Abstract
Peatlands subjected to sulfate deposition have been shown to produce less methane, believed to be due to competitive exclusion of methanogenic archaea by sulfate-reducing bacteria. Here, we address whether sulfate deposition produces impacts on a higher microbial group, the testate amoebae. Sodium sulfate was applied to experimental plots on a Scottish peatland and samples extracted after a period of more than 10 years. Impacts on testate amoebae were tested using redundancy analysis and Mann–Whitney tests. Results showed statistically significant impacts on amoebae communities particularly noted by decreased abundance of Trinema lineare, Corythion dubium, and Euglypha rotunda. As the species most reduced in abundance are all small bacterivores we suggest that our results support the hypothesis of a shift in dominant prokaryotes, although other explanations are possible. Our results demonstrate the sensitivity of peatland microbial communities to sulfate deposition and suggest sulfate may be a potentially important secondary control on testate amoebae communities.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Peatlands are exposed to sulfate deposition from both anthropogenic sources, primarily fossil fuel burning, and natural sources, primarily volcanoes. Recent studies have shown that deposition of sulfate on peatlands leads to a reduction in methane production [31, 46] and emission [9, 11]. This suppression of methane emission may be a highly important process in terms of global climate. Sulfate emissions currently reduce wetland CH4 flux by around 8% and could contribute to a 50% reduction in the northern wetland CH4 flux following a large Icelandic eruption [10, 12]. The cause of this methane suppression is believed to be the competitive exclusion of methanogenic archaea (MA) by sulfate-reducing bacteria (SRBs). An increase in sulfate reduction simultaneous with inhibition of methane efflux has been demonstrated, supporting this hypothesis [8]. However, to date, no studies have directly investigated the impact of sulfate deposition on peatland microbial communities. Here, we explore whether sulfate deposition might produce impacts on a higher microbial group, potentially relating to the inferred ecological shift in MA and SRB communities. This study focuses on testate amoebae, a polyphyletic group of protists, which constitute a large proportion of microbial biomass in Sphagnum peatlands (Gilbert et al. [14] estimate 14% and Mitchell et al. [27] estimate up to 30%). Testate amoebae are a particularly suitable object for study due to the presence of a solid shell (the test), which allows taxa to be identified to species level without resorting to molecular techniques. The decay-resistant test also allows testate amoebae to be identified after death, enabling longer term processes to be studied. Some peatland palaeoecological records show testate amoebae community changes coincident with volcanic tephra deposition [7, 36]. One hypothesis for these changes is that they are related to volcanogenic sulfate deposition. Testate amoebae include both taxa that are directly bacterivorous and taxa that predate other microorganisms as well as consume fungi and particulate organic matter; some taxa are mixotrophic [15]. The testate amoebae community response is therefore likely to be complex. In this study, we use an experimental approach to test the impact of sulfate deposition on testate amoebae communities of a natural peatland.
Site and Methods
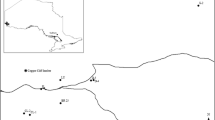
Experiments were conducted on Moidach More, an ombrotrophic peatland in Morayshire, northeast Scotland (UK grid reference NJ0241, 57°27′ N, 3°36′ W, 275 m asl, Fig. 1). Vegetation of the site includes Sphagnum species (Sphagnum magellanicum, Sphagnum recurvum, and Sphagnum capillifollium), Trichophorum cespitosum, Erica tetralix, and Calluna vulgaris [9]. The site receives little ambient sulfate deposition (ca. 5 kg ha−1 year−1 SO 2−4 ). Experiments were conducted on an uncut area towards the west of the site. Twenty 2 × 2 m plots were established in three adjacent blocks. Sodium sulfate was applied at three concentrations over a period of 18 months, commencing in June 1997. Measurements of methane flux and related environmental data were carried out at regular intervals until December 1998 and then occasionally until late 2003 [11]. Experimental setup is described in detail by Gauci et al. [9]. Samples for the present study were extracted from control plots and plots subjected to the heaviest sulfate treatment (95 kg ha−1 year−1 SO 2−4 ) in April 2008. This level of deposition is equivalent to the upper end of the range of anthropogenic deposition or what might be expected in northern peatland areas following a large Icelandic volcanic eruption. A high sampling intensity was used to account for fine-scale spatial variability in testate amoebae communities [25]. Twenty-five samples were extracted from each of three pairs of treatment plots and control, yielding a total of 150 samples. Plots are referred to by their block (1, 2, or 3) and their treatment: control (A) or treated (B).
