Abstract
The morphology of endolithic colonization in a limestone escarpment and surrounding rocky debris (termed float) at a high-altitude arid site in central Tibet was documented using scanning electron microscopy. Putative lichenized structures and extensive coccoid bacterial colonization were observed. Absolute and relative abundance of rRNA gene signatures using real-time quantitative polymerase chain reaction and phylogenetic analysis of environmental phylotypes were used to characterize community structure across all domains. Escarpment endoliths were dominated by eukaryotic phylotypes suggestive of lichenised associations (a Trebouxia lichen phycobiont and Leptodontidium lichen mycobiont), whereas float endoliths were dominated by bacterial phylotypes, including the cyanobacterium Chroococcidiopsis plus several unidentified beta proteobacteria and crenarchaea. Among a range of abiotic variables tested, ultraviolet (UV) transmittance by rock substrates was the factor best able to explain differences in community structure, with eukaryotic lichen phylotypes more abundant under conditions of greater UV-exposure compared to prokaryotes. Variously pigmented float rocks did not support significantly different communities. Estimates of in situ carbon fixation based upon 14C radio-labelled bicarbonate uptake indicated endolithic productivity of approximately 2.01 g C/m2/year−1, intermediate between estimates for Antarctic and temperate communities.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Porous sedimentary rocks such as limestone and sandstone are a major feature of terrestrial environments worldwide. Exposed outcrops are characterized by escarpments (steeply elevated slopes) that over time become surrounded by rocky debris that detach from the scarp due to weathering; these accumulations of debris are termed float. The pore spaces of such rocks provide a sheltered niche for microbial colonization, and such microorganisms are termed endoliths. Endolithic microbial colonization of porous rocks such as limestone and sandstone has been recorded as a feature of several hot and cold arid environments ranging from the Antarctic Dry Valleys to hot deserts such as the Negev in Israel [6]. Endolithic colonization has also been reported for rocky outcrops in alpine and semi-arid areas [10, 22]. Endoliths are assumed to be the main source of primary production in hyper-arid environments where plants are rare or infrequently encountered [9]. They have also been implicated in geobiological phenomena such as bioweathering of rock [2]. Endoliths have also gained attention from astrobiologists due to their potential as analogs for possible life on Mars [15].
Relatively few studies have reported the microbial composition of endolithic communities. The pioneering work of Imre Friedmann led to observations that endolithic colonization by the cyanobacterium Chroococcidiopsis was ubiquitous in deserts worldwide, although relatively infrequent in polar regions [6]. Friedmann identified two compositional classes of Antarctic endoliths: cyanobacteria-dominated and lichenised [7]. Endolithic-lichenised communities typically comprised the chlorophyte Trebouxia and unidentified fungal symbionts [7]. It has been postulated that the absence of endolithic eukaryotic lichens in hot deserts was due to greater environmental stresses in these locations [6].
The advent of molecular tools to resolve community molecular diversity in culture-independent studies has allowed resolution of far greater diversity than was previously appreciated by morphological and cultivation studies [11]. A polymerase chain reaction (PCR) study of Antarctic sandstone endoliths using ‘universal’ 16S/18S rRNA gene primers revealed complex communities of algae, fungi, and bacteria [4]. A lichen-dominated community comprised around 70% algal and fungal phylotypes, while the dominant phylotypes in a cyanobacterial endolithic community were identified as cyanobacteria, α-proteobacteria, and deinococci. Archaea were not detected despite the use of primers specifically targeting this domain. Other studies of endoliths in non-extreme environments have focused upon the phylum Cyanobacteria. They revealed a broad range of filamentous and unicellular cyanobacterial taxa [17, 22]. A study of endoliths from limestone, sandstone, and granite cliffs in a semi-arid habitat showed that archaea, bacteria, and algae were present but varying widely in abundance with different locations and rock types [26]. Fungal phylotypes were not reported. Together, these studies suggest that considerable variation in endolith community composition from different climatic and geographic locations is likely.
Large expanses of high-altitude cold desert occur on the Tibetan plateau at altitudes above 4,500 m. Potential endolithic habitats occur in extensive outcrops of porous limestone rock that occur with extensive float deposition around escarpments. An investigation of endolithic microbiology in this region is of considerable relevance in that such as study may help address issues of biogeographic variation and climatic effects on endolithic communities. Here, we report a study of morphology, multi-domain phylogenetic diversity, and productivity for Tibetan endolithon.
