Abstract
Although organic-aggregate-associated bacteria play a pivotal role in microbial food webs and in the cycling of major elements, their community composition and diversity have not been extensively studied, especially in shallow freshwater systems. This study is among the first to explore intra-lake horizontal heterogeneity of organic-aggregate-associated bacterial community composition (OABC) in the large, shallow, and eutrophic Lake Taihu. During November 2006, samples were collected at four locations representing different trophic states and food web structures. Regional variability of OABC and diversity were studied by amplified ribosomal DNA restriction analysis and comparative analysis of four large 16S ribosomal RNA clone libraries. Our results demonstrate that OABC were numerically dominated by members of the β-proteobacteria (19.2–38.6%), Bacteroidetes (3.6–20.0%), and α-proteobacteria (11.5–19.2%) groups. The dominance of the Bacteroidetes group was related to algae-based aggregates. Horizontal heterogeneity of OABC exists within habitats, suggesting that the trophic state of the water and the physicochemical properties of organic aggregates (OA) play a key role. Diverse bacterial communities found on OA were substantially different from free-living ones. Comparative statistical analyses of the habitats of OA-associated bacteria highlight the potential ecological importance of the exchange between OABC and the surrounding planktonic community. Lastly, we found at least 45% of sequences closely related to ones previously found in soils, sludge, sediments, and other habitats. This demonstrates that microorganisms from terrestrial and sediment habitats are an important component of OA.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Microscopic and macroscopic aggregates are an abundant component of aquatic ecosystems. The occurrence and ecological importance of macroscopic aggregates (also known as marine snow) in the pelagic environment has been extensively studied for more than 30 years [1, 21, 26, 44], and several studies on macroscopic aggregates in deep-lake and lotic ecosystems, known as lake snow and river snow, have been reported in recent years [7, 18, 19]. It has also been suggested that aggregates serve as transient microhabitats suitable for anaerobic processes within an oxygenated water column [34]. Abundant bacteria, rich nutrients, and high metabolic activity of attached microorganisms made the aggregates “hotspots” in energy fluxing, biogeochemical cycling, and food web dynamics [2, 6, 9, 33, 43]. Attached bacteria are often larger and more abundant and are capable of having much higher cellular uptake rates of sugars and amino acids than free-living bacteria [43]. Comparative phylogenetic evidence based on 16S ribosomal DNA (rDNA) sequences of aggregate-associated and free-living bacteria indicated that organic-aggregate-associated bacterial communities (OABC) differ from those inhabiting the surrounding water column [14, 38]. Although there are differences between aggregate-associated and free-living bacterial communities, the release of amino acids into the surrounding water and high detachment rates of aggregate-associated bacteria suggest substantial exchange between the two communities [20, 51].
Compared with pelagic systems in which low hydrodynamic stress allows macroscopic (>500 μm) organic aggregates to form, aggregate formation in shallow turbid systems experiencing strong wind mixing exhibits pronounced differences [43]. Wind-driven sediment resuspension and intense restructuring of the aggregates are major features of shallow systems [43]. As a result, microaggregates (5–500 μm) can dominate shallow systems. Aggregate-associated microbial decomposition is likely to be significant in shallow systems and can lead to the solubilization and mineralization of particulate organic matter, which can supply phytoplankton with inorganic nutrients [43].
While organic aggregates in marine, deep-lake, and lotic systems have been well characterized, little is known about the OABC and their ecological importance in large shallow lakes. To better understand the importance of OABC in these systems, we investigated the spatial heterogeneity of OABC in a shallow eutrophic subtropical lake (Lake Taihu), the third largest lake in China. In contrast to smaller lakes, Taihu has significant geographic environmental gradients, and it harbors two distinct ecological habitats (i.e., macrophyte- and phytoplankton-dominated lake habitats). For these reasons, Taihu provides an ideal model system for investigating the influences of environmental parameters on OABC [36, 55]. Several studies have been reported on the composition and dynamics of pelagic and total bacterial communities in Taihu [52, 54–57]. This is the first attempt to explore the composition of OABC in this large subtropical shallow lake. The major aim of the present study was to explore and characterize the OABC in Lake Taihu and to assess (1) whether the OA-associated and free-living differ in composition and (2) whether horizontal heterogeneity of OABC exists within habitats having different trophic states and food web dynamics.
