Abstract
The aim of this study was to evaluate the resistance to ten antimicrobial agents and the presence of bla TEM1 gene of Gram-negative bacteria isolated from three natural oligotrophic lakes with varying degrees of anthropogenic influence. A total of 272 indigenous bacteria were recovered on eosin methylene blue medium; they were characterized for antimicrobial resistance and identified taxonomically by homology search and phylogenetic comparisons. Based on 16S ribosomal RNA sequences analysis, 97% of the isolates were found to be Gram-negative bacteria; they belonged to 11 different genera. Members of the genera Acinetobacter, Enterobacter, and Pseudomonas predominated. Most of the bacteria were resistant to at least one antimicrobial. The incidence of resistance to β-lactams, chloramphenicol, and mercury was high, whereas resistance to tetracycline, aminoglycosides, and nalidixic acid was low. There was a great frequency of multiple resistances among the isolates from the three lakes, although no significant differences were found among the disturbed and reference lakes. The ampicillin resistance mechanism of 71% of the isolates was due to the gene bla TEM1 . Our study suggests that multiresistant Gram-negative bacteria and the bla TEM1 gene are common in freshwater oligotrophic lakes, which are subject to different levels of anthropogenic inputs.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Antimicrobial resistance has recently been recognized as a worldwide ecological problem [33, 34, 56, 60]. The emergence and rapid dissemination of antimicrobial resistance among bacterial pathogens are responsible for the high frequency of failure to effectively treat infections worldwide; this is one of the major challenges for public health in the modern world [56]. It is well documented that antimicrobial resistance is a direct consequence of antibiotic overuse and misuse in human and veterinary medicine, aquaculture, and agriculture; this increasing selective pressure exerted on these bacteria can result in the propagation of resistant bacteria in diverse environments [1, 9, 61]. Antibiotic resistance can originate from gene mutations or by horizontal transfer between phylogenetically diverse bacteria [50, 65]. Besides, newly acquired resistance genes may be maintained in new populations in the absence of antibiotic selection pressure [2]. Bacterial antimicrobial resistance is an outcome of evolution and is a natural phenomenon, although it has been accelerated by several human-related activities.
Overall, a high frequency of bacterial resistance to various antimicrobials is well documented in most clinical isolates [25, 39]. Moreover, this phenomenon has also been reported in wild animal populations and natural water samples [4, 16, 33, 35, 47, 53]. These studies demonstrate that resistance can also be maintained without antibiotic selective pressures; however, the extent to which these environmental factors affect resistance is not fully understood.
β-Lactams are among the most frequently used antimicrobials, and resistance to this class of agents is often observed [14, 24, 37]. β-Lactamases, the enzymes that hydrolyze β-lactam antibiotics, are the main source of resistance to these drugs. Genes for β-lactamases may be found on chromosomes, plasmids, transposons, and integrons [23, 64]. TEM-1 β-lactamase gene is common among Gram-negative bacteria; it is one of the main causes of bacterial resistance to β-lactam antibiotics [14, 37, 43].
Considering that few studies have focused on antimicrobial resistance in natural oligotrophic lakes, from tropical regions, and that the human activities can influence the incidence of resistance, we evaluated the resistance to ten antimicrobials and the occurrence of the bla TEM1 gene in culturable Gram-negative bacteria from lakes exposed to varying levels of anthropogenic inputs.
Methods
Study Area
Three natural lakes situated in the middle stretch of the Rio Doce basin, in Brazil, were chosen: Dom Helvécio and Gambazinho lakes, located in a protected area (Parque Estadual do Rio Doce, PERD, 19°29′24″–19°48′18″ S and 42°28′18″–42°38″30″ W, Fig. 1); which is the largest remnant of the Atlantic Forest in the state of Minas Gerais, and Jacaré Lake (9.8 m; 1.03 km2), located in the area surrounding the PERD, which is directly impacted by intermittent loads of untreated domestic sewage and by human and agricultural activity (sport-fishing club, eucalyptus plantations). Dom Helvécio Lake is the deepest (32.5 m) lake in Brazil and the largest (6.87 km2) lake of the middle Rio Doce Lake System (251 lakes with distinct trophic states). Moreover, it is open to the public for recreational activities, (fishing, rowing boats, and swimming) and is less impacted by human activity than Jacaré Lake, very likely a direct consequence of its resilience, provided by its size. Gambazinho Lake (10.3 m; 0.1 km2) is not open to tourists thus exhibiting less disturbed conditions, with restricted access to authorized staff and researchers. Therefore, we consider the Jacaré Lake as the most disturbed by human activity, followed by Dom Helvécio and Gambazinho lakes, the least disturbed reference lake. A particularly important feature of the majority of these lakes is the spreading of exotic species, namely mollusks (e.g., Melanoides tuberculatus) and fishes, e.g., piranha, tucunaré among others, which constitute a real threat to local/regional biodiversity conservation. Among the lakes situated within the PERD, Gambazinho Lake is the only exception among 30 lakes not having exotic fish species [32].