Materials and Methods
Field Sampling
The field location was a large limestone outcrop in central Tibet near the town of Gertze (N32°11.546′, E84°12.001′–N32°12.992′, E84°11.926′, altitude 4,651–4,953 m). The region is characterized as a cold desert [12], with an arid landscape experiencing long winter periods where temperatures remain well below freezing. Brief summers are typically arid although snowmelt and rainfall do create localized areas of moisture sufficiency. Sampling was carried out in August 2004. Escarpments (scarp) were surrounded at their base by extensive deposits of small boulders and rocks derived from the escarpment (termed ‘float’ by geologists). This float was the most abundant feature of the surrounding terrain and extended up to approximately 100 m from the base of the escarpment as a dense rocky pavement. A visual inspection in the field revealed float-comprised rocks with a distinct gray, pink, or white pigmentation, and these were treated as separate substrates in the field for sampling purposes. Colonized float and escarpment rock was visually detected in the field after fracturing with a geologists’ hammer and observing the typical green band of microbial growth. For a rough estimate of colonization frequency, float substrates were examined for colonization (presence/absence) at 1 m intervals along a 20 × 1 m belt transect and assessed for colonization. For estimates of colonization in escarpment, 20 samples were chiseled from the rock to a depth of approximately 5 cm (beyond the maximum colonization depth for limestone scarp endoliths) at approximately 1-m intervals along a parallel transect at heights of approximately 1–5 m depending on accessibility (precise intervals between sampling in such terrain was not possible). The use of belt transects is the most favored approach to sample lithic microbial colonization frequency [28]. Three colonized rock samples from each substrate (judged to be the most representative based upon visual examination) were collected aseptically, placed in sterilized (gamma irradiated) zip-lock bags and stored at ambient temperature (at or near freezing) in darkness until processed (approximately 2 weeks drive from the remote field location). Upon return to the laboratory, rock samples were re-fractured to expose ‘fresh’ colonized surfaces.
Microscopy
Microscopic examination was carried out using scanning electron microscopy (Stereoscan 440, Leica, Cambridge, UK). Colonized limestone fractures were fixed in 2.5% glutaraldehyde for 8 h, air-dried overnight, and then gold-sputter-coated for 30 s (SCD 005, BAL-TEC, Lichtenstein) prior to visual examination.
Recovery of Environmental DNA
Total DNA was recovered by lysis in CTAB (cetyltrimethylammonium bromide) with lysozyme and RNAse, followed by phenol:chloroform extraction at 60°C. Genomic DNA was checked for quality by electrophoresis in 1% agarose gels and quantified by spectrophotometry (Smartspec Plus, Bio-Rad, Hercules, CA, USA).
Real-time Quantitative PCR (qPCR)
PCR amplification was quantified in real-time (Applied Biosystems prism 7000, Foster City, CA, USA) by flourometric monitoring with SYBR Green 1 dye (Invitrogen, Carlsbad, CA, USA). All standard curves were constructed using plasmids from cloned rRNA genes (Qiagen, La Jolla, CA, USA) for Archaea, Bacteria, and Eukarya. The number of copies in standards was calculated using the Zbio.net on-line converter (http://www.molbiol.ru/eng/scripts/01_07.html). Slopes of the standard curves generated were −3.03, −3.21, and −3.28 for Archaea, Bacteria, and Eukarya, respectively. All three standard curves achieved a high correlation coefficient (>0.99). Quantification of genes in each sample was performed in triplicate. Dissociation curves were studied for each run to ensure the threshold cycle (Ct) reflected efficient and specific amplification. The absolute copy number of genes was obtained by interpolation from the respective standard curves.
Denaturing Gradient Gel Electrophoresis (DGGE)
16S rRNA genes were PCR amplified using primers specific to Archaea, Bacteria, and cyanobacteria as previously described [13]. Eukaryotic 18S rRNA genes were PCR amplified using universal eukaryal primers as previously described [29]. Standardized DNA loads (2 μg amplified DNA) were separated by denaturing gradient gel electrophoresis (DGGE) [16] in a urea/formamide denaturing gradient in a 7% acrylamide gel, run at 150 V in 1X TAE buffer (pH 8.0) at 60°C (DGGE-2001, CBS Scientific, Solana Beach, CA, USA). DGGE conditions were optimized as follows: Archaea: 0–75% gradient, 7 h electrophoresis; Bacteria: 0–85%, 9 h; Eukarya: 0–65%, 8 h; Cyanobacteria: 0–85%, 8.5 and 10 h. All visible bands were excised, soaked overnight in deionized water at 4°C followed by re-amplification and purification (GFX, Amersham, Bucks, UK).