Materials and Methods
Site Description and Sampling
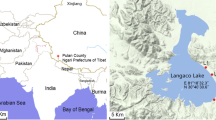
Lake Taihu (30° 05′–32° 08′–N and 119° 08′–121° 55′–E) is located in the delta of the Yangtze River in Eastern China. It has a large surface area (2,338 km2) and is shallow (mean depth = 1.9 m) [36]. The maximum lake length is 68.5 km and the width is 56 km (Fig. 1). The shallowest region, which has a mean depth of <1.5 m, is about 452 km2 and situated mostly in East Taihu, while the area with depths >2.5 m is in the north and west part of the lake, occupying 197 km2. Freshwater input is dominated by the northern and western watersheds, which discharge via the eastern basin and East Taihu Bay. The hydrology and nutrient inputs to the lake result in a trophic gradient characterized by hypereutrophic conditions in the northern part and mesotrophic conditions in its southeastern part [12].
Comparison of the quantitative distribution of clones affiliated with selected phylogenetic groups in the noncyanobacterial organic-aggregate-associated bacterial clone libraries from four locations of Lake Taihu. Clones affiliated with Acidobacteria, Verrucomicrobia, Nitrospira, and Gemmatimonadetes are included in “Others.” The sampling sites are located in the areas of River Mouth of Liangxi River (site 0), Meiliang Bay (site 3), Lake Center (site 8), and East Taihu (site 24) with contrasting physicochemical properties. The sampling sites are described in more detail in Table 1
Samples were collected at four stations representing different trophic states and food web structures. More detailed ecological characterizations of the four stations can be found elsewhere [31, 54, 55]. One sampling station (site 0) is located in a highly eutrophic area near the river mouth of Liangxi River; the eutrophication is due to nitrogen and phosphorus from domestic wastewater discharged from the river (Fig. 1). The second sampling station (site 3) is located in Meiliang Bay, which is also highly enriched with nitrogen and phosphorus. This bay, at the northern end of Taihu, has a surface area of approximately 100 km2. It experiences intensive blooms of algae, dominated by Cyanobacteria, during summer and autumn. The third sampling station (site 8) is located in the open lake (Fig. 1), where the water is less enriched with nitrogen and phosphorus but exposed to frequent wind mixing [54]. The fourth sampling station (site 24) is located in East Taihu, which is characterized by submersed macrophyte communities and clear water, with relatively low phytoplankton concentrations.
Samples were collected from surface water (top 50 cm) with 5-l plastic bottles cleaned with 75% ethanol on November 14, 2006. The samples were immediately transported to the laboratory and processed. A 46-ml subsample was transferred in autoclaved PP tubes (Greiner Bio-one GmbH, Germany) containing 4 ml of prefiltered (pore size, 0.2 μm) glutaraldehyde (final concentration 2% v/v). Samples were then stored in a refrigerator at 4°C until slides for enumeration and image analysis were prepared. To concentrate the aggregates, samples were centrifuged (5,000×g) for 10 min, modified after [7]. The precipitates were resuspended in autoclaved particle-free 0.85% NaCl solution to increase the concentration 200-fold. They were then used to count the abundances of bacteria associated with OA or stored frozen at −80°C until DNA extraction. Water depth, water temperature, pH, Secchi depth, and coverage by macrophytes were measured on location. Conductivity, dissolved oxygen, total nitrogen (TN), dissolved inorganic nitrogen (DIN), total phosphorus (TP), dissolved inorganic phosphorus, total suspended solids (TSS), dissolved organic carbon, chlorophyll a (Chl a), and the OA-associated TP and TN were measured according to standard methods [23].
Enumeration of Bacteria and Organic Aggregates
For bacterioplankton and OA abundance, a well-mixed subsample was diluted with particle-free distilled water (1:10) and stained with 4′,6′-diamidino-2-phenylindole (DAPI, Sigma) at a final concentration of 2 μg/ml for 10 min [25]. In order to minimize destruction of aggregates, vacuum filtration was achieved by a simple hand pump (MityVac, USA) with the vacuum press <10 mmHg. After staining, samples were filtered onto black polycarbonate filters (0.2-μm pore size, 25-mm diameter, Poretics™) and embedded in nonfluorescent immersion oil (Cargille type A, Cargille Laboratories, Inc., USA). Bacterioplankton cells and OA were enumerated using a Zeiss Axiovert 135 M epifluorescence microscope equipped with an HBO 100-W mercury lamp and a filter set (excitation: G 365, emission: BP 445/50, Zeiss filter set 49; Zeiss, Germany). A minimum of 20 fields and at least 400 bacterioplankton cells and 100 organic aggregates (diameter >5 μm) were counted per sample. For the abundance of bacteria associated with OA, the resuspended OA solution was diluted 200-fold and treated with formaldehyde, dispersant, and ultrasound [48], followed by the standard epifluorescence microscopic count method for the bacterioplankton.