Samples approximately 30 × 30 × 50 mm depth were extracted from randomly selected positions covering the surface area of each plot. To minimize influence of vegetation structure on testate amoebae communities, samples were extracted from a single moss species, S. magellanicum. A variety of environmental data were collected to allow evaluation of any differences between plots that are unrelated to the experimental treatments. The main environmental controls on testate amoebae communities are wetness, acidity, and nutrient status [1, 33, 42]. Data relevant to all these parameters were collected. The pH of the samples was determined by suspending 2 cm3 of surface peat in 50 ml of deionized water and measuring pH using a Jenway 3320 pH meter after 1 h. Loss on ignition (LOI), which may be a proxy for nutrient status [34], was determined by drying peat samples at 105°C, weighing, incinerating at 550°C and then re-weighing. Depth to water table (DWT) was measured by making a small hole adjacent to the sampling point and measuring the depth to the water table after leaving for at least 2 h to equilibrate.
Testate amoebae preparation used a slightly modified version of the method of Hendon and Charman [19]. The upper 50 mm of ten stems of S. magellanicum were separated from other bryophytes and used in testate amoebae sample preparation. The volume of the sample was measured by displacement in water. Samples were boiled for 10 min to disaggregate and a Lycopodium inoculum added to allow calculation of test concentration [39, 45]. The sample was filtered at 300 μm with the fine fraction retained. Back-filtering with a finer sieve was not used, as this is liable to lead to the loss of some smaller tests (e.g., Cryptodifflugia oviformis, Trinema lineare) and amoebae concentrations were high rendering this unnecessary. Samples were stained to allow differentiation of living from dead amoebae. Samples were centrifuged to concentrate and then stored in water. Slides were prepared by mixing a drop of the preparation with glycerol. A count of 150 tests was aimed for (mean = 163), higher than the total advocated by Payne and Mitchell [35], as changes in amoebae community due to the experimental additions may be subtle. Taxonomy generally followed the scheme of Charman et al. [4] with a few minor exceptions such as splitting of the Corythion–Trinema type. Species abundances were converted to biomass using the approach outlined by Gilbert et al. [13]. Biovolumes were approximated by assuming geometrical shapes [24] based on dimensions in the published literature or estimates under the microscope and converted to carbon biomass using the conversion factor 1 μm3 = 1.1 × 10−7 μg C [48].
The data were collated and six multivariate datasets calculated: (1) relative abundances of taxa as a percentage of total number of tests; (2) relative abundances of taxa considering only living individuals; (3) abundance of taxa as concentrations of all tests; (4) abundance of taxa as concentration considering live individuals only; (5) estimated biomass based on all individuals; and (6) estimated biomass based on living individuals. In addition, five univariate datasets were also calculated: (7) overall test concentration; (8) concentration of living amoebae; (9) live individuals as a percentage of total tests; (10) species richness; (11) total estimated biomass based on all individuals; and (12) total estimated biomass based on live individuals. The impact of the treatments in the univariate data was tested using Mann–Whitney tests in PAST ver. 1.84 [17]. The multivariate data structure was investigated using principal components analysis (PCA), and the impact of the treatments in the multivariate data was tested using redundancy analysis (RDA). A series of RDAs were used to test the impact of a nominal variable for experimental treatment both on its own and with various combinations of the environmental data (pH, DWT, and LOI) introduced as co-variables. Significance was assessed using Monte Carlo permutation tests (999 permutations restricted for experimental design). Species data were Hellinger transformed [23, 37]. All ordination analyses were carried out in Canoco ver. 4.53 [40].