DNA Sequencing and Phylogenetic Analysis
Purified DGGE band amplicons were sequenced using the BigDye Terminator Cycle Sequencing kit (ABI 3730 Genetic Analyzer, Applied Biosystems, Foster City, CA, USA). All sequences generated by this study have been deposited in the National Center for Biotechnology Information (NCBI) GenBank database under accession numbers FJ489993 to FJ490055. Approximate phylogentic affiliations were determined by BLAST searches of the NCBI GenBank database (http://www.ncbi.nlm.nic.gov/). Screening for possible chimeric sequences was made using Chimera_Check available on the Ribosome Database Project website (http://rdp.cme.msu.edu.html). Multiple alignments were created with reference to selected GenBank sequences using Clustal X v.1.81 [25]. Maximum likelihood analysis was performed using PAUP* 4.0b8 [24]. Bayesian posterior probabilities [20] and bootstrap values (1,000 replications) are shown for branch nodes supported by more than 50% of the trees.
Measurement of Substrate Characteristics
On-site measurement of photosynthetically active radiation (PAR), ultraviolet (UV)-A, and UV-B radiation was carried out at solar noon using a Li-Cor LI-I400 datalogger (Li-Cor Inc, Nebraska) for PAR and a UV-UVX radiometer (UVP Inc, Upland, CA, USA) for UV irradiance. For light transmittance studies, thin sections corresponding to the depth of endolithic colonization in each substrate were prepared (Petro-Thin Thin Sectioning System, Buehler, Lake Bluff, IL, USA) and polished using silica carbide powder of 180, 300, and 600 grit. Rock sections were positioned on light sensors, and light transmission measurements were recorded at solar noon. All measurements were repeated for three independent replicates.
For rock geochemical composition, elemental mapping was performed using energy dispersive microanalysis (EDS; Oxford Instruments, INCAx-sight EDS Detectors, INCA Energy Software, Oxford, UK) under an extra high tension of 20 kV. Three random points were analyzed for each rock sample at a magnification of ×100. Five iterations were performed automatically for each read. Moisture content and total organic content of samples was measured gravimetrically after heating to 105°C and 450°C overnight, respectively, and weighing to constant weight. All measurements were repeated for three independent replicates.
Statistical Analysis
Parametric one-way and two-way analysis of variance and analysis of similarities tests were performed using Statistical Package for the Social Sciences v13.0 (LEAD Technologies Inc., Schaumburg, IL, USA). Significant differences were recorded at probability levels of p < 0.05 or better. All multivariate analyses were performed using PRIMER 6.1.5 (Primer-E Ltd, Plymoyth, UK). The BEST: Bio-Env procedure was employed for ranking the effect of abiotic variables on biotic data patterns. Under BEST analysis, abiotic factors similarity matrices were created by Euclidean distance measurement, whereas biotic data similarity matrices were created by Bray–Curtis similarity measurement. Global tests were carried out to assess the significance level of the Spearman coefficient (ρ) generated by BEST. Raw data was tested by draftsman plot to assess the need for transformation before analysis.
In Situ 14C Bicarbonate Fixation Assay
All productivity estimates commenced at solar noon. 3.7 MBq of 14C-sodium carbonate (Sigma; 0.3959 GBq/mmol) was dissolved in 10 ml dH2O. Twelve float samples with 100% colonization coverage were broken into small pieces of approximately 1 g (these were later weighed after drying in the laboratory). Samples were hydrated for 30 min in sterile distilled prior to the start of the experiment. To initiate the reaction, 200 µl of 14C solution was painted onto the rock surface. Incubations were performed in triplicate for 40, 80, and 160 min. Dark controls were prepared by wrapping in aluminum foil. After incubation, 500 µl of acetic acid was added in order to drive off excess carbonate. Transport to the laboratory was at ambient temperature in darkness. Immediately prior to analysis, 10 ml of scintillation fluid (EcoLume, MP Biomedicals, Santa Ana, CA, USA) was added before determination of incorporated radioactivity (Beckman LS 6500 Scintillation System, Fullerton, CA, USA).
Results
Endolithic Colonization
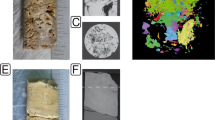
The endolithon comprised a distinct green band of colonization 1–1.2 mm below escarpment surfaces and 1–5 mm below float surfaces (Fig. 1). In a few samples, a pink layer (which rapidly decolourized upon exposure of colonized rock) was also observed above the green layer. Colonization of rock face on escarpments was patchy (seven out of 20 transect locations), but 100% of float was colonized (n = 60). Scanning electron microscopy revealed that colonized rock was characterized by two distinct morphologies that were present in all samples and also in pink layers where present. Relatively large spheres encapsulated within a polygonal lattice were interpreted as lichenized structures where fungal hyphae were viewed as in immediate contact with large coccoid algal cells (Fig. 1). These occurred among an extensive mass of Chroococcidiopsis-like smaller coccoid cells observed throughout colonized surfaces within rock (Fig. 1). A copious extracellular secretion was evident, and this may have obscured other morphotypes. The secretion also made accurate estimate of abundance for different morphotypes impossible.