Scanning Electron Microscopy
The procedure for scanning electron microscopy (SEM) followed the protocol described by Paerl and Shimp [35]. Subsamples of 50-ml surface water were fixed overnight in 2% v/v glutaraldehyde (final concentration) buffered with 0.1 M sodium cacodylate at 4°C. Two milliliters of each sample except site 24 (10 ml) were filtered through a 5-μm-pore-size polycarbonate membrane (25-mm diameter, Millipore). The filters were washed with 0.1-M sodium cacodylate buffer, serially dehydrated in 10%, 25%, 50%, 75%, and 100% tert-butanol solutions (10 min in each stage), followed by vacuum freeze drying for 1 h, and viewed on a scanning electron microscope (JSM 6300, JEOL).
DNA Extraction, PCR Amplification, and Cloning
Total DNA for OA-associated microorganisms was extracted using proteinase K, sodium dodecyl sulfate, and cetyltrimethylammonium bromide concomitant with chloroform extraction and isopropanol precipitation according to the previous protocol [58]. To reduce inhibition by humic acid during polymerase chain reaction (PCR) amplification, the crude DNA extracts were diluted to 50-fold.
Amplifications of 16S ribosomal RNA (rRNA) genes for the clone library construction of OA-associated samples from the four stations were performed using the eubacterial forward primer 8F (5′-AGAGTTTGATCMTGGCTCAG-3′) and universal reverse primer 1492R (5′-GGTTACCTTGTTACGACTT-3′) [32], which are targeted to universally conserved regions to permit the amplification of an approximately 1,500-bp fragment. PCR amplification was performed in 50-μl reaction mixtures containing 1× PCR buffer, 1.5 mM MgCl2, 200 μM dNTPs, 0.5 μM of each primer, 1.5 U of Taq polymerase (MBI Fermentas, Germany), and about 10 ng diluted DNA extract. Amplification was carried out in a thermocycler (PTC 200 gradient cycler, MJ Research) using a touchdown program: denaturation at 94°C for 5 min, 13 cycles of denaturation at 94°C for 1 min, annealing at 65°C (the temperature was decreased by 1°C every cycle until the touchdown temperature of 53°C was reached) for 1 min and extension at 72°C for 1.5 min. Twenty-two additional cycles were carried out at an annealing temperature of 53°C, followed by a final extension at 72°C for 10 min. PCR products were purified immediately with the E.Z.N.A.® Cycle-Pure Kit (Omega), and the 16S rRNA gene fragments were cloned into pGEM-T Easy vector (Promega) following the manufacturer’s instructions. The randomly chosen clones were amplified directly from cells using vector primers T7 and Sp6 to check the sizes of the inserts and abandon false-positive clones.
Amplified Ribosomal DNA Restriction Analysis
Amplified ribosomal DNA restriction analysis (ARDRA) was used to characterize the 16S rRNA gene diversity within the collection of 1,030 positive clones from the four clone libraries. The PCR products were digested simultaneously with two restriction enzymes HaeIII and HindIII (MBI Fermentas, Germany). The digestions were performed for 3 h at 37°C in 20-μl reaction volumes containing 10-μl PCR products, 2-μl 10× buffers, 7.5-μl sterile Milli-Q water, and 0.5 μl (10 U/μl each) of the restriction enzymes. The digestion products were run on a 2.5% agarose gel in 1× Tris–borate–EDTA buffer at 4 V cm−1 for 3 h. Gels were stained with SYBR Green I (1:10,000 dilution; Amresco), made visible by UV transillumination, and photographed with an Omega 10™ gel documentation system (Ultra-Lum Inc., USA). The images were analyzed with Bionumerics software version 4.0 (Applied Maths, Kortrijk, Belgium) according to BioNumerics Quick Guide. Gel photographs were entered into a BioNumerics database, and the background noise was removed from the densitometric curves. After normalization, the bands in the sample lanes were recognized first with the software’s automatic band search, followed by manual corrections. The ARDRA patterns were used to construct a dendrogram by using the unweighted pair group method with arithmetic mean clustering algorithm using the Pearson correlation coefficient along with the fine optimization option. Bacterial isolates that produced the same ARDRA pattern were grouped together and a lane or cluster of lanes with less than 5% distance to the adjacent lane(s) was defined as a primary operational taxonomic unit (pre-OTU).