Results
A total of 31 taxa were encountered in the 150 samples. The most abundant taxa were Archerella flavum (30.5% of total count), Corythion dubium (10.2% of total), Euglypha strigosa (9.6% of total), and Nebela tincta type (7.8% of total). Some differences between the treatments and controls are apparent in the total abundance of taxa within plots (Table 1). Higher abundances of E. strigosa, Placocista spinosa type and Hyalosphenia papilio are apparent in the treated plots (although the later is absent in area 2). Consistently lower abundances of Euglypha rotunda type and T. lineare are apparent in the treated plots, although abundance of the former taxon is very low. Differences between the treated and untreated samples are apparent but are not particularly marked in the PCA plot (Fig. 2). For mid-values of axis 1, treated samples generally have higher scores than untreated samples on axis 2; there are more treated than untreated samples at the highest values on axis 1.
Analysis of univariate data showed significant difference between treated and untreated plots for proportion of living tests and concentration of live amoebae (P < 0.05) but not for total test concentration, number of species, and testate amoebae biomass based on live and all individuals (in the later case, the relationship is only marginally insignificant, P = 0.06).
The RDA results show that the experimental treatment explains a significant proportion of the variance with all but one of the multivariate datasets (Table 2). pH and LOI did not explain a significant proportion of the variance independent of the other variables (probably due to limited range) and were therefore excluded from analyses. Most variance is explained when considering all tests (either as concentration or percentage); 3.1% of variance is explained by the treatment variable, and this is slightly reduced to 2.8% when DWT is partialled out. The weakest relationships are produced when using the estimated biomass data, perhaps due to the inevitable approximations in these calculations [2] or the comparatively small size of some of the most sensitive taxa. The relationship between the treatment and the species data is not significant when calculating biomass on the basis of live individuals alone.
Figure 3 shows the ordination plot with percentage data based on all tests; plots based on other datasets are similar and are not presented. Taxa known to be hydrophilous (A. flavum and Amphitrema wrightianum) are negatively correlated with DWT, while taxa such as Heleopera petricola, Assulina muscorum, and Euglypha cristata are positively correlated, indicating they are more xerophilous (although the overall water table range is quite limited). The treatment variable is positively correlated with H. papilio, Arcella arenaria type, and to a lesser extent C. oviformis, and negatively correlated with T. lineare, E. rotunda type, and less distinctly C. dubium and Trinema complanatum. It is notable that these latter taxa are similar, all small Euglyphid species. Post hoc Mann–Whitney tests showed significant differences (P < 0.05) in relative abundance of all these taxa between treated and untreated samples.
Redundancy analysis of testate amoebae data based on relative abundance of all tests. Showing selected major species and significant environmental variables. Species codes: AFLAV, Archerella flavum; TLIN, Trinema lineare; EROT, Euglypha rotunda type; TCOMP, Trinema complanatum; CDUB, Corythion dubium; AMUS, Assulina muscorum; ECRIS, Euglypha cristata; HPET, Heleopera petricola; ESTRI, Euglypha strigosa; COVI, Cryptodifflugia oviformis; AARE, Arcella arenaria type, Hyalosphenia papilio; AWRI, Amphitrema wrightianum; NGRIS, Nebela griseola
Discussion
The results demonstrate a significant impact of sulfate deposition on testate amoebae communities. The univariate data analysis shows that the experimental treatments reduce the concentration of live amoebae and percentage of live amoebae, suggesting a less active amoebae community. This is in parallel with studies of the impact of nutrient enrichment on peatland testate amoebae. Mitchell [24] and Gilbert et al. [13, 14] found nutrient enrichment (with N and P, N and P, K, Ca, and N, P, K, and Ca) and CO2 enrichment [27] reduced the contribution of testate amoebae to microbial biomass. Although there was no measurable impact on estimated biomass here, we attribute this to the large errors involved in biomass estimates based on taxon assemblage data and the small size of many of the most sensitive taxa. The significant changes in proportion of living individuals support the value of this simple index in testate amoebae-based biomonitoring [43, 44].
A 3.1% of variance is explained by the treatment variable with the percentage data, and this relationship is highly significant (P = 0.001). Although this seems a small proportion, in the context of inherently noisy testate amoebae data, this is far from irrelevant. By comparison, DWT, the strongest environmental control, explains 7.6% of variance with the other environmental data partialled out (P = 0.001). This result shows a distinct impact of sulfate application on amoebae community structure. The impact of treatment on amoebae emerges equally strongly in the RDA when using data based on concentration or percentages, showing that there are absolute changes in the abundance of amoebae taxa, not simply relative changes in abundance.
