phylogenetic relationships, based on 16S rRNA gene, of epiphytic bacterial communities from non-polluted (left) and polluted sites (right). The designations in bold correspond to different ribotypes obtained from sequenced clones. The number of clones for each ribotype is indicated in parenthesis, and asterisks show exclusive ribotypes for each site. The names in italic correspond to sequences retrieved from the NCBI database. Values on the nodes indicate the bootstrap percentages. I, CFB; II, Proteobacteria; IIa, γ-Proteobacteria; IIb, α-Proteobacteria; III, Planctomycetes; IV, Verrucomicrobiae; and V, Cyanobacteria
The comparison between non-polluted and polluted sites showed a total of 16 ribotypes present in non-polluted and 24 in polluted sites. Of them, 11 and 17 unique ribotypes were found in non-polluted and polluted sites, respectively. Seven ribotypes were shared for both sites, of which 17.7% of clones (five ribotypes; 14 clones) corresponded to α-Proteobacteria and 13.9% (two ribotypes, 11 clones) corresponded to CFB.
Discussion
This study demonstrates that the composition and structure of the studied epiphytic bacterial communities are strongly dependent on the algal host, on the time of sampling and, to a lesser degree, on copper pollution.
Before discussing specific points it is important to highlight that classical–ecological indexes such as richness and diversity, useful to characterize communities of macro-organisms were, at least for the bacterial communities reported in this study, much less informative. The number of OTUs and Shannon’s index values were not significantly different between the bacterial communities living on different macroalgae. It is possible that the same T-RFs correspond to separate taxa, therefore similar diversity and richness values could be produced by different bacterial species compositions. Unfortunately, the T-RFLP method is not appropriate to make reliable taxonomic assignments but, however, is quite suitable to make bacterial community structure comparisons. So far, the most robust alternative to study the structure of bacterial communities is to use a combination of tools, including parametric, nonparametric, and phylogenetic analyses [3].
The main factor controlling the composition and structure of these epiphytic bacterial communities is algal host. Our results are in agreement with previous studies in the interaction of marine microorganisms and sessile eukaryotes (e.g. coral or sponges), that demonstrated an important degree of host specificity, including the recruitment of different epibiotic communities by different host species [17, 36]. The nature of the interactions between macroalgae and their bacterial communities is practically unknown, and physical and chemical mechanisms have been proposed to regulate the phenomenon of epibiosis; 1) some algae slough-off external cells and parts of cell wall, cleaning themselves from epibionts [27]; 2) chemical signals from the host, from the first colonizing bacteria, or both, can act as deterrent or attractors for other bacteria or epibionts mediating bacteria settlement on algal surfaces [31]; and 3) cell walls and cuticles of seaweeds are chemically and structurally complex, and the proportion of their components change among algal species.
The processes regulating the dynamics of epiphytic bacterial communities are also hardly understood. Although some evidence shows that variability in the quantity and type of epibiosis on a given host is caused by the seasonal availability of microbial colonizers in the environment [36], physiological changes in the host or additional interactions such as allelopathy and symbiosis, may also influence community structure and bacterial function [33]. Algae are organisms living in changing environments, in particular those inhabiting intertidal habitats, which makes reasonable to suggest that their overall physiology and particularly its chemical signaling behavior exhibits temporal, but also species-dependent fluctuations [16]. The results of this study show that bacterial communities associated with macroalgae are dynamic and, in Ulva spp. and L. nigrescens, display significant variations in time. The variations in time are mainly due to changes in the relative abundances of a stable fraction of the community represented by the dominant T-RFs and a minor proportion of a variable fraction of the community represented by the presence of T-RFs with low relative abundances. These results are in agreement with previous observations on bacterial communities from U. australis, where samples collected in different seasons shared almost 60% of the OTUs [35]. Seasonal variations in community structure of bacteria have been previously reported for other microbial communities from sediments, seawater, freshwater, and soils [19, 28].