Sampling and Bacterial Isolation
Two samplings efforts were carried out during the dry season in two different years (July 2003 and September 2005). Three replicates (500 mL bottles) were taken from each lake at each sampling time from a depth of 1 m at a central station (limnetic zone). To assess water conditions, selected physical and chemical variables were measured on each date. Water temperature, pH, dissolved oxygen concentration (DO), and oxidation/reduction potential, conductivity, turbidity, and total dissolved solids (TDS) were measured in situ with a multiprobe (Horiba, model U-22) and total alkalinity determined by titration [45]. Concentrations of chlorophyll-a, soluble reactive silicate, total nitrogen (TN), total phosphorus (TP), ammonium-, nitrite-, nitrate-nitrogen, and soluble reactive phosphorus were measured at four depths (100%, 10%, and 1% surface light, and aphotic zone), as previously described [18, 45].
Bacteria were isolated by plating 100 μL of each water sample directly on eosin methylene blue agar plates (EMB, Difco). EMB culture medium is quite selective for Gram-negative bacteria [31]. The total number of colony-forming units (CFU) was determined after incubation at 30°C, for up to 48 h. The isolates were further streaked onto the surface of tryptone soy agar (Oxoid) and checked for purity, prior to subsequent molecular and phenotypic analyses. Subsequently, the isolates were stored in glycerol at −70°C.
Antimicrobial Susceptibility Testing
Minimum inhibitory concentrations (MIC) were determined by the agar dilution method in accordance with the National Committee for Clinical Laboratory Standards (NCCLS) guidelines [48], in Mueller–Hinton medium (Difco). Ten antimicrobial agents were selected as representatives of the drugs commonly used in the treatment of human and animal infections caused by Gram-negative bacteria: ampicillin, amoxicillin–clavulanic acid, tetracycline, chloramphenicol, nalidixic acid, amikacin, gentamicin, kanamycin, streptomycin, and the heavy metal salt, mercury bichloride. All antibiotics were obtained from Sigma Chemical Co. and mercury bichloride was obtained from the Merck Co. The data were interpreted according to MIC breakpoints, as recommended by the NCCLS or by the British Society for Antimicrobial Chemotherapy (BSAC) [3, 44]. In the absence of specific NCCLS and BSAC recommendations for some bacterial genera, the resistance breakpoints defined by NCCLS for Enterobacteriaceae were applied for Aeromonas spp. [8], and MIC breakpoints for non-Enterobacteriaceae were applied for Chryseobacterium spp. and Stenotrophomonas spp. [26, 62]. The susceptibility breakpoint for mercury bichloride was established as 4 µg/mL based on previous study [46].
Multiple Antibiotic Resistance Index
The multiple antibiotic resistance (MAR) index of the samples were calculated by the equation a/(b × c), where a represents the aggregate antibiotic resistance score of all isolates from the sample, b is the number of antibiotics, and c is the number of isolates from the sample [27].
DNA Extraction
Genomic DNA was prepared from a loopful of cells grown in nutrient broth for 18 h. The cell pellet was re-suspended in 500 μL of TE buffer (0.1 mol L−1 Tris–HCl pH 8; 0.001 mol L−1 EDTA). The cells were lysed by addition of 30 μL of SDS 20% and 3 μL of Proteinase K (20 mg mL−1). The DNA was purified as previously described [10].
16S Ribosomal RNA Gene Amplification
The complete 16S ribosomal RNA (rRNA) gene was amplified by touchdown polymerase chain reaction (PCR). Reactions were carried out with the conserved primers PA (5′-TCCTGGCTCAGATTGAACGC-3′), modified from Kuske et al. [30], and with U2 (5′-ATCGGYTACCTTGTTACGACTTC-3′), as described by Lu et al. [42]. Polymerase chain reaction mixtures (20 μL) consisted of 0.4 mM of each dNTP, 0.5 µM of each primer, 1 U of Taq DNA polymerase (Phoneutria, Minas Gerais, Brazil), and 40 ng of bacterial DNA. PCR was conducted with a Mini-cycler ™ PTC-100 (MJ Research Inc. Waltham, MA, USA) with an initial cycle of 10 min at 94°C, followed by 28 cycles of denaturation at 94°C for 1 min, annealing beginning at 53°C and ending at 45°C for 1 min, and extension at 72°C for 3 min. The annealing temperature was lowered 1°C every 2 cycles until it reached 45°C; this annealing temperature was maintained until the end of the cycling process. Cycles were finished by an elongation step of 10 min at 72°C.
Sequencing and Phylogenetic Analysis of 16S rDNA
The 16S rRNA gene partial sequence was made utilizing the primers PA and E926R (5′-CCGICIATTIITTTIAGTTT-3′) [63]. The sequences of PCR products were analyzed automatically by using standard protocols with a DYEnamic ET dye terminator kit (Amersham Biosciences) and the MegaBACE 1000 capillary sequencer (Amersham Biosciences). Each sequence was repeated at least three times in forward and reverse directions for every bacterial isolate. The 16S rRNA gene sequences were analyzed, checked for quality, aligned, and analyzed using Phred v.0.20425 [13], Phrap v.0.990319 [20], and Consed 12.0 [19] softwares.
The 16S rRNA gene sequences were assigned to the taxonomical hierarchy (proposed in release 6.0 of the nomenclatural taxonomy of Garrity and Lilburn by the CLASSIFIER function in the Ribosomal Database Project (RDP)). Sequences were also compared against sequences held in RDP using SEQUENCE_SIMILARITY and against sequences held in GenBank using BLASTN, to search for similar homologous sequences of 16S rRNA gene partial sequences of our 272 isolates. Identification was defined as a 16S rRNA gene sequence similarity of >97% with that of a sequence deposited in the EMBL database [11]. The nucleotide sequences generated were deposited in the GenBank database with accession numbers EU260122 to EU260373.