The relationships are stronger when considering all individuals than considering only living individuals. The number of live individuals counted in some samples is very low (as few as three amoebae), possibly related to boiling in sample preparation. With such low counts, the amoebae community will be poorly characterized [35]. A further factor contributing to the weaker relationships when only live individuals are considered is likely to be the length of time, which elapsed between experimental treatments and sample extraction. It is quite possible that the amoebae community over the period of several years represented by the full test community has been more affected by the experimental additions than the testate amoebae community currently living at the site. Nevertheless, the fact that the treatment variable is still highly significant even when just considering living amoebae shows a long-lasting impact, consistent with the observations of prolonged methane flux suppression [11].
Determining the relationship between the experimental treatments and the amoebae community changes is complex, as a group testate amoebae have wide food preferences, including bacteria, particulate organic matter, microalgae, cyanobacteria, plant cells, other protists, fungi, and micro-metazoa [6, 15, 50]. Ecologically meaningful interpretation of species changes is difficult, as comparatively little is known of the autecology of individual taxa. Gilbert et al. [15] located published information on feeding preferences for only 33 species (out of perhaps 2,000 described species [28]). The degree of specificity in food source is also largely unknown. Gilbert et al. [16] showed Nebela collaris (sensu lato) to feed on a wide variety of material ranging from diatoms to fungal spores. Other taxa may have much more specific food requirements; in an aquatic system, Nishibe et al. [32] found that Penardochlamys sp. preyed exclusively on cyanobacteria of the genus Microcystis. Furthermore, food preferences may well be seasonally variable (e.g., [18]).
The RDA plot shows a positive relationship between treatment and abundance of H. papilio, Arcella arenaria, and C. oviformis and a negative relationship with E. rotunda type, C. dubium, T. complanatum, and T. lineare. T. lineare, T. complanatum, and E. rotunda are believed to be bacterivorous and C. dubium to prey on bacteria and fungi [15]. H. papilio has been noted to feed on fungi, microalgae, ciliates, and metazoa [15]. We are not aware of any information on the feeding habits of C. oviformis or A. arenaria, although another Arcella species (Arcella gibbosa) has been noted to feed on bacteria, microalgae, fungi, and flagellates.
It is notable that the species that appear to be deleteriously impacted by sulfate additions are among comparatively few testate amoebae species, which are largely bacterivorous. By contrast, taxa that respond positively have less specific feeding preferences. This pattern is unlikely to be a coincidence. We are not aware of any previous research specifically relating testate amoebae and MA or SRB. As testate amoebae are most abundant in upper peats while archaea are largely constricted to deeper layers of the peat [47], it is unlikely that testate amoebae are major predators of MA. Previous research does however indicate that other wetland protists predate SRB (and indeed methanotrophs [29, 30]).
The lack of research on how testate amoebae fit into the microbial foodweb in peatlands means that we cannot fully explain the mechanism that relates sulfate addition to changes in testate amoebae communities observed in this study. However, it is certainly tempting to conclude a relationship between the decline in bacterivorous testate amoebae and the putative decline in methanogens. The mechanism for this is unlikely to be as simple as these species preferentially consuming archaea over bacteria; it is more probable that the interaction is indirect through other organisms. It is even possible that sulfate deposition somehow promotes the predation of these taxa. Methanogenic endosymbionts have been widely reported from protists (e.g., [20, 41], including wetland ciliates [38]), although as far as we are aware of, there has been no record of methanogenic symbionts in testate amoebae. It is interesting to speculate that some of the apparent association between methane flux suppression and testate amoebae community change could be related to predation of ciliates with methanogenic symbionts by testate amoebae.