Sequencing and Phylogenetic Analysis
Representative clone inserts from each of the unique pre-OTUs in each clone library were sequenced on an automated DNA capillary sequencer (model 3730; Applied Biosystems) using the primer 8F and the ABI Prism BigDye terminator sequencing kit v3.1 (Applied Biosystems). All partial 16S rDNA sequences were edited manually using the software BioEdit (version 7.0.9). To check the accuracy of ARDRA patterns, sequences were aligned with ClustalW and grouped together based on sequence similarity. And sequences with 97% sequence similarity to any other were treated as a single phylotype (or OTU). All assembled sequences were examined for chimeric artifacts using the CHIMERA_CHECK software of the Ribosomal Database Project II (RDP) [29]. Twenty-seven chimerical sequences and 15 DNA sequences from plastids (or chloroplast) were excluded from further analysis. The remaining sequences were compared to the 16S rDNA sequences of the closest organisms from the GenBank and RDP databases to obtain a preliminary phylogenetic affiliation of the clones. The phylogenetic analyses were performed with the Molecular Evolutionary Genetics Analysis (MEGA) software package version 4.0 [46] (available at http://www.megasoftware.net). Evolutionary history was inferred using the minimum evolution method [40]. The robustness of tree topologies was confirmed by maximum parsimony analysis with 500 bootstrap replications. Evolutionary distances were computed using the maximum composite likelihood method [45] and neighbor-joining algorithm was used to generate the initial tree.
Calculation of Diversity Indices and Clone Library Similarity Analysis
For each clone library, the ARDRA- and sequencing-based distribution of clones in different OTUs was used to estimate the 16S rDNA library size, coverage, richness, and evenness. These diversity indices were calculated by using the free software Species Prediction and Diversity Estimation (Chao and Shen, http://chao.stat.nthu.edu.tw/softwareCE.html). Species richness was estimated using the nonparametric Chao estimator S =BS obs + (a 2/2b), where S obs is the observed number of OTUs; a is the number of OTUs observed just once and b is the number of OTUs observed twice [10]. Coverage is calculated as C = 1 − (s/n), where s is the number of unique OTUs and n is the number of individuals in the subsample. The bacterial diversity and dominance of each cloning library was estimated by using Shannon’s diversity index (H'), a summary variable that incorporates the richness and evenness of phylotypes (OTUs), and the Simpson’s dominance index, respectively. For the diversity index calculation, the following equation was used: \(H\prime = - \sum {\left( {p_i \times \ln p_i } \right)} \), where p i = n i /N, n i is the number of clones in the ith OUT and N is the total number of clones in the sample. Here, the higher value indicates higher diversity. Sørensen similarity index was used for calculating the similarity degree at community level [11]. Furthermore, to evaluate how well our clone libraries represent the true diversity of the sample, nonparametric richness estimators (S Chao1) were calculated progressively as more clones were sequenced described by Kemp and Aller [24].
Statistics
Canonical correspondence analysis (CCA) was applied for revealing relationship between the members of OABC and the major environmental variables controlling the compositions of different members. All data were log(x + 1)-transformed. The CCA was performed with the software CANOCO 4.5 (SCIENTIA Software) by using unimodal method because detrended correspondence analysis run on species variables indicated that the length of the first axis was >2. The significance of the first ordination and canonical axes together was assessed in permutation tests with 499 unrestricted Monte Carlo permutations.
Nucleotide Sequence Accession Numbers
The 429 partial 16S rRNA gene sequences determined in the present study were deposited in the GenBank with the following accession numbers: EU272936 to EU273248 and EU373097–EU373211.
Results
Environmental Characterization
Key abiotic, chemical, and biological parameters measured at the four sites of Lake Taihu are presented in Table 1. From north to southeast, the nutrient concentrations at the sampling stations showed a well-defined gradient. For example, the concentrations of TP and TN in the river mouth of Meiliang Bay were about 48 and ten times that of in East Taihu, respectively. The Lake Center had the highest value of TSS (168.47 mg l−1). Regression analysis revealed that OA-associated TP was significantly correlated with TP, TN, and DIN (P < 0.05). Only East Taihu was covered (coverage >80%) by a mixed community of floating leafed species (mainly Nymphoides peltata) and submersed macrophytes (mainly Potamogeton malaianus, Elodea nuttallii). The Chl a concentrations determined at the four sampling stations varied from 1.79 μg l−1 in the macrophyte-dominated East Taihu to 13.39 μg l−1 in the algae-dominated Meiliang Bay. Although the Chl a concentration in the center of the lake was not very high, there was a substantial amount of brown senescent and dead algal material in the water column at the time of sampling.