a Field location in central Tibet, with sampling site indicated by box. b Section through a limestone rock, with typical green band of endolithic colonization indicated by the arrow (scale bar 10 mm). c Scanning electron micrograph of typical endolithic colonized surface in limestone from central Tibet. Arrows indicate L, putative lichen structures, and C, Chroococcidiopsis-like morphotypes (scale bar 5 μm)
qPCR-defined Abundance of Phylotypes
We used real-time quantitative PCR to estimate abundance across all domains. By calibrating our PCR individually against archaeal, bacterial, and eukaryal amplicons, we were able to establish absolute and relative abundance for each domain in each sample. The results were striking despite large variations between some replicates (Table 1). Scarp rocks supported between approximately one- and fourfold fewer rRNA gene copies overall than float rocks. Scarp rocks were dominated by eukaryal phylotypes, with bacteria and archaea comprising less than half the total copy number. Conversely, float rocks were all dominated by prokaryotic phylotypes, although significant variation between the abundance of archaeal and bacterial phylotypes occurred between different float samples (Table 1).
DGGE-defined Diversity Among Archaea, Bacteria, and Eukarya
We used DGGE to separate environmental phylotypes. Banding patterns revealed a large number of band migration categories among Bacteria [23] and Eukarya [16], with relatively few for Archaea [6] (Supplementary on-line material Fig. S1). All DGGE bands were sequenced including co-migrating bands in order to establish phylotype identity. A total of 63 phylotypes were distinguished based upon sequence data. Phylogenetic analyses revealed that phylotypes could be assigned to approximately 14 lineages. Each substrate supported a relatively restricted community of four to 11 phylogenetically distinct phylotypes. All substrate types supported phylotypes identified as the lichen phycobiont Trebouxia jamesii (Fig. 2), a lichen mycobiont of the genus Leptodontidium (Fig. 3) and sequences corresponding to Trebouxiophyceae plastids. Multiple phylotypes of unidentified β-proteobacterium (Fig 4), the cyanobacterium Chroococcidiopsis (Fig 5) and a crenarchaeote (Fig. 6) were also ubiquitous. Other bacterial phylotypes were encountered only in individual substrate samples. Estimates of Shannon’s diversity index were below 0.9 for all samples, indicating a relatively depauperate microbial community. There was no significant difference in Shannon’s diversity index between substrate types. We also assessed diversity for separately extracted green and pink layers from a float sample. Eukaryal (T. jamesii and Leptodontidium) and archaeal phylotype compositions were identical for both layers. The lower (green) layer supported additional Chroococcidiopsis-, Deinococcus-, Hymenobacter-like, and plastid phylotypes that were not recovered in the pink layer.
Phylogenetic relationships among endolithic algal phylotypes. Tree topologies are supported by bootstrap values for 1,000 replications, shown for branches supported by more than 50% of the trees. Scale bar represents nucleotide changes per position. Sequence identifiers (in bold) for phylotypes generated by this study are followed by a list of samples from which a given phylotype was recovered (bracketed: scarp = escarpment, GF = gray float, PF = pink float, WF = white float, G = green layer, PL = pink layer). All sequences can be mapped to original denaturing gradient gel electrophoresis banding positions (Supplementary on-line supporting material Fig. S1) by using these sequence codes
Phylogenetic relationships among endolithic fungal phylotypes. Tree topologies are supported by bootstrap values for 1,000 replications, shown for branches supported by more than 50% of the trees. Scale bar represents nucleotide changes per position. Sequence identifiers (in bold) for phylotypes generated by this study are followed by a list of samples from which a given phylotype was recovered (bracketed: scarp = escarpment, GF = gray float, PF = pink float, WF = white float, G = green layer, PL = pink layer). All sequences can be mapped to original denaturing gradient gel electrophoresis banding positions (Supplementary on-line supporting material Fig. S1) by using these sequence codes
Phylogenetic relationships among endolithic bacterial phylotypes. Tree topologies are supported by bootstrap values for 1,000 replications, shown for branches supported by more than 50% of the trees. Scale bar represents nucleotide changes per position. Sequence identifiers (in bold) for phylotypes generated by this study are followed by a list of samples from which a given phylotype was recovered (bracketed: scarp = escarpment, GF = gray float, PF = pink float, WF = white float, G = green layer, PL = pink layer). All sequences can be mapped to original denaturing gradient gel electrophoresis banding positions (Supplementary on-line supporting material Fig. S1) by using these sequence codes
Phylogenetic relationships among endolithic cyanobacterial phylotypes. Tree topologies are supported by bootstrap values for 1,000 replications, shown for branches supported by more than 50% of the trees. Scale bar represents nucleotide changes per position. Sequence identifiers (in bold) for phylotypes generated by this study are followed by a list of samples from which a given phylotype was recovered (bracketed: scarp = escarpment, GF = gray float, PF = pink float, WF = white float, G = green layer, PL = pink layer). All sequences can be mapped to original denaturing gradient gel electrophoresis banding positions (Supplementary on-line supporting material Fig. S1) by using these sequence codes