Some abiotic factors such as temperature, salinity, dissolved oxygen, pH, and the presence of specific metals have been also reported as agents able to modify the structure of bacterial communities in marine environments [30]. In the study area selected for this work, it has been well-documented that high copper concentration is the main differential factor between the coastal zone in and around Chañaral, where the metal concentrations are higher than normal and do not vary significantly throughout the year [1, 18, 30]. Indeed, copper has a differential effect on these bacterial communities. For example, epiphytic communities associated to Ulva spp. from non-polluted and polluted sites were significantly different. However, it is important to note that Ulva compressa and U. intestinalis are two species hardly distinguishable in field, and the proportion of each one in our study sites could be influencing the differences observed in the community structure. Nevertheless, our results indicate a probable selective pressure exerted by the long-lasting copper enrichment on members of the bacterial community available to colonize Ulva in polluted sites. In the case of L. nigrescens, which occurs naturally only in non-polluted sites (the algal samples from the impacted sites came from individuals transplanted from a non-polluted site 1 year before this study began), the community structures were highly similar in all sites (Electronic Supplementary Material, Table S3) and no significant differences between epiphytic communities from pristine and polluted sites were observed (data not shown). Furthermore, the lack of differences in structure between Lessonia epiphytic communities observed in samples exposed to different levels of copper suggests a low degree of bacterial colonization or lack of settlement of new bacterial populations on Lessonia fronds in polluted sites (Electronic Supplementary Material, Table S3 and S4). In addition, differences between these two algal hosts are probably due to different physiological responses to copper exposure. Each host may respond to environmental stress produced by copper by exudating different kinds of metabolites or ligands, thus modifying the availability and therefore, the toxicity of metals, providing a micro-environment that favors or selectively inhibits the growth of different bacterial species [20]. When Lessonia is exposed to copper, it produces concentrations of copper complexing ligands around three times higher than controls. In similar conditions, however, the release of ligands is not stimulated in Ulva [2]. Therefore, Lessonia has the capacity to rapidly respond to copper excess by producing organic ligands that increase the complexing capacity of the water, which in turn attenuates the level of labile (i.e. toxic) copper [2]. This cascade of effects may allow the existence of similar bacterial communities in pristine and polluted sites.
In Ulva communities, α-Proteobacteria and Bacteroidetes (Cytophaga-Flavobacteria-Bacteroides) were the dominant groups in hosts from polluted and non-polluted sites. In pristine environments, α-Proteobacteria is the dominant group among epiphytic communities of U. australis [35]. Bacteria assigned to CFB have been reported as the main modulators of morphogenesis in various species of Ulva [22]. Therefore, it is not surprising that they were the dominant members in these communities. Albeit, bacteria classified at higher taxonomic levels were essentially the same in polluted and non-polluted environments, their specific composition is different, as shown by T-RFLP’s and ribotypes obtained with clonal analyses. In this context, we observed a group of low abundance but specific to polluted sites; Verrucomicrobiae is represented by three different sequences with relatively low percentage of similarity (∼92%) to sequences available in databases. The closest genus for these sequences was Rubritalea, a little known chemoheterotrophic marine bacteria isolated from sediments and sponges that has never been reported before in association with macroalgae. The ecological information available in the literature is scarce, and additional efforts are required to establish the relationship between Rubritalea and copper-enriched environments. Furthermore, BLAST analysis of sequences of bacteria epiphytic on Ulva revealed mostly heterotrophic bacteria, which could be potentially using polysaccharides or dissolved organic matter produced by algae as carbon source [32], required in intertidal oligotrophic environments. Regarding the latter, the nature of exudates produced by marine macroalgae is almost unknown, and their chemical identification and characterization are required for a better understanding of the mechanisms underlying the interactions between algal hosts and their epiphytic communities.
Finally, the results of clonal analyses generally agree with T-RFLP results regarding total number of ribotypes present in the epiphytic community on Ulva. However, the clonal approach discloses the presence of a high number of exclusive clones for each site, which suggests that copper excess could favor growth of specific OTUs and inhibit others, as indicated by different bacterial community structures associated to the same algal host, but growing in different environmental conditions. We conducted additional BLAST analyses in order to find copper resistance mechanisms associated to genera identified in this study. However, we did not find a relation between clones from polluted sites and specific-resistance mechanisms, since these appear to be widely distributed in clones from both types of communities.