Bacterial Community Analysis
The Unifrac metric method (http://bmf.colorado.edu/unifrac) was used to compare bacterial communities from the distinct lakes using phylogenetic information [41]. The phylogenetic tree was generated using the neighbor-joining method with MEGA 3.1, using Kimura 2P distance measures. These data were used to compare bacterial communities, testing statistical differences among all samples, by using the unweighted pair group method (UPGMA) and principal coordinate analysis (PCA). Cluster Environments option of Unifrac was used to determine which environments in the tree had similar microbial communities and PCA option was used to find the most important axes along which our samples vary. Jackknifing was used to support UPGMA clustering results and significance tests were also performed, as previously described [40].
bla TEM1 Gene Amplification
Isolates that had an MIC for ampicillin ≥16 µg/mL, for Pseudomonas and Acinetobacter isolates, and ≥32 µg mL−1 for Enterobacteriaceae and other isolates were screened for the bla TEM1 gene. Each isolate was subjected to PCR amplification using primers TEM_F (5′-AAAGATGCTGAAGATCA-3′) and TEM_R (5′-TTTGGTATGGCTTCATTC-3′) described by Speldooren et al. [57]. In all PCR experiments, a blank control (PCR mixture without DNA), the Escherichia coli C282 [blaTEM-1] and E. coli ATCC 25922 were used as a positive and negative control, respectively [6]. Reaction mixtures (20 µL) contained 0.2 mM of each dNTP, 0.1 µM of each primer, 1 U of Taq DNA polymerase (Phoneutria), and 40 ng of bacterial DNA. The temperature profile was as follows: initial cycle of 5 min at 94°C, followed by 40 cycles of denaturation at 94°C for 40 s, annealing at 42°C and 30 s, and extension at 72°C for 1 min 30 s, and a final cycle with an elongation step of 10 min at 72°C.
Statistical Analyses
Data analysis was carried out with the SAS software package [54] with a completely randomized analysis of variances for equally replicated treatments (P > 0.05).
To correlate multiple resistance to the presence of the bla TEM1 gene, the observations were classified simultaneously according to two attributes, the MAR index [low (0.3–0.6) and high (0.7–1.0)] and the occurrence of bla TEM1 gene [presence (+) or absence (−)]. The frequencies in the different categories were arranged in a two-way table (known as the 2 × 2 contingency table). Then, a Fisher exact probability was used as a test for systematic association of attributes [59].
The software NTSYS-PC version 2.2 (Exeter Software. Co., New York, USA) was used for PCA of environmental variables. The blot was redrawn from a PCA compiled through NTSYS-PC. Linear regression was performed to confirm PCA results.
Statistical comparisons of results from anthropogenically disturbed (Jacaré and Dom Helvécio) and undisturbed reference (Gambazinho) lakes were done to determine differences in bacterial resistance.
Results
Abiotic Features of the Lakes
The physical and chemical characteristics of the lakes are given in Table 1. In July 2003, the lakes exhibited practically isothermal conditions while high variation of along the water column temperature was observed in September 2005. The maximum difference in temperature between the top and bottom layers in September was 4.7°C, while in Gambazinho Lake, this difference was only 0.9°C.
Anoxic conditions were only observed in Dom Helvécio Lake in 2003. There were marked differences for oxidation–reduction potential among the lakes in 2005, with the lowest levels in Dom Helvécio Lake and substantially higher values in 2003. Significant differences were also detected in turbidity (P < 0.01) and TDS (P < 0.01) concentrations among the lakes with values ranging from as low as 0.03 in Jacaré, a disturbed lake out of the PERD, to 10 in Gambazimho, an undisturbed lake. Conductivity usually did not fluctuate greatly except for Gambazinho Lake. According to the Salas and Martino [55] model, the lakes were classified as oligotrophic for both years.
Identification and Phylogenetic Analysis of the Isolates
The total number of isolates recovered on EMB culture medium in each lake per sampling ranged from 1.4 × 102 CFU/ml to 4.9 × 102 CFU/ml. A total of 272 randomly selected bacterial isolates (51, 46, 39 and 48, 45 and 43 from Jacaré, Dom Helvécio, and Gambazinho lakes in 2003 and 2005, respectively) were identified by partial 16S rRNA gene sequence analysis, and more than 98% were affiliated to the genus level. Most of the identified bacteria (97%) were Gram-negative.
Analysis of the bacterial isolates revealed a predominance of γ-Proteobacteria (89%), belonging to nine genera: Acinetobacter (34.2%), Enterobacter (26.1%), Pseudomonas (13.2%), Aeromonas (3.3%), Erwinia (3.3%), Stenotrophomonas (2.9%) Morganella (2.2%), Serratia (0.7%) and Moraxella (1.5%; Fig. 2). Among these, high relative abundance was detected for Enterobacter (Jacaré Lake, 2003 sampling), Acinetobacter (Jacaré and Dom Helvécio lakes, sampling 2005), and Pseudomonas (Gambazinho Lake, sampling 2005; Fig. 2). The other bacterial isolates were β-Proteobacteria (0.5%), flavobacteria (7.5%), bacilli (2.5%), and actinobacteria (0.5%). Other specific genera also found were such as Aquitalea, Chryseobacterium, Lactococcus, Staphylococcus, and Arthrobacter (Fig. 2). Bacteria belonging to the genera Aquitalea, Lactococcus, Staphylococcus, Serratia, Arthrobacter, and Stenotrophomonas were isolated in smaller proportions. Only five isolates fell into unclassified genera and were assigned to families of Enterobacteriaceae and Pseudomonadaceae. Among the 14 genera identified, Acinetobacter, Enterobacter, and Pseudomonas constituted 74% of the isolates.