Phylogenetic relationships among endolithic archaeal phylotypes. Tree topologies are supported by bootstrap values for 1,000 replications, shown for branches supported by more than 50% of the trees. Scale bar represents nucleotide changes per position. Sequence identifiers (in bold) for phylotypes generated by this study are followed by a list of samples from which a given phylotype was recovered (bracketed: scarp = escarpment, GF = gray float, PF = pink float, WF = white float, G = green layer, PL = pink layer). All sequences can be mapped to original denaturing gradient gel electrophoresis banding positions (Supplementary on-line supporting material Fig. S1) by using these sequence codes
Effect of Substrate Variables
A range of abiotic substrate variables were measured including moisture content, total organic content, chemical composition, and transmittance of PAR, UV-A, and UV-B radiation (Supplementary on-line material Table S1). Moisture and organic content values were below 1% for all colonized rock samples. For all colonized samples, ambient irradiance (PAR, UV-A, and UV-B) was reduced by more than 95% at the depth of colonization. It was noteworthy that colonization of scarp samples occurred at depths within the rock substrate where UV and PAR transmittance were significantly higher (PAR 4.9%, UV-A 2.7%, and UV-B 2%) than that of colonized float substrates (PAR 2.3–3.3%, UV-A and UV-B all <1%; Supplementary on-line material Table S1). At this high-altitude site, ambient irradiance is extremely high with measured values at solar noon on the date of sampling of 2,216 μM/m2/s (PAR), 2.53 mW/cm2 (UV-A) and 1.27 W/cm2 (UV-B). Differences in transmittance of both PAR and UV irradiance among substrate types significantly affected colonization. A BEST analysis utilizing multiple ranked correlation analysis to rank the effect of abiotic variables on community structure was conducted. This analysis revealed that a single variable (UV-A) was best able to explain community structure (ρ = 0.602), followed by the two variables UV-A plus UV-B (ρ = 0.595), and the combination of PAR, UV-A, and UV-B (ρ = 0.511). The influence of PAR alone was not significant (ρ = 0.196). The chemical composition of substrates was not significant in influencing community structure. No obvious explanation for color differences among float rocks was evident from geochemical data.
In Situ Productivity Estimates
In order to gain an estimate of potential in situ productivity for endolithic communities under moisture-sufficient conditions, we assessed in situ 14C bicarbonate fixation in the field for fragments of colonized pink float. Carbon fixation equivalent to a primary productivity of 0.9 mg C/g colonized rock/h−1 (SD = 0.23) was calculated. Based upon field observations, we applied an estimated colonization volume of 1% of the mass for any given rock, occurrence of 10 kg rock/m2 of ground surface and an estimate of 6 months per year when conditions would be warm and wet enough to allow photosynthesis, for a duration of 12 h/day at the rate measured in our study at solar noon. From this, we propose a very approximate productivity rate for high-altitude limestone float terrain of 2.01 g C/m2/year−1 (SD = 0.43). This calculation is based upon the approach previously used to calculate productivity of polar endoliths and hypoliths [3, 8].
Discussion
The macroscopic appearance of Tibetan endoliths (a green band of colonization) resembled those recovered from alpine Europe [22], North America [14] and also for ‘infrequent’ cyanobacterial endoliths in the Antarctic Dry Valleys [7]. No similarity in colonization morphology could be discerned between Tibet endoliths and the green/white/black layered lichen endoliths of the Antarctic [7]. At the microscopic level, the Tibetan endoliths appeared to represent a hybrid between polar lichenized endoliths and alpine cyanobacterial endoliths—where we recorded a green cyanobacterial layer also colonized by lichenized eukaryotes. This may reflect adaptation due to the intermediate climatic nature of the Tibetan high-altitude tundra between alpine and polar regions. We therefore suggest that the Tibetan endoliths may represent a distinct form of endolithic colonization with traits of both polar and alpine lichens. We identified structures that were interpreted as lichens where fungal hyphae formed a lattice around algal cells. This appeared more structured than the fungal–algal association recorded for Antarctic endoliths [7]. No reports of lichenized structures are given in other studies of endoliths. This close association probably accounted for the lack of algal phylotypes encountered in the absence of fungal phylotypes and vice versa in our study.
The relatively small scale of the transects employed in this study was intended to give a baseline indication of diversity rather than to infer landscape-scale ecology. Nonetheless, our colonization frequency data indicated that float was a more readily colonized substrate than escarpment, and this may relate to the higher UV stress recorded for escarpment rocks, plus other unmeasured factors such as water availability over temporal scales. There is limited ecological data from other studies of endoliths with which to compare colonization frequencies. Frequency of endolithic colonization at a temperate North American location was estimated at 45% [14], whilst our recent observations of Antarctic endoliths indicate colonization frequency for sandstone float of less than 5% [18]. This strongly suggests that colonization frequency may be related to environmental stress.