An alternative mechanism to a change in methanogens/SRBs is that sulfate deposition directly or indirectly modifies the chemical environment, such that it becomes more suitable for some testate amoebae taxa than for others. While we cannot exclude this possibility, we cannot see a clear mechanism whereby this might occur. A further possibility is that impacts are due to the sodium applied with the sulfate. We think this is unlikely as: (1) the quantity of Na applied is very small, (2) Na+ was not shown to be a significant variable in a recent ecological study [33]; and (3) Gauci et al. [11] showed no methane suppression in control plots with NaCl applied, suggesting that there is at least no impact on the microbial community involved with methanogenesis. We suggest that our results provide some circumstantial support for the hypothesis of a shift from methanogens to SRBs and that this produces consequent impacts throughout the microbial food web.
These experimental results suggest that sulfate may be an important environmental control on testate amoebae communities. Where sulfates have been measured in ecological studies, sulfate is correlated with major testate amoebae species gradients (e.g., [49]). Opravilova and Hajek [33] and Mitchell et al. [26] have shown sulfate to be a small but statistically significant independent environmental control on amoebae communities. A contrary result was found by Lamentowicz et al. [22], although this study was focused on a single site and therefore has limited environmental gradients. Taken together, our experimental results and the previous ecological survey results suggest that sulfate may be underestimated as a control on amoebae communities. It would certainly be useful to analyze sulfate more regularly in ecological studies of testate amoebae and particularly interesting to analyze testate amoebae in peatlands along a gradient of anthropogenic sulfate deposition. It would be interesting to repeat this study with a greater number of plots and to see if impacts are still detectable with lower levels of sulfate application. Studies combining analyses of testate amoebae with analyses of other microbial groups (e.g., [21]) might help unravel the mechanism of impact. It is perhaps worth noting that saltmarshes (which have significant sulfate input) have notably different testate amoebae communities from ombrotrophic peatlands (which generally do not), although clearly there are also many other differences in these ecosystems [5].
Testate amoebae are increasingly widely used in palaeoecological studies to provide a proxy-record of hydrological change [3, 28]. Inherent in this work is the assumption that testate amoebae community change is primarily driven by peatland hydrological change and therefore by climate. These results suggest that sulfate pollution may also be an important (albeit much weaker) control. This might complicate hydrological reconstruction in peatlands subject to sulfate deposition.
References
Booth RK (2002) Testate amoebae as paleoindicators of surface-moisture changes on Michigan peatlands: modern ecology and hydrological calibration. J Paleolimnol 28:129–348
Charriere F, Pavillon N, Colomb T, Depeursinge C, Heger TJ, Mitchell EAD, Marquet P, Rappaz B (2006) Living specimen tomography by digital holographic microscopy: morphometry of testate amoeba. Optics Express 14:7005–7013
Charman DJ (2001) Biostratigraphic and palaeoenvironmental applications of testate amoebae. Quat Sci Rev 20:1753–1764
Charman DJ, Hendon D, Woodland W (2000) The identification of testate amoebae (Protozoa: Rhizopoda) in peats. Quaternary Research Association Technical Guide Series, Cambridge
Charman DJ, Roe HM, Gehrels WR (2002) Modern distribution of saltmarsh testate amoebae: regional variability of zonation and response to environmental variables. J Quat Sci 17:387–409
Coûteaux M-M (1984) Relationships between testate amoeba and fungi in humus microcosms. Soil Biol Biochem 17:339–345