Comparison of Bacterial Genera in the Communities
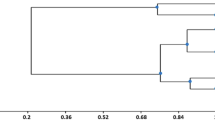
The distribution of bacterial genera differed considerably across the three lakes in the samples collected in 2003 and 2005. Gambazinho and Dom Helvécio lakes (sampling, 2003) had a considerably higher number of genera than found in Jacaré Lake (Fig. 2). In 2005, the three lakes had similar bacterial communities (Pseudomonas, Acinetobacter, Enterobacter, and Stenotrophomonas), differing only in the number of isolates. This conclusion was based on the UPGMA tree and PCA, in which the isolates from the three lakes clustered together (Figs. 3 and 4); similarity among them was well supported by jackknife values (P < 0.001). The node that groups these isolates (N3, Fig. 3) was recovered 100% of the times (considering the small sample composed of 39 sequences). In PCA, the first principal component separated the bacterial communities isolated in 2005 from those isolated in 2003. Also, samples isolated in 2003 were more diverse, with more variation among them (Fig. 4). The highest similarity was observed between bacterial communities from Jacaré and Dom Helvécio lakes sampled in 2005, dominated by Acinetobacter isolates. Bacterial communities isolated from the three lakes in 2003 were significantly different from those isolated in 2005, as shown by Unifrac significance tests (P ≤ 0.05). In general, the variation in the number of genera was higher between samples from the same lake at different years than between samples from different lakes.
PCA ordination diagram (covariance two-dimensional plot of the dataset) of genera abundance from the three lakes sampled in different periods, and with 16 measured environmental variables. Axis scaling is for sample scores. One unit is equivalent to 20 for environmental variables and 40 for genera scores. Note that factor 2 only accounts for a minor part of the variance
PCA
PCA was also useful to analyze the samples in environmental space, correlating the axes along which variation occurred with environmental variables. The PCA of the dataset produced two factors, 1 (pH) and 2 (temperature), that accounted for 67.47% and 28.86% of the variance, respectively, or 96.33% of the total (Fig. 4). Except for the Enterobacter and pH (Pearson (n) = 0.856, P < 0.001); Chryseobacterium and temperature (Pearson (n) = −0.907, P < 0.05); Enterobacteriacea and D.O. (Pearson (n) = −0.958, P < 0.05); and Enterobacteriaceae and redox (Pearson (n) = −0.910 P < 0.05) no other correlations were found between environmental variables and the other genera studied. Linear regression analysis of these three environmental variables and specific genera confirmed these findings (not shown).
Antimicrobial Resistance Analyses
The Gram-negative bacteria that were isolated from the three oligotrophic lakes in the 2 years (2003 and 2005) were characterized for their antibiotic resistance phenotype. Resistance profiles were different among lakes and sampling periods.
Bacteria isolates resistant to three or more of the antimicrobial agents were designated as multiple antimicrobial resistant (ranging from 3 to 10 antimicrobials); they were recovered from all samples at high percentages. A high frequency of multiresistant bacteria was seen for most of the isolates from all the three lakes: Jacaré Lake (76% in 2003 and 100% in 2005), Dom Helvécio Lake (62% in 2003 and 85% in 2005), and Gambazinho Lake (64% in 2003 and 77% in 2005), but these percentages were not statistically different between the years. Overall, the MAR indices did not present any significant difference (P = 0.6873) among the lakes in all sampling periods, ranging from 0.44 to 0.52.
Pseudomonas, Acinetobacter, and Enterobacter Isolates
Pseudomonas, Acinetobacter, and Enterobacter isolates represented the majority of bacterial isolates retrieved from water samples. Two percent of these isolates were susceptible to all tested antimicrobial agents, 7% were resistant to a single antimicrobial and 82% were multiresistant. This high incidence of resistance was found in isolates from all lakes (Table 2). The most common resistance phenotypes detected were to ampicillin (90%), amoxicillin–clavulanic acid (77%), and chloramphenicol (76%).
Pseudomonas isolates exhibited the highest MICs, and they had the highest antimicrobial resistance percentage to most of the antimicrobials (except for the aminoglycosides). Pseudomonas and Acinetobacter isolates presented the lowest MICs and the lowest percentage resistance to amikacin and gentamicin (Table 2). Otherwise, resistance to amikacin and gentamicin was higher for Enterobacter isolates; 90% of the isolates were at or below ≥128 and 64 μg mL−1, respectively (Table 2). Enterobacter and Acinetobacter isolates from Gambazinho Lake were less resistant than those from Jacaré and Dom Helvécio, mainly for the antimicrobials ampicillin, amoxicillin–clavulanic acid, nalidixic acid, and chloramphenicol.