Bacterial and Organic Aggregate Abundance and Morphology
Along the sampling stations, OA abundance ranged form 1.4 to 7.5 × 105 ml−1, and the abundance of free-living and OA-associated bacteria varied from 4.8 to 14.1 × 106 ml−1 and 3.6 to 22.8 × 106 ml−1, respectively (Table 1). About 43% to 58% of total bacteria were found associated with OA. East Taihu showed the lowest abundance of OA, OA-associated bacteria, and free-living bacteria. The highest bacterioplankton and OA-associated bacterial abundance were recorded in Meiliang Bay and the central region of the lake. Regression analysis revealed that OA-associated bacterial abundance was significantly correlated with TSS (P < 0.05). Clear morphological differences can be observed on SEM micrographs for OA and OA-associated bacteria collected from the four sampling stations (Fig. 2). The size of most OA ranged from 10 to 200 μm, and they consisted predominantly of debris of Cyanobacteria and zooplankton (in Meiliang Bay and Lake Centre), unidentifiable components (in River Mouth), and diatom frustules and detritus of macrophytes or charophyta (in East Taihu; Fig. 2). Intense colonization of mucilage surrounding cyanobacterial cells was observed by DAPI stain and SEM.
Clone Library Coverage and Diversity Analysis
For each of the four stations, comprehensive clone libraries were built with careful consideration for ARDRA resolution and sequence quality. In total, 223 OTUs were obtained from 852 clones using 97% similarity threshold to classify different OTUs. The S Chao1 accumulation curves approached an asymptote, which indicated that the sampling of our clone libraries was satisfactory (Fig. 3 A1–D1). Then, richness (Chao), dominance (Simpson index), and biodiversity coverage of the clone libraries were calculated (Table 2). The predicted value of Chao1 at each sampling station varied between 122.1 and 174.9, with an average of 142.1. Interestingly, the estimated richness (average = 121) for the two lake stations (Meiliang Bay and Lake Center) that experienced cyanobacterial blooms is less than that (average = 149) of the other two stations, which suggested that the richness of the OA-associated bacterial community may have been negatively affected by the conditions created by the cyanobacterial bloom.
Left panels: predicted number of phylotypes based on SChao1 versus size of subsamples of four libraries derived from natural OA-associated bacterial communities. Each point is the mean of ten replicate subsamples of the library. Right panels: the corresponding phylotype frequency distribution for each library. Phylotype richness estimates reach an asymptotic maximum for all three libraries, indicating that these libraries were large enough to yield stable and unbiased estimates of phylotype richness. A1–2: River Mouth, B1–2: Meiliang Bay, C1–2: Lake Center, D1–2: East Taihu
The reciprocal Simpson index was much lower in the two bloom libraries compared with the other two libraries, revealing a more uneven distribution of the phylotypes (OTUs) for the bloom communities. However, when all cyanobacterial clones were excluded from the analyses, the reciprocal Simpson index for the two bloom libraries increased to values that were much higher than those of other two libraries (Table 2), suggesting that the distribution of noncyanobacterial clones between different OTUs was more even. The higher H’ values representing species richness and evenness of the two bloom libraries (cyanobacterial clones were excluded from analysis) confirmed the Simpson index analysis (Table 2).
Bacterial Community Structure
Only five of the 223 identified OTUs (2.2%) were found at all locations. In contrast, 167 OTUs (74.9%) were only found at one of the sampled locations; 37 OTUs (16.6%) were found in two of the locations and 14 OTUs (6.3%) at three locations. The frequencies of the 167 OTUs (found only at one of the sampled locations) were generally (89.2%) very low, with one or two representative clones; a few (10.2%) OTUs contained from three to nine clones per location, and only one OTU yielded 16 clones. In general, most OTUs had not more than three clones per OTU, representing 81.7%, 86.8%, 86.7%, and 80.0% of the total OTUs from River Mouth, Meiliang Bay, Center Lake, and East Taihu libraries, respectively (Fig. 3 A2–D2).
The community assemblages were compared at both the community fingerprinting level and clone library level using Sørensen index of similarity [11]. The similarity values obtained from comparisons of the shared OTUs based on community fingerprinting ranged from 17.8% to 31.8%, which was similar to the values (22.4% to 39.0%) obtained from comparative analyses of the four noncyanobacterial 16S rRNA gene clone libraries studied (Table 3). However, when Cyanobacteria were counted, the homogeneity reached up to 60%. The most similar clone libraries were from the two cyanobacterial bloom systems, Meiliang Bay and Lake Center, and this was also corroborated by community-level ARDRA fingerprinting.
Taxonomic Groups and Their Distribution
Among 852 clones, a total of 429 nonchimerical partial (700 bp) 16S rRNA gene sequences from the four studied bacterial libraries were acquired. There were 225 clones belonging to the Cyanobacteria phylum, and few of them were from the clone library of River Mouth. Members of Microcystis spp. were found almost only in Meiliang Bay and Lake Center, whereas members of Synechococcus spp. were retrieved mostly from Lake Center and East Taihu (Fig. S1_A). In addition, 110 clones (48.9%) from all of the studied locations affiliated with the Cyanobacteria phylum could not be related to any known genus. All the clones were excluded from the subsequent analysis because this study mainly focused on the composition of heterotrophic bacterial communities associated with OA.