References
Andrade S, Moffett J, Correa JA (2006) Distribution of dissolved species and suspended particulate copper in an intertidal ecosystem affected by copper mine tailings in northern Chile. Mar Chem 101:203–212
Andrade S, Pulido MJ, Correa JA (2009) The effect of organic ligands exuded by intertidal seaweeds on copper complexation. Chemosphere 78:397–401
Bohannan B, Hughes J (2003) New approaches to analyzing microbial biodiversity data. Curr Opin Microbiol 6:282–287
Castilla JC (1983) Environmental impact in sandy beaches of copper mine tailings at Chañaral, Chile. Mar Pollut Bull 14:459–464
Castilla JC, Nealler E (1978) Marine environmental impacts due to mining activities of El Salvador copper mine, Chile. Mar Pollut Bull 9:67–70
Clarke KR (1993) Non-parametric multivariate analyses of changes in community structure. Aust J Ecol 19:117–143
Clarke KR, Warwick RM (1994) Change in marine communities: an approach to statistical analysis and interpretation, 2nd edn. Plymouth Marine Laboratory, Plymouth
Correa JA, Lagos NA, Medina MH, Castilla JC, Cerda M, Ramírez M, Martínez E, Faugueron S, Andrade S, Pinto R, Contreras L (2006) Experimental transplants of the large kelp Lessonia nigrescens (Phaeophyceae) in high-energy wave exposed rocky intertidal habitats of northern Chile: experimental, restoration and management applications. J Exp Mar Biol Ecol 335:13–18
Dobretsov SV, Qian PY (2002) Effect of bacteria associated with the green alga Ulva reticulata on marine micro- and macrofouling. Biofouling 18:217–228
Dryden CL, Gordon AS, Donat JR (2004) Interactive regulation of dissolved copper toxicity by an estuarine microbial community. Limnol Oceanogr 49:1115–1122
Egan S, Thomas T, Holmström C, Kjelleberg S (2000) Phylogenetic relationship and antifouling activity of bacterial epiphytes from the marine alga Ulva lactuca. Environ Microbiol 2:343–347
Egan S, James S, Holmström C, Kjelleberg S (2001) Inhibition of algal spore germination by the marine bacterium Pseudoalteromonas tunicata. FEMS Microbiol Ecol 35:67–73
Egan S, Torsten T, Kjelleberg S (2008) Unlocking the diversity and biotechnological potential of marine surface associated microbial communities. Curr Opin Microbiol 11:219–225
Engel S, Jensen PR, Fenical W (2002) Chemical ecology of marine microbial defense. J Chem Ecol 28:1971–1985
Guppy R, Bythell JC (2006) Environmental effects on the bacterial diversity in the surface mucus layer of the reef coral Montastraea faveolata. Mar Ecol Prog Ser 328:133–142
Hellio C, Marechal JP, Veron B, Bremen G, Clare AS, Le Gal Y (2004) Seasonal variation of antifouling activities of marine algae from the Brittany Coast (France). Mar Biotechnol 6:67–82
Hentschel U, Hopke J, Horn M, Friedrich A, Wagner M, Hacker J, Moore B (2002) Molecular evidence for a uniform microbial community in sponges from different oceans. Appl Environ Microbiol 68:4431–4440
Lee MR, Correa JA, Zhang H (2002) Effective metal concentrations in porewater and seawater labile metal concentrations associated with copper mine tailings disposal into the coastal waters of the Atacama region of northern Chile. Mar Pollut Bull 44:956–976
Lipson DA (2007) Relationships between temperature responses and bacterial community structure along seasonal and altitudinal gradients. FEMS Microbiol Ecol 59:418–427
Lombardi AT (2000) Copper complexation by cyanophyta and chlorophyta exudates. Phycologia 39:118–135
Longford S, Tujula NA, Crocetti G, Holmes AJ, Holmström C, Kjelleberg S, Steinberg PD, Taylor MW (2007) Comparison of diversity of bacterial communities associated with three sessile marine eukaryotes. Aquat Microb Ecol 48:217–229
Marshall K, Joint I, Callow ME, Callow JA (2006) Effect of marine bacterial isolates on the growth and morphology of axenic plantlets of the green alga Ulva linza. Microb Ecol 52:302–310
Massieux B, Boivin ME, van den Ende FP, Langenskiöld J, Marvan P, Barranguet C, Admiraal W, Laanbroek HJ, Zwart G (2004) Analysis of structural and physiological profiles to assess the effects of Cu on biofilm microbial communities. Appl Environ Microbiol 70:4512–4521
Matsuo Y, Suzuki M, Kasai H, Shizuri Y, Harayama S (2003) Isolation and phylogenetic characterization of bacteria capable of inducing differentiation in the green alga Monostroma oxyspermum. Environ Microbiol 5:25–35
Medina M, Andrade S, Faugeron S, Lagos N, Mella D, Correa JA (2005) Biodiversity of rocky intertidal benthic communities associated with copper mine tailing discharges in northern Chile. Mar Pollut Bull 50:396–409