Dwyer RB, Mitchell FJG (1997) Investigation of the environmental impact of remote volcanic activity on north Mayo, Ireland, during the mid-Holocene. Holocene 7:113–118
Gauci V, Chapman SJ (2006) Simultaneous inhibition of CH4 efflux and stimulation of sulphate reduction in peat subject to simulated acid rain. Soil Biol Biochem 38:3506–3510
Gauci V, Dise N, Fowler D (2002) Controls on suppression of methane flux from a peat bog subjected to simulated acid rain sulfate deposition. Glob Biogeochem Cycles 16:1004
Gauci V, Matthews E, Dise N, Walter B, Koch D, Granberg G, Vile M (2004) Sulfur pollution suppression of the wetland methane source in the 20th and 21st centuries. Proc Natl Acad Sci U S A 101:12583–12587
Gauci V, Dise N, Blake S (2005) Long-term suppression of wetland methane flux following a pulse of simulated acid rain. Geophys Res Lett 32:L12804
Gauci V, Blake S, Stevenson DS, Highwood EJ (2008) Halving of the northern wetland CH4 source by a large Icelandic volcanic eruption. J Geophys Res 113:G00A11
Gilbert D, Amblard C, Bourdier G, Francez A (1998) The microbial loop at the surface of a peatland: structure, functioning and impact of nutrients inputs. Microb Ecol 35:83–93
Gilbert D, Amblard C, Bourdier G, Francez AJ (1998) Short-term effect of nitrogen enrichment on the microbial communities of a peatland. Hydrobiologia 373(374):111–119
Gilbert D, Amblard C, Bourdier G, Francez A-J, Mitchell EAD (2000) Le regime alimentaire des Thécamoebiens (Protista, Sarcodina). Année Biol 39:57–68
Gilbert D, Mitchell E, Amblard C, Bourdier G, Francez A-J (2003) Population dynamics and food preferences of the testate amoeba Nebela tincta major-bohemica-collaris complex (Protozoa) in a Sphagnum peatland. Acta Protozool 42:99–104
Hammer Ø, Harper DAT, Ryan PD (2001) PAST: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontol Electron 4. http://palaeo-electronica.org/2001_1/past/issue1_01.htm
Han B-P, Wang T, Lin Q-Q, Dumont HJ (2008) Carnivory and active hunting by the planktonic testate amoeba Difflugia tuberspinifera. Hydrobiologia 596:197–201
Hendon D, Charman DJ (1997) The preparation of testate amoebae (Protozoa: Rhizopoda) samples from peat. Holocene 7:199–205
Inoue J-I, Noda S, Hongoh Y, Ui S, Ohkuma M (2008) Identification of endosymbiotic methanogen and ectosymbiotic spirochetes of gut protists of the termite Coptotermes formosanus. Microbes Environ 23:94–97
Krashevska V, Bonkowski M, Maraun M, Ruess L, Kandeler E, Scheu S (2008) Microorganisms as driving factors for the community structure of testate amoebae along an altitudinal transect in tropical mountain rain forests. Soil Biol Biochem 40:2427–2433
Lamentowicz L, Lamentowicz M, Gabka M (2008) Testate amoebae ecology and a local transfer function from a peatland in western Poland. Wetlands 28:164–175
Legendre P, Gallagher ED (2001) Ecologically meaningful transformations for ordination of species data. Oecologia 129:271–280
Mitchell EAD (2004) Response of testate amoebae (Protozoa) to N and P fertilization in an Arctic wet sedge tundra. Arct Antarct Alp Res 36:77–82
Mitchell EAD, Borcard D, Buttler A, Grosvernier P, Gilbert D, Gobat J-M (2000) Horizontal distribution patterns of testate amoebae (Protozoa) in a Sphagnum magellanicum carpet. Microb Ecol 39:290–300
Mitchell EAD, Buttler A, Grosvernier P, Rydin H, Albinsson C, Greenup AL, Heijmans MMPD, Hoosbeek MR, Saarinen T (2000) Relationships among testate amoebae (Protozoa), vegetation and water chemistry in five Sphagnum-dominated peatlands in Europe. New Phytol 145:95–106
Mitchell EAD, Gilbert D, Buttler A, Grosvernier P, Amblard C, Gobat J-M (2003) Structure of microbial communities in Sphagnum peatlands and effect of atmospheric carbon dioxide enrichment. Microb Ecol 16:187–199
Mitchell EAD, Charman DJ, Warner BG (2008) Testate amoebae analysis in ecological and paleoecological studies of wetlands: past, present and future. Biodivers Conserv 17:2115–2137
Murase J, Frenzel P (2008) Selective grazing of methanotrophs by protozoa in a rice field soil. FEMS Microbiol Ecol 65:408–414