The qPCR data indicated that scarp supported markedly less overall abundance of phylotypes than float. Despite this, the predominantly eukaryotic nature of the scarp community implies larger cell mass than the mainly bacterial float, and so this may not necessarily reflect great differences in standing biomass. The fact that precise copy number for rRNA genes among all environmental phylotypes was unknown creates limitations for these data in that they cannot be directly indicative of biomass or provide accurate between-domain comparisons. This is an inherent problem in the application of universally primed qPCR to environmental samples. Relative abundance estimates for archaea, bacteria, and eukarya in the endolithon were strikingly different between scarp and float substrates, although all supported what may be keystone endolithic phylotypes. This suggests that whilst the communities were likely founded by the same organisms, local niche conditions favored proliferation of eukaryal lichens (Trebouxia and Leptodontidium) in scarp and prokaryotes (largely Chroococcidiopsis and an unidentified crenarchaeon) in floats. Our findings indicate that this reflects measurable differences in UV stress between scarp and float. We do not discount that other factors such as period of exposure (i.e. time since creation of float or exposure of limestone scarp by geological weathering) may also be important. The observed plasticity in domain abundance between niches is interesting since it implies that wide ranges in the frequency of occurrence for phototrophs and heterotrophs, eukaryotes, and prokaryotes are possible within endolithic communities.
The overall diversity encountered in endolithon of scarp and float substrates from this study can be considered as restricted relative to other microbial niches. This concurs with a recent study of the Rocky Mountain endolithon in the USA where relatively low diversity communities were recorded [26]. Our study appears to be the first to elucidate both mycobiont and phycobiont of non-polar endolithic lichens at the molecular level, since the phylotypes in our study affiliated most closely to Leptodontidium sp. and T. jamesii, both of which are known lichen associates in various environments. Some plastid phylotypes recovered were phylogenetically closely affiliated to Stichococcus bacillaris recovered from Alpine endolithon [22], although their study did not record any associated fungal component to the endolithon. This chlorophyte alga shares a familial relationship with Trebouxia (also a member of the Trebouxiophyceae), and since we did not observe free-living algal morphotypes, it is concluded that the Stichococcus-like phylotype from our study probably also represents a lichen mycobiont. It is noteworthy that this phylotype occurred in two such geographically separated environments. This was also observed for free-living algal phylotypes in Antarctic and Rocky Mountain endolithon [26]. This implies that a relatively ubiquitous inoculum for potential endolithic colonization probably exists.
Among the bacteria, it is interesting that phylogenetically distinct Chroococcidiopsis sp. phylotypes occurred in each substrate. All were affiliated with other strains from lithic habitats in arid environments, and so it is possible that a relatively diverse inoculum for this taxon seeds endolithic niches, and founder effects may be a significant force in determining the dominant strain in a given niche. It is not possible to determine from 16S rRNA gene sequences whether these reflect ecotypes adapted to slightly different niche conditions, although this has been postulated for cyanobacteria in hyper-arid deserts [19]. The only other cyanobacterial phylotype recovered indicated a Phormidium-like taxon, and this occurred in only one sample. The cyanobacterial population in Tibetan endoliths therefore lacked the relatively high cyanobacterial diversity recorded for Alpine and North American endoliths [17, 22]. Indeed, the near-monoculture of Chroococcidiopsis was more similar to cyanobacterial populations recorded for endoliths in the Antarctic Dry Valleys [18] and hypoliths in the driest deserts on Earth [19, 28]. This may indicate that the cumulative impacts of environmental stress in Tibet are similar to those in polar and hyper-arid deserts.
Other phylotypes all affiliated with heterotrophic clades. It is interesting that most phylotypes were phylogenetically similar to those recovered from other extreme environments, including polar regions and hot springs. Desiccation and radiation-tolerant taxa were indicated by the Deinococci phylotypes. No anoxygenic phototrophic bacterial phylotypes were encountered, although they were recovered from Antarctic and Rocky Mountain endoliths [4, 26]. Archaeal phylotypes formed a very closely related group among the ‘uncultured’ Crenarchaeota. At this time, very little is known about representatives of this group, other than that they account for a growing percentage of archaeal phylotypes isolated from environmental samples. A relatively restricted group of heterotrophic bacteria and archaea appear to be supported in the endolithon, which may reflect the extreme environmental stress and limited organic substrate availability from slow-growing lichen phycobiont and cyanobacterial photoautotrophy. We discount specificity of the PCR primers and their separation using DGGE as a significant source of under-estimation in bacterial or archaeal diversity, since we have used the same experimental approach has yielded some of the highest diversity estimates recorded for other extreme habitats [13].