Resistance to mercury was observed in all the genera, and it was generally associated with at least two other resistance markers. The highest percentage of mercury resistance was observed for Pseudomonas (78%), followed by Enterobacter (51%). Resistance to multiply antimicrobials exhibited was variable among the isolates.
Other Gram-Negative Bacteria
Other Gram-negative isolates, Serratia, Moraxella, Stenothrophomonas, Erwinia, Aeromonas, Aquitalea, Morganella, and Chryseobacterium, were recovered from the three lakes in smaller numbers than were Enterobacter, Acinetobacter, and Pseudomonas isolates. For most of these isolates, with the exception of the Enterobacteria, no standard MIC breakpoints have been established.
Among Enterobacteria and Aeromonas isolates, the highest resistance percentages were detected against ampicillin, chloramphenicol, and gentamicin (Table 2). Against Chryseobacterium isolates, almost all of the antimicrobials tested (except for nalidixic acid) seem to be active compounds, with high MIC50 and MIC90 values (Table 2). All Stenotrophomonas isolates were resistant to all of the aminoglycosides, with MIC50 and MIC90 at or below ≥128 μg mL−1 (Table 2).
Antibiotic resistance was different among the different genera. Seventy-one percent of these Gram-negative bacterial isolates were multiresistant, and all Stenotrophomonas isolates displayed multiresistance. More than 65% of these isolates were resistant to ampicillin, chloramphenicol, gentamicin, and amikacin. Ninety-three percent of these bacteria, for which the breakpoint for ampicillin was not available, presented high MIC50 values (≥128 μg mL−1), suggesting resistance to this antibiotic (Table 2). Only one isolate of Aquitalea genus was obtained; it exhibited low MICs to almost all antibiotics tested, with the exception of the β-lactams.
Presence of the bla TEM Gene in the Gram-Negative Bacteria
Ampicillin-resistant bacteria were predominant, and we therefore considered it important to examine them for the bla TEM gene. A total of 232 ampicillin-resistant Gram-negative isolates were screened for the bla TEM gene. The PCR-amplified bla TEM revealed that ampicillin-resistant isolates were strongly associated with this gene as well as to multiple antimicrobial resistance for Jacaré and Dom Helvécio lakes in the first year of sampling (P < 0.001). This gene was found in all the genera. Among the 165 isolates harboring the bla TEM gene, there were 73 Acinetobacter, 40 Enterobacter, 24 Pseudomonas, 11 Chryseobacterium, five Stenotrophomonas, four Erwinia, two Morganella, two Moraxella, one Aquitalea, one Serratia, one Aeromonas, and one representative from the Pseudomonadaceae family.
Discussion
Considerable concern exists about antibiotic-resistant pathogenic bacteria in clinical environments. Aquatic ecosystems which received wastewater, pathogenic and opportunistic bacteria, could serve as a reservoir of resistance genes, eventually inserted into mobile elements [5]. On the other hand, understanding about the community structure of antibiotic resistant bacteria and potential abiotic factors controlling species abundances in natural tropical lakes are still poorly studied. The present study reports on the antibiotic resistance potential of Gram-negative bacteria from natural lakes, despite a lack of known prior exposure to antibiotics.
The bacterial isolates originated from natural oligotrophic lakes that were under the same climatic and isothermal conditions during the sampling periods. Mixture of the water column, facilitated mainly by wind force [52], promoted homogeneous distribution of organisms and nutrients along the water column exhibiting the well-known warm monomictic pattern [6]. Therefore, our data should be representative of the entire water column.
The relatively low number of colony-forming units per milliliter obtained in this study could be due to the use of selective culture medium (EMB), since our purpose was the isolation only of Gram-negative bacteria. Another reason could be related to oligotrophic environment, according to Pagioro et al. [51]. This environment exhibits a lower density of bacteria.
Analysis of 16S rRNA gene sequences identified almost all bacteria at the genus level. We could have used either MacConkey or EMB media for the isolation of Gram-negative bacteria, but MacConkey medium has more nutrients than EMB. Although our experimental design favored isolation of Gram-negative bacteria (97%), it allowed the isolation of some (3%) Gram-positive bacteria that were naturally present in the lakes, including Arthrobacter, Staphylococcus, and Lactobacillus isolates. As expected, the isolation medium was highly selective for the recovery of Gram-negative bacteria (Aeromonas, Acinetobacter, Aquitalea, Enterobacter, Erwinia, Morganella, Serratia, Moraxella, Pseudomonas, Stenotrophomonas, and Chryseobacterium). It is known that these bacteria are ubiquitous in the environment, although most studies on antimicrobial resistance have been performed using strains of clinical origin [35, 49]. Besides that, multiple antimicrobial resistance has been found to be a common phenomenon among Gram-negative bacteria [15, 42, 43].
It is known that the composition of bacterioplankton communities in lakes and oceans varies in time and space [38, 58]. Significant differences were found in bacterial composition among the lakes studied in 2003, when compared to those observed in 2005, as shown by Unifrac analysis, which allows the comparison of small number of isolates [41]. Furthermore, bacterial composition revealed that the Jacaré Lake showed their own unique predominant bacteria in both sampling periods, whereas the samples collected from Gambazinho and Dom Helvécio lakes only presented unique predominant bacteria in the 2003 sampling period. The predominance of Acinetobacter, Enterobacter, and Pseudomonas suggest that these bacteria may be important inhabitants in tropical lakes; they have also been reported in previous studies [4, 12, 23]. Although Aeromonas is considered a waterborne bacterium [17], it was found in small numbers likely because it is not easily isolated on EMB and requires selective media. Most of the bacteria that were isolated are opportunistic pathogens.