As determined by the Basic Local Alignment Search Tool and the RDP classifier, most noncyanobacterial clones from the libraries were affiliated with the β-proteobacteria (32.7%), α-proteobacteria (15.0%), Bacteroidetes (12.3%), γ-proteobacteria (9.1%), Planctomycetes (8.6%), Actinobacteria (4.6%), δ-proteobacteria (3.3%), Firmicutes (2.7%), and ɛ-proteobacteria (2.4%) groups. Clones affiliated with Acidobacteria, Verrucomicrobia, Nitrospira, and Gemmatimonadetes were also found in low numbers (<10 clones per phylum; Fig. 1). In addition, 34 clones (5.4%) could not be affiliated with any known bacterial group.
In total, 392 clones were affiliated with the phylum Proteobacteria, which was the most abundant phylum in the four libraries; representing 72.2%, 53.1%, 55.5%, and 62.3% of the noncyanobacterial clones, respectively (Fig. 1). The Proteobacteria are divided into five subphylums, and four of these (α-, β-, γ-, and δ-proteobacteria) were detected in all four libraries. Members of the subphylum β-proteobacteria were frequently encountered in all four OA-associated bacterial clone libraries, corresponding to 38.6%, 19.2%, 32.8%, and 35.6% of the clones from River Mouth, Meiliang Bay, Center Lake, and East Taihu libraries, respectively (Fig. 1). About 35.9% of the phylotypes (OTUs) were most closely related to phylogenetic groups containing cultured representatives, i.e., Comamonadaceae, Burkholderiaceae, and Alcaligenaceae (Fig. 4b). Most of the sequences affiliated with Burkholderiales, in which the family of Comamonadaceae and Burkholderiaceae were dominant. Phylogenetically, more than a half (53.8%) of the phylotypes of β-proteobacteria belonged to already-known freshwater clusters, e.g., Rhodoferax sp. BAL47, LiUU-5-340.2, and Polynucleobacter necessaries (represented 18% of all β-proteobacteria clones; Fig. 4b). Sequences affiliated with α-proteobacteria accounted for 16.1%, 11.5%, 11.7%, and 19.2% of the noncyanobacterial clones, respectively, and grouped mainly in the family Sphingomonadaceae containing cultured representatives (Figs. 1 and 4a). Many of the 16S rDNA sequences affiliated with α-proteobacteria belong to previously described freshwater clusters, e.g., LD12, LiUU-9-283, Novosphingobium subarctica, and Brevundimonas intermedia (Fig. 4a). Compared with the other three clone libraries, the relative abundance of γ- and δ-proteobacteria in Meiliang Bay library was much higher, but no ɛ-proteobacteria were found (Figs. 1 and S1_B). Among all the phylotypes affiliated with γ-, δ-, and ɛ-proteobacteria, only four phylotypes affiliated with γ-proteobacteria contained cultured representatives; the others had no pure cultured representatives. Interestingly, all the sequences affiliated with δ-proteobacteria closely related to sequences previously obtained from sediment, soil, sludge, or biofilm samples and not from pelagic freshwater ones.
Minimum evolution trees inferred by analysis of partial 16S rDNA sequences (700 bp) obtained from four clone libraries for River Mouth (a*), Meiliang Bay (b*), Lake Center (c*), and East Taihu (d*) in Lake Taihu. Numbers of clones in each OTU are given in parentheses. Brackets following clone name indicate clusters previously reported by Crump and Hobbie [13], Eiler and Bertilsson [15], Hahn [22], Wu et al. [56], and Zwart et al. [59]. Separate phylogenetic trees are shown for the (a) α-proteobacteria (Escherichia coli, DQ118017 was used as out-group), b β-proteobacteria (Geobacter bremensis, U96917 was used as out-group), c Bacteroidetes (C. akagii CK58, AJ237755 was used as out-group). The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) is shown next to the branches. Branches corresponding to partitions reproduced in less than 50% bootstrap replicates are collapsed. The evolutionary distances were computed using the maximum composite likelihood method [45] and were in the units of the number of base substitutions per site. The neighbor-joining algorithm was used to generate the initial tree. Phylogenetic analyses were conducted in MEGA4 [46]
Seventy-seven clones affiliated with the phylum Bacteroidetes accounted for 12.3% of the total sequenced clones (Fig. 1). Bacteroidetes was the most commonly represented phylum in the OA-associated bacterial library of Meiliang Bay characterized by a Cyanobacteria-dominated algal bloom at the time of sampling. A further separation into classes within Bacteroidetes is clearly visible in the phylogenetic tree in Fig. 4c. The majority of the 16S rRNA gene sequence fragments of the phylum Bacteroidetes belong to the class Sphingobacteria. Only 15 clones were affiliated to the class Flavobacteria, but no sequence related to the class Bacteroidetes. Fourteen clones could not be affiliated to any class of the phylum Bacteroidetes. In general, 44% clones affiliated with Bacteroidetes belong to previously described freshwater clusters (Fig. 4c) [13, 59].