The major difference between scarp and float rocks related to UV transmittance, and this was the most significant factor affecting community structure. The eukaryotic lichens dominated the relatively high-UV scarp, whilst prokaryotic colonization was reduced compared to float. This may reflect relatively high-UV tolerance among lichen taxa [5, 21]. The cyanobacterium Chroococcidiopsis that dominated float rocks is also a highly radiation-tolerant taxon [1], and so uncertainties remain over the environmental drivers for different taxa comprising scarp and float communities and any photo-protective role that they may impart to the community. We recorded no difference in moisture content between substrates upon collection, although it is possible to envisage different long-term moisture regimes exist between escarpment and float. It is possible that since Chroococcidiopsis is common in the world’s driest and hottest deserts [27, 28] that this reflects greater adaptation to moisture stress or relatively warmer temperatures in float communities. Alternatively, the differences may reflect temporal effects. Given that polar endolithic lichen communities are known to be very slow growing and also long-lived [23], their prevalence in escarpment may reflect longer duration of colonization in this substrate.
The in situ productivity estimates for Tibet endolithon were approximately twofold higher than estimates made for Antarctic endoliths on the basis of laboratory measurements [8]. They were also approximately twofold higher than in situ estimates for polar hypoliths [3]. Conversely, productivity estimates for endoliths in a temperate North American location suggest an upper limit of productivity some tenfold higher than Tibetan endoliths [14]. Although all of these estimates contain numerous assumptions about the period during a year when photosynthesis is possible, they suggest a possible broad trend of declining productivity with increasing environmental stress. These data suggest that under field conditions, the high-altitude Tibetan endolithic community may be significantly more productive than lithic polar communities and yet still several orders of magnitude less productive than endoliths in temperate regions. Nevertheless, given the sparse aboveground vegetation at high altitudes and the vast expanses of limestone substrate in Tibet, endoliths are likely to be a significant source of carbon input at high altitudes. The scope of our field study did not allow for comparative testing between escarpment and float communities, and so we are unable to draw conclusions at this stage about any differences that may exist between them in terms of their productivity.
In conclusion, we have characterized the microbial diversity of Tibetan endoliths across all domains and identified that whilst escarpments support lichen-dominated communities, those in float are largely prokaryotic. These high-altitude tundra endoliths appears distinct, with features reminiscent of both polar and alpine endoliths. We identified that levels of transmitted UV irradiance were best able to explain diversity patterns, although we do not discount effects from other environmental drivers on temporal scales. Productivity estimates were intermediate between polar and temperate endoliths and may warrant an upward revision of productivity estimate for high-altitude tundra ecosystems.
References
Billi D, Friedmann EI, Hofer K, Grilli-Caiola M, Ocampo-Friedmann R (2000) Ionizing-radiation resistance in the desiccation-tolerant cyanobacterium Chroococcidiopsis. Appl Environ Microbiol 66:1489
Budel B, Webber B, Kuhl M, Pfanz H, Sultemeyer D, Wessels D (2004) Reshaping of sandstone surfaces by cryptoendolithic cyanobacteria: bioalkalization causes chemical weathering in arid landscapes. Geobiology 2:261
Cockell CS, Stokes MD (2004) Widespread colonization by polar hypoliths. Nature 431:414
de la Torre JR, Goebel BR, Friedmann EI, Pace NR (2003) Microbial diversity of cryptoendolithic communities from the McMurdo dry valleys, Antarctica. Appl Environ Microbiol 69:3858
Edwards HGM, Cockell CS, Newton ES, Wynn-Williams DD (2004) Protective pigmentation in UV-B-screened Antarctic lichens studied by Fourier transform Raman spectroscopy: an extremophile bioresponse to radiation stress. J Raman Spectrosc 35:463
Friedmann EI (1980) Endolithic microbial life in hot and cold deserts. Orig Life Evol Biosph 10:223
Friedmann EI (1982) Endolithic microorganisms in the Antarctic cold desert. Science 215:1045
Friedmann EI, KAppen L, Meyer MA, Nienow JA (1993) Long-term productivity in the cryptoendolithic microbial community of the Ross desert, Antarctica. Microb Ecol 25:51