The impact of the dissemination of antimicrobial resistant bacteria and their resistance genes in natural aquatic environments has been of major concern [2, 29]. Many pathogens and commensals are common in aquatic environments. The emergence of bacterial resistance in aquatic environments can bring increased risk to human and animal health [26]. A high frequency of Gram-negative resistant bacteria was detected in the three lakes; most of the isolates (98%) were resistant to at least one of the antimicrobials. Most of them demonstrated resistance to ampicillin, amoxicillin–clavulanic acid, and chloramphenicol and were more susceptible to aminoglycosides. Similar results have been reported for Acinetobacter spp. isolated from various aquatic sources (sewage, freshwater aquaculture, and intestinal contents) by Guardabassi [21]. However, the percentage resistance that we observed was higher mainly for amoxicillin associated with clavulanic acid, a β-lactamase inhibitor. A predominance of ampicillin-resistant Gram-negative bacteria was also reported in the rivers of the USA by Ash et al. [4] and Enterobacteriaceae isolated from pristine freshwater [36]. In contrast, Goñi-Urriza and coworkers [17] reported much lower frequencies of ampicillin-resistant and multiple antimicrobial resistant bacteria found in an urban effluent.
The MAR index is an excellent tool that permits us to analyze the dissemination of bacteria resistance in a given population. When isolates are exposed to high risk sources of contamination where antibiotics are often used, a MAR index value >0.2 is observed. When antibiotics are seldom or never used, a MAR index value less than or equal to ≤0.2 is observed [28]. MAR indices of the lakes studied were >0.4, which was unexpected; to the knowledge of the authors, the lakes had not previously been exposed to antimicrobial agents.
It is well established that most Gram-negative bacteria are resistant to β-lactams, due to β-lactamase production [14]; molecular detection of the genes encoding these enzymes has been used to investigate the dissemination of resistance in clinical isolates [14, 40]. In view of the high percentage of bacteria resistant to β-lactams in our samples and the clinical importance of this class of antibiotics, we were prompted to analyze the occurrence of the bla TEM gene among the different isolates. The bla TEM1 gene was detected in most of the isolates resistant to ampicillin recovered from our samples. The proportion of isolates in this collection of β-lactam-resistant bacteria carrying the bla TEM1 gene is comparable to that of other studies [7]. We conclude that the bla TEM1 gene is widespread in clinical as well as from natural oligotrophic lake isolates.
Although most of the bacteria inhabiting the three natural oligotrophic lakes are multiresistant and harbor the bla TEM1 gene, our results did not reflect the anthropogenic disturbance level between disturbed and reference lakes. From an evolutionary perspective, these observations make it tempting to speculate that resistant environmental bacteria pre-exist to known industrialized antibiotics: Hall and Barlow [22] estimated that β-lactamases arose over two billion years ago, predating the divergence of Gram-positive and Gram-negative bacteria. Further studies to address these issues would be helpful in understanding the dynamics of the resistance and the role of human activities in this complex phenomenon.
References
Agerso Y, Sandvang D (2005) Class 1 integrons and tetracycline resistance genes in alcaligenes, Arthrobacter, and Pseudomonas spp. isolated from pigsties and manured soil. Appl Environ Microbiol 71:7941–7947
Alonso A, Sanchez P, Martinez JL (2001) Environmental selection of antibiotic resistance genes. Environ Microbiol 3:1–9
Andrews JM (2001) BSAC standardized disc susceptibility testing method. J Antimicrob Chemother 48:43–57
Ash RJ, Mauck B, Morgan M (2002) Antibiotic resistance of Gram-negative bacteria in rivers, United States. Emerg Infect Dis 8:713–716
Baquero F, Martínez JL, Cantón R (2008) Antibiotics and antibiotic resistance in water environments. Curr Opin Biotechnol 19:260–265
Barbosa FAR, Ppadisãk J (2002) The forgotten lake stratification pattern: atelomixis, and its ecological importance. Verh Int Verein Limnol 28:1385–1395
Brizas L, Zarazaga M, Saenz Y, Ruiz-Larrea F, Torres C (2002) Beta-lactamases in ampicillin-resistant Escherichia coli isolates from foods, humans, and healthy animals. Antimicrob Agents Chemother 46:3156–3163
Castro-Escarpulli G, Figueras MJ, Aguilera-Arreola G, Soler L, Fernandez-Rendon E, Aparicio GO, Guarro J, Chacon MR (2003) Characterisation of Aeromonas spp. isolated from frozen fish intended for human consumption in Mexico. Int J Food Microbiol 84:41–49