Only 4.6% of the total clones were affiliated with the phylum Actinobacteria, and most of them were phylogenetically related to the freshwater clusters Urk0-14, ML-5-51.2, ACK-M1, acII-D, Med0-06, CL500-29, and Sta2-30 [49, 56, 59] (Fig. S1_C). Interestingly, the phylotypes affiliated to Actinobacteria in the Lake Center library accounted for 57.1% of all Actinobacterial phylotypes from all the four libraries.
A total of 54 clones affiliated with Planctomycetes accounted for 8.6% of the total clones, and most of them had no closely related cultured representatives (Fig. 1, Fig. S1_D). Firmicutes, Acidobacteria, Verrucomicrobia, Nitrospira, and Gemmatimonadetes were minor components in the libraries studied; they varied from 0.3% to 2.7%. Firmicutes were absent in East Taihu library (Fig. S1_D). Acidobacteria were found in all of the libraries, but Verrucomicrobia were absent from the nutrient-contaminated location (River Mouth). Nitrospira were only found in Lake Center and East Taihu libraries (Fig. S1_C). Only two clones from Meiliang Bay and the Lake Center were affiliated with the phylum Gemmatimonadetes (Fig. S1_D). Finally, 34 clones (accounting for 5.4% of all clones) from the four libraries could not be affiliated to any known group. Some of them are shown in the phylogenetic tree in Fig. S1.
Statistics
Results of CCA illustrated that the differences in OABC between the four sampling sites are related to the four most important environmental factors (TP, TN, pH, and water temperature). TP and TN contributed significantly to variance in OABC (P < 0.05; Fig. 5). The eigenvalues of the first and second axis were 0.746 and 0.656, respectively, and the two axes explained 71.4% of the observed variation in OABC.
Discussion
The sampling method of aggregates is of vital importance for further analysis. While single aggregates in the sea and in deep lakes often were collected by scuba diving [43], the high concentration of small particles in Lake Taihu requires other methods. Unfortunately, at present time, we have no method which can completely separate the inorganic material from organic material in turbulent shallow lakes. In this study, we chose the centrifugation method to collect our OA samples following the protocol of Böckelmann et al. [7]. There are two reasons. Firstly, there were abundant new-formed small cyanobacterial colonies in the water samples from Meiliang Bay and Lake Center. According to our previous study using DAPI and scanning electronic microscopy, we found that most of the new-formed small cyanobacterial colonies almost had no associated heterotrophic bacteria. Using the centrifugation method, we removed majority of this type of algal colonies suspended in the aqueous phase after centrifugation. Because our research was focused on the associated heterotrophic bacteria, by doing so, we were able to study an important category of attached bacteria. Secondly, our research objects were the community structure of associated bacteria. Organic materials, not the inorganic ones, were intensively colonized by bacteria in the surface water of Lake Taihu (data not shown). Although the sampling procedure integrated over many different particle types including organic and inorganic particles, the collected inorganic components had little effect on the analysis of aggregate-associated bacterial community composition. So we called our collected samples as organic aggregates for emphasizing the habitat of the associated bacteria, just as Böckelmann et al. [7] did.
Although the investigation was a momentary picture of the lake based on one sampling date, our present preliminary study provides insights into the ecological importance of OA-associated bacterial communities. We found that bacterial colonization was highly variable between the aggregates sampled from the four locations with different food web structures; usually, colonization was highest on cyanobacterial aggregates. SEM identified a wide variety of bacterial morphologies, suggesting a diverse community associated with organic aggregates, especially associated with cyanobacterial aggregates (Fig. 2). This was confirmed by phylogenetic analyses of 16S rRNA genes, where bacteria affiliated with nine phyla were identified.