Murase J, Noll M, Frenzel P (2006) Impact of protists on the activity and structure of the bacterial community in a rice field soil. Appl Environ Microbiol 72:5436–5444
Nedwell DB, Watson A (1995) CH4 production, oxidation and emission in a UK ombrotrophic peat bog: influence of SO 2−4 from acid rain. Soil Biol Biochem 27:893–903
Nishibe Y, Manage P, Kawabata Z, Nakano S (2004) Trophic coupling of a testate amoeba and Microcystis species in a hypertrophic pond. Limnology 5:71–76
Opravilova V, Hajek M (2006) The variation of testacean assemblages (Rhizopoda) along the complete base-richness gradient in fens: a case study from the Western Carpathians. Acta Protozool 35:191–204
Payne RJ, Mitchell EAD (2007) Ecology of testate amoebae from mires in the Central Rhodope Mountains, Greece and development of a transfer function for paleohydrological reconstruction. Protist 158:159–171
Payne RJ, Mitchell EAD (2009) How many is enough? Determining adequate count totals for ecological and palaeoecological studies of testate amoebae. J Paleolimnol (in press)
Payne RJ, Blackford JJ (2008) Volcanic impacts on peatlands: Palaeoecological evidence from Alaska. Quat Sci Rev 27:2012–2030
Rao CR (1995) A review of canonical coordinates and an alternative to correspondence analysis using Hellinger distance. Qüestiió 19:23–63
Schwarz MVJ, Frenzel P (2005) Methanogenic symbionts of anaerobic ciliates and their contribution to methanogenesis in an anoxic rice field soil. FEMS Microbiol Ecol 52:93–99
Stockmarr J (1971) Tablets with spores used in absolute pollen analysis. Pollen Spores 13:615–621
Ter Braak C, Šmilauer P (1997-2004) CANOCO for Windows. Biometris-Plant Research, The Netherlands
Tokura M, Ohkuma M, Kudo T (2000) Molecular phylogeny of methanogens associated with flagellated protists in the gut and with the gut epithelium of termites. FEMS Microbiol Ecol 33:233–240
Tolonen K, Warner B, Vasander H (1994) Ecology of testaceans (Protozoa: Rhizopoda) in mires in Southern Finland: II multivariate analysis. Archiv Protistenkund 144:97–112
Vickery E (2006) Monitoring ombrotrophic peatland damage and restoration using protozoa as indicator organisms. PhD thesis, University of Plymouth, Plymouth, UK
Vickery E, Charman DJ (2004) Biomonitoring of peatland restoration using testate amoebae. In: Verhoeven JTA, Dorland E, Coemans M (eds) 7th INTECOL international wetlands conference, vol. Book of abstracts, Utrecht, The Netherlands, 25–30 July 2004
Warner BG (1990) Testate amoebae (Protozoa). Methods in quaternary ecology. Geoscience, Canada 5, Geological Association of Canada, pp. 65–74
Watson A, Nedwell DB (1998) Methane production and emission from peat: the influence of anions (sulphate, nitrate) from acid rain. Atmos Environ 32:3239–3245
Weijers JWH, Schouten S, van der Linden M, van Geel B, Sinninghe Damst JS (2004) Water table related variations in the abundance of intact archaeal membrane lipids in a Swedish peat bog. FEMS Microbiol Lett 239:51–56
Weisse T, Muller H, Pinto-Coelho RM, Schweizer A, Springmann D, Baldringer G (1990) Response of the microbial loop to the phytoplankton spring bloom in a large prealpine lake. Limnol Oceanogr 35:781–794
Woodland W, Charman D, Simms P (1998) Quantitative estimates of water tables and soil moisture in Holocene peatlands from testate amoebae. Holocene 8:261–273
Yeates GW, Foissner W (1995) Testate amoebae as predators of nematodes. Biol Fertil Soils 20:1–7
Acknowledgements
RJP was supported by a Humanities Research Fellowship from the University of Manchester. Fieldwork was funded by the University of Manchester. Thanks to Moray Estates and Scottish Natural Heritage for permission to work on Moidach More. Figure 1 was drawn by Graham Bowden. Comments from three anonymous reviewers helped improve the paper.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Payne, R., Gauci, V. & Charman, D.J. The Impact of Simulated Sulfate Deposition on Peatland Testate Amoebae. Microb Ecol 59, 76–83 (2010). https://doi.org/10.1007/s00248-009-9552-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-009-9552-6