Friedmann EI, Ocampo R (1976) Endolithic blue-green algae in dry valleys—primary producers in Antarctic desert ecosystem. Science 193:1247
Gerrath JA, Mathes U, Larson DW (2000) Endolithic algae and cyanobacteria from cliffs of the Niagara Escarpment, Ontario, Canada. Can J Bot 78:807
Hugenholtz P, Goebel BM, Pace NR (1998) Impact of culture-independent studies on emerging phylogenetic view of bacterial diversity. J Bacteriol 180:4765
Kuhle M (1990) The cold deserts of high Asia (Tibet and contiguous mountains). GeoJournal 20:319
Lau CY, Jing H, Aitchison JC, Pointing SB (2006) Highly diverse community structure in a remote central Tibetan geothermal spring does not display monotonic variation to thermal stress. FEMS Microbiol Ecol 57:80
Mathes-Sears U, Gerrath JA, Larson DW (1997) Abundance, biomass and productivity of endolithic and epilithic lower plants on the temperate-zone cliffs of the Niagara Escarpment, Canada. Int J Plant Sci 158:451
McKay CP (1993) Relevance of Antarctic microbial ecosystems to exobiology. In: Friedmann EI (ed) Antarctic microbiology. Wiley-Liss, New York, p 593
Muyzer G, de Waal EC, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59:695
Norris T, Castenholz RW (2006) Endolithic photosynthetic communities within ancient and recent travertine deposits in Yellowstone national Park. FEMS Microbiol Ecol 57:470
Pointing SB, Chan Y, Lacap DC, Lau CY, Jurgens J, Farrell RL (2009) Highly specialized microbial diversity in hyper-arid polar desert. Proc Natl Acad Sci USA (in press)
Pointing SB, Chan Y, Lacap DC, Lau CY, Jurgens J and Farrell RL (2009) Highly specialized microbial diversity in hyper-arid polar desert. Proc Natl Acad Sci (USA). doi:10.1073/pnas.0908274106
Rannala B, Yang ZH (1996) Probability distribution of molecular evolutionary trees: a new method of phylogenetic inference. Mol Ecol 43:304
Sanchoz LG, de la Torre R, Horneck G, Ascaso C, de los Rios A, Pintado A, Wierzchos J, Schuster M (2007) Lichens in space: results from the 2005 LICHENS experiment. Astrobiology 7:443
Sigler WV, Bachofen R, Zeyer J (2003) Molecular characterization of endolithic cyanobacteria inhabiting exposed dolomite in central Switzerland. Environ Microbiol 5:618
Sun HJ, Friedmann EI (1999) Growth on geological time scales in the Antarctic cryptoendolithic microbial community. Geomicrobiol J 16:193
Swofford DL (2001) PAUP*: phylogenetic analysis using parsimony (*and other methods) version 4.0b8. Sinauer Associates, Sunderland
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The Clustal X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl Acids Res 24:4876
Walker JJ, Pace NR (2007) Phylogenetic composition of Rocky Mountain endolithic microbial ecosystems. Appl Environ Microbiol 73:3497
Warren-Rhodes K, Rhodes KL, Pointing SB, Boyle L, Dungan J, Liu S, Zhou P, McKay CP (2007) Lithic cyanobacterial ecology across environmental gradients and spatial scales in China’s hot and cold deserts. FEMS Microbiol Ecol 61:470
Warren-Rhodes K, Rhodes KL, Pointing SB, Ewing S, Lacap DC, Gómez-Silva B, Amundson R, Friedmann EI, McKay CP (2006) Hypolithic cyanobacteria, dry limit of photosynthesis and microbial ecology in the hyperarid Atacama Desert, Chile. Microb Ecol 52:389
White TJ, Burns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Gelfand DH, Sninsky JJ, White TJ, Innis A (eds) PCR protocols, a guide to methods and applications. Academic, San Diego, p 315
Acknowledgements
The authors are grateful to the Tibet Ministry of Geology for fieldwork assistance and to the Hong Kong Research Grants Council (grant numbers HKU 7375/05M and HKU 7733/08M) and Stephen Hui Trust Fund for financial support.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
Denaturing gradient gel electrophoresis (DGGE) profiles for limestone endolithon (scarp, gray float [GF], pink float [PF], white float [WF], pink layer [PL], and green layer [GL]) using polymerase chain reaction primers specific to eukarya (a), bacteria (b), cyanobacteria (c), and archaea (d). Numbers beside each gel indicate the denaturant gradient, numbers specific to each band are the phylotype identifiers (same as used in phylogenetic trees), asterisks in bacteria DGGE indicate plastid phylotypes, white arrows in bacteria DGGE (b) indicate cyanobacterial phylotypes and white arrows in eukaryal DGGE (a) indicate fungal phylotypes. (DOC 849 kb)
Table S1
Abiotic variables for limestone scarp and float colonized by endolithic microorganisms. (DOC 104 kb)
Rights and permissions
About this article
Cite this article
Wong, F.K.Y., Lau, M.C.Y., Lacap, D.C. et al. Endolithic Microbial Colonization of Limestone in a High-altitude Arid Environment. Microb Ecol 59, 689–699 (2010). https://doi.org/10.1007/s00248-009-9607-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-009-9607-8