Morán AC, Hengst MB, De la Iglesia R, Andrade S, Correa JA, González B (2008) Changes in bacterial community structure associated with coastal copper enrichment. Environ Toxicol Chem 27:2239–2245
Nylund GM, Pavia H (2005) Chemical versus mechanical inhibition of fouling in the red alga Dilsea carnosa. Mar Ecol Prog Ser 299:111–121
Osgood MP, Boylen CW (1990) Seasonal variations in bacterial communities in Adirondack streams exhibiting pH gradients. Microb Ecol 20:211–230
Riquelme C, Rojas A, Flores V, Correa JA (1997) Epiphytic bacteria in a copper-enriched environment in Northern Chile. Mar Pollut Bull 34:816–820
Stauber JL, Andrade S, Ramírez M, Adams M, Correa JA (2005) Copper bioavailability in a coastal environment of northern Chile: comparison of bioassay and analytical speciation approaches. Mar Pollut Bull 50:1363–1372
Steinberg PD, de Nys R (2002) Chemical mediation of colonization of seaweed surfaces. J Phycol 38:621–629
Stewart FJ, Fritzen CH (2004) Bacteria–algae relationships in Antarctic sea ice. Antarct Sci 16:143–156
Strom S (2008) Microbial ecology of ocean biogeochemistry: a community perspective. Science 320:1043–1045
Tait K, Joint I, Daykin M, Milton DL, Williams P, Cámara M (2005) Disruption of quorum sensing in seawater abolishes attraction of zoospores of the green alga Ulva to bacterial biofilms. Environ Microbiol 7:229–240
Tujula NA, Crocetti GR, Burke C, Thomas T, Holmström C, Kjelleberg S (2009) Variability and abundance of the epiphytic bacterial community associated with a green marine Ulvacean alga. ISME J. doi:10.1038/ismej.2009.107
Wahl M (2008) Ecological lever and interface ecology: epibiosis modulates the interactions between host and environment. Biofouling 24:427–438
Webster NS, Bourne D (2007) Bacterial community structure associated with the Antarctic soft coral, Alycyonium antarcticum. FEMS Microbiol Ecol 59:81–94
Yannarell AC, Triplett EW (2005) Geographic and environmental sources of variation in lake bacterial community composition. Appl Environ Microbiol 71:227–239
Acknowledgments
This study is part of the research program FONDAP 1501 0001 funded by CONICYT, to the Center for Advanced Studies in Ecology & Biodiversity (CASEB) Program 7. Additional support was provided by the Millennium Scientific Initiative through the Millennium Nucleus EMBA grant P/04-007-F; the grants Marine Genomics-CONICYT, FUNDACION ANDES (C-13851), PBCT RED-12, and ICA grant (to JAC). M.B. Hengst was supported by a CONICYT Ph.D. fellowship. We deeply thank to two anonymous referees for their contributions to improve an early version of this paper. Thanks to Jessica Beltran, Carolina Camus, and Alejandra González for their assistance in the field.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Figure S1
Average values of Richness (up) and Shannon diversity indexes (low) for epiphytic bacterial communities of Ulva, Lessonia, and Scytosiphon, obtained from T-RFLP profiles with MspI endonuclease digestion. The asterisk indicates significant differences between richness values for communities from Scytosiphon with respect to Ulva and Lessonia for algae from Palito 200, obtained with a posteriori Tukey’s test. Diamond, Ulva; square, Lessonia; and triangle, Scytosiphon. PA, Pan de Azúcar; Z, Zenteno; C, Canal; P, Palito 200; and A, Achurra (DOC 31.5 kb)
Table S1
One-way ANOSIM results for comparisons between different sampling periods and localities, from richness, diversity, and evenness values obtained from T-RFLP data digested with MspI, for epiphytic communities associated to Ulva and Lessonia (DOC 38.0 kb)
Table S2
Summary of the two-way crossed ANOSIM test based on Bray–Curtis similarity matrices derived from abundance data square-root transformed to determine the effect of month and copper level on the community structure of bacterial epiphytes associated with Ulva and Lessonia, during 2005 and 2006 (DOC 31.0 kb)
Table S3
Comparisons of epiphytic communities of Lessonia from pristine and copper-polluted sites. Data from T-RFLP were obtained by digestions with HhaI and MspI, and similarity values between sites were obtained with square-root transformed data by SIMPER analysis (DOC 42.0 kb)
Table S4
T-RFs contribution to similarity of epiphytic community structure of Lessonia in pristine and copper-polluted sites, obtained by SIMPER analysis (DOC 80.5 kb)
Rights and permissions
About this article
Cite this article
Hengst, M.B., Andrade, S., González, B. et al. Changes in Epiphytic Bacterial Communities of Intertidal Seaweeds Modulated by Host, Temporality, and Copper Enrichment. Microb Ecol 60, 282–290 (2010). https://doi.org/10.1007/s00248-010-9647-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-010-9647-0