Díaz-Cruz MS, Alda MJL, Barceló D (2003) Environmental behavior and analysis of veterinary and human drugs in soils, sediments and sludge. Trends Anal Chem 22:340–351
Dolzani L, Tonin E, Lagatolla C, Monti-Bragadin C (1994) Typing of Staphylococcus aureus by amplification of the 16S–23S intergenic spacer sequences. FEMS Microbiol Lett 119:167–174
Drancourt M, Bollet C, Carlioz A, Martelin R, Gayral JP, Raoult D (2000) 16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates. J Clin Microbiol 38:3623–3630
Drucker VV, Panasyuk EY (2006) Potentially pathogenic bacteria in a microbial community of Lake Baikal. Hydrobiologia 568:267–271
Ewing B, Green P (1998) Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res 8:186–194
Fluit AC, Visser MR, Schmitz FJ (2001) Molecular detection of antimicrobial resistance. Clin Microbiol Rev 14:836–871
George AM (1996) Multidrug resistance in enteric and other gram-negative bacteria. FEMS Microbiol Lett 139:1–10
Gilliver MA, Bennett M, Begon M, Hazel SM, Hart CA (1999) Antibiotic resistance found in wild rodents. Nature 401:233–234
Goñi-Urriza M, Capdepuy M, Arpin C, Raymond N, Caumette P, Quentin C (2000) Impact of an urban effluent on antibiotic resistance of riverine Enterobacteriaceae and Aeromonas spp. Appl Environ Microbiol 66:125–132
Golterman HL, Clymo RS, Ohnstad MAM (1978) Methods for chemical analysis of fresh waters. Blackwell Scientific, Oxford
Gordon D, Abajian C, Green P (1998) Consed: a graphical tool for sequence finishing. Genome Res 8:195–202
Green P. (1994) PHRAP documentation. http://www.phrap.org
Guardabassi L, Dalsgaard A, Olsen JE (1999) Phenotypic characterization and antibiotic resistance of Acinetobacter spp. isolated from aquatic sources. J Appl Microbiol 87:659–667
Hall BG, Barlow M (2004) Evolution of the serine β-lactamases: past, present and future. Drug Resist Updat 7:111–123
Hazen TC (1988) Fecal coliforms as indicators in tropical waters: a review. Int J Toxic Assess 3:461–47
Henriques I, Moura A, Alves A, Saavedra MJ, Correia A (2006) Analysing diversity among beta-lactamase encoding genes in aquatic environments. FEMS Microbiol Ecol 56:418–429
Hujer KM, Hujer AM, Hulten EA, Bajaksouzian S, Adams JM, Donskey CJ, Ecker DJ, Massire C, Eshoo MW, Sampath R, Thomson JM, Rather PN, Craft DW, Fishbain JT, Ewell AJ, Jacobs MR, Paterson DL, Bonomo RA (2006) Analysis of antibiotic resistance genes in multidrug-resistant Acinetobacter sp. isolates from military and civilian patients treated at the Walter Reed Army Medical Center. Antimicrob Agents Chemother 50:4114–4123
Kirby JT, Sader HS, Walsh TR, Jones RN (2004) Antimicrobial susceptibility and epidemiology of a worldwide collection of Chryseobacterium spp.: report from the SENTRY Antimicrobial Surveillance Program (1997–2001). J Clin Microbiol 42:445–448
Krumperman PH (1983) Multiple antibiotic resistance indexing Escherichia coli to identify risk sources of faecal contamination of foods. Appl Environ Microbiol 46:165–170
Krumperman PH (1985) Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl Environ Microbiol 46:165–170
Kummerer K (2004) Resistance in the environment. J Antimicrob Chemother 54:311–320
Kuske CR, Barns SM, Busch JD (1997) Diverse uncultivated bacterial groups from soils of the arid southwestern United States that are present in many geographic regions. Appl Environ Microbiol 63:3614–3621
Laitinen S, Nevalainen A, Kotimaa M, Liesivuori J, Martikainen P (1992) Relationship between bacterial counts and endotoxin concentrations in the air of wastewater treatment plants. Appl Environ Microbiol 58:3774–3776
Latini AO, JrM P (2004) Reduction of a native fish fauna by alien species: an example from Brazilian freshwater tropical lakes. Fish Manag Ecol 11:71–79
Levy S (1997) Antibiotic resistance: an ecological imbalance. In Antibiotic resistance: origins, evolution, selection and spread. In Ciba Foundation Symposium 207
Levy SB (2001) Antibiotic resistance: consequences of inaction. Clin Infect Dis 33:124–129
Levy SB (2005) Antibiotic resistance—the problem intensifies. Adv Drug Deliv Rev 57:1446–1450
Lima-Bittencourt CI, Cursino L, Gonçalves-Dornelas H, Pontes DS, Nardi RMD, Callisto M, Chartone-Souza E, Nascimento AMA (2007) Multiple antimicrobial resistance in Enterobacteriaceae isolates from pristine freshwater. Gen Mol Res 6:510–521
Livermore DM (1995) Beta-lactamases in laboratory and clinical resistance. Clin Microbiol Rev 8:557–584
Lobova TI, Maksimova EY, Popova LY, Pechurkin NS (2002) Geographical and seasonal distribution of multiple antibiotic resistance of heterotrophic bacteria of Lake Shira. Aquat Ecol 36:299–307
Lodise TP Jr, Lomaestro B, Drusano GL (2007) Piperacillin-tazobactam for Pseudomonas aeruginosa infection: clinical implications of an extended-infusion dosing strategy. Clin Infect Dis 44:357–363
Lozupone C, Knight R (2005) UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71:8228–8235
Lozupone C, Hamady M, Knight R (2006) UniFrac—an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics 7:371
Lu JJ, Perng CL, Lee SY, Wan CC (2000) Use of PCR with universal primers and restriction endonuclease digestions for detection and identification of common bacterial pathogens in cerebrospinal fluid. J Clin Microbiol 38:2076–2080
Mabilat C, Courvalin P (1990) Development of “olygotyping” for characterization and molecular epidemiology of TEM-lactamase in members of the family Enterobacteriaceae. Antimicrob Agents Chemother 34:2210–2216
MacGowana AP, Wise R (2001) Establishing MIC breakpoints and the interpretation of in vitro susceptibility tests. J Antimicrob Chemother 1:17–28
Mackereth FJH, Heron J, Talling JF (1978) Water analysis: some revised methods for limnologists. Freshwater Biological Association Scientific Publication, 36, Titus Wilson & Sons Ltd, Kendal, p 117p
Nascimento AMA, Chartone-Souza E (1999) Re-evaluation of antibiotic and mercury resistance in Escherichia coli populations isolated in 1978 from Amazonian rubber tree tappers and Indians. Res Microbiol 150:407–411
Nascimento AMA, Cursino L, Gonçalves-Dornelas H, Reis A, Chartone-Souza E, Marini MA (2003) Antibiotic-resistant Gram-negative bacteria in birds from the Brazilian Atlantic forest. Condor 105:358–361
National Committee for Clinical Laboratory Standards (NCCLS) (2001) Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. ASM, Washington
Ndip RN, Dilonga HM, Ndip LM, Akoachere JF, Nkuo Akenji T (2005) Pseudomonas aeruginosa isolates recovered from clinical and environmental samples in Buea, Cameroon: current status on biotyping and antibiogram. Trop Med Int Health 10:74–81
O’Brien TF (2002) Emergence, spread, and environmental effect of antimicrobial resistance: how use of an antimicrobial anywhere can increase resistance to any antimicrobial anywhere else. Clin Infect Dis 34:78–84
Pagioro TA, Velho LFM, Lansac-Tôha FA, Pereira DG, Nakamura AKS, Perenha MCZ, Santos VD (2003). Influência do grau de trofia sobre os padrões de abundância de bactérias e protozoários planctônicos em reservatórios do Estado do Paraná. In: Rodrigues, L.; Agostinho, A. A.; Gomes, L. C. & Thomaz, S. M. (eds). Anais do Workshop Produtividade em Reservatórios e Bioindicadores. Nupélia. Maringá. 281p.
Petrucio MM, Barbosa FAR, Furtado ALS (2006) Bacterioplankton and phytoplankton production in seven lakes in the middle Rio Doce basin, south-east Brazil. Limnologica 36:192–203
Pontes DS, Lima-Bittencourt CI, Azevedo MSP, Chartone-Souza E, Nascimento AMA (2007) Phenotypic and genetic analysis of Enterobacter spp. from a Brazilian oligotrophic freshwater lake. Can J Microbiol 53:983–991
Institute SAS (1987) SAS/STAT guide for personal computers, version 6. SAS Institute, Cary, NC
Salas HJ, Martino P (1991) A simplified phosphorus trophic state model for warm-water tropical lakes. Water Res 25:341–350
Singer RS, Ward MP, Maldonado G (2006) Can landscape ecology untangle the complexity of antibiotic resistance? Nat Rev Microbiol 4:943–952
Speldooren V, Heym B, Labia R, Nicolas-Chanoine MH (1998) Discriminatory detection of inhibitor-resistant beta-lactamases in Escherichia coli by single-strand conformation polymorphism-PCR. Antimicrob Agents Chemother 42:879–884
Stabili L, Cavallo RA (2004) Biodiversity of culturable heterotrophic bacteria in the southern Adriatic Sea Italian coastal waters. Sci Mar 68:31–41
Steel RGD, Torrie JH (1980) Principles and procedures of statistics: a biometrical approach. McGraw-Hill, Toronto, Ontario, Canada
Summers AO (2002) Generally overlooked fundamentals of bacterial genetics and ecology. Clin Infect Dis 34:85–92
Teuber M (2001) Veterinary use and antibiotic resistance. Curr Opin Microbiol 4:493–499
Valdezate S, Vindel A, Loza E, Baquero F, Cantón R (2001) Antimicrobial susceptibilities of unique Stenotrophomonas maltophilia clinical strains. Antimicrob Agents Chemother 45:1581–1584
Watanable K, Kodama Y, Harayama S (2001) Design and evaluation of PCR primers to amplify 16S ribosomal DNA fragments used for community fingerprinting. J Microbiol Methods 44:253–262
Weldhagen GF (2004) Integrons and beta-lactamases—a novel perspective on resistance. Int J Antimicrob Agents 23:556–562
White DG, Mcdermott PF (2001) Emergence and transfer of antibacterial resistance. J Dairy Sci 84:151–155
Acknowledgments
We thank FAPEMIG and CNPq for providing financial support. DSP, FRS, and AMAN were supported by CNPq.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Pontes, D.S., Pinheiro, F.A., Lima-Bittencourt, C.I. et al. Multiple Antimicrobial Resistance of Gram-Negative Bacteria from Natural Oligotrophic Lakes Under Distinct Anthropogenic Influence in a Tropical Region. Microb Ecol 58, 762–772 (2009). https://doi.org/10.1007/s00248-009-9539-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-009-9539-3