Many studies have demonstrated that the β-proteobacteria are abundant in most freshwater environments [15, 17] and they are also believed to share the ability to degrade complex organic macromolecules with the Bacteroidetes [28]. In our study, sequences affiliated with the clusters obtained from two cyanobacterial blooms in Swedish lakes [15] were abundant, e.g., the LiUU-9-115.2 and LiUU-9-283.2 clusters of α-proteobacteria; the LiUU-3-128, LiUU-5-340.2, and LiUU-5-131 clusters of β-proteobacteria (Fig. 4a, b). Both α- and β-proteobacteria were dominant in the mucilage of Cyanobacteria during bloom events [16, 30]. One explanation for this is that several members of the Proteobacteria attach to Cyanobacteria and cause cell lyses and death of some cyanobacterial species [16, 30]. The β-proteobacteria also constituted the numerically most important bacterial group in lotic microbial aggregates (river snow) obtained from the Elbe River [7] and lake snow aggregates from Lake Constance [41, 50]. These findings are consistent with our results. In contrast, our present study showed sequences affiliated with the Polynucleobacter necessarius cluster of β-proteobacteria to be a major group dwelling on OA (Fig. 4b). Furthermore, three subclusters (PencB, PencC, and PencD) with minimum 16S rRNA gene similarities of >95.0% were detected. The Polynucleobacter community inhabiting Taihu Lake was well characterized by fluorescence in situ hybridization (FISH) with Polynucleobacter-specific probes [52] and by other cultivation-independent methods [55], and even a Polynucleobacter strain (isolate MWH-TaW3) was isolated from this lake [22]. It is stated that all detected Polynucleobacter cells (detected by FISH) appeared as single freely suspended cells [52]. This statement contrasts with findings presented here. Although we cannot exclude the coprecipitated free-living ones by the centrifugation method, we argue that such big percentage of Polynucleobacter were not mainly from the pelagic ones. The Polynucleobacter accounted for 5.9% of the total OA-associated bacterial clones in our study, while they only represented 1.1% of the total free-living clones reported by Wu et al. [55]. One explanation may be that specific Polynucleobacter cells have adapted well to the anaerobic microhabitats inside of OA. Recently, obligate endosymbionts of at least two species of ciliate have been found; the 16S rDNA sequences of these endosymbionts are >99% similar to that of its closest free-living Polynucleobacter bacterium [47]. Thus, another explanation for these conflicting results is that the Polynucleobacter sequences obtained here are from unknown endosymbionts of ciliate or other protozoa. P. necessarius bacteria, detected by cultivation-independent methods, are found globally in the pelagic zone of many lakes across the entire range of trophic states [59]. It appears that they depend upon autochthonous rather than allochthonous substrates, but little is known of their ecological properties [22, 53].
Previous findings have highlighted the importance of Bacteroidetes, formerly known as the Cytophaga–Flavobacteria–Bacteroides phylum, during phytoplankton blooms [15, 39]. In our study, members of the Bacteroidetes formed a substantial component of the bacterial communities associated with algae-based OA (Fig. 1). Previous studies have demonstrated that bacterioplankton belonging to this group is abundant in lakes associated with cyanobacterial blooms [15] and in marine habitats [17]. In addition, they have also been shown to represent a significant part of microbial assemblages frequently found on macroscopic organic matter particles, such as marine snow [14, 38], lake snow [41], and river snow [7]. Many studies also showed that Bacteroidetes were dominant in particle-associated communities in freshwater systems [4, 42]. The high abundance of Bacteroidetes in the free-living assemblages and detritus-associated communities is related to their physiological characteristics. Surface-dependent gliding motility is an important and widespread characteristic of these bacteria. Second, Bacteroidetes are chemoorganotrophic and efficiently degrade a variety of high-molecular-weight compounds such as protein, cellulose, pectin, and chitin, typical components of the high molecular mass fraction of DOM [28]. In the present study, about 56% of the clones affiliated with Bacteroidetes had no close freshwater clusters representatives. These Bacteroidetes might be able to exploit complex organic macromolecules such as cyanobacterial exudates or exopolymers. More direct evidence has shown that the abundance of a Bacteroidetes species capable of lysing Cyanobacteria cells was tightly coupled to the development of a bloom event [37]. Previously observed exoenzyme activities associated with naturally occurring aggregates [21, 44] may be due to the specific colonization of newly formed phytodetrital aggregates by members of these bacterial groups [14]. Accordingly, it is not surprising that phylotypes affiliated with Bacteroidetes were especially abundant in the samples derived from cyanobacterial aggregates from Lake Taihu in this study. Although it is not always possible to predict phenotypic properties solely on the basis of its phylogenetic position [14], together with previous studies, our data suggest that these populations may be responsible for microbial-mediated particle decomposition in the organic aggregates.
Actinobacteria form one of the dominant fractions in freshwater bacterioplankton communities, accounting for as much as 70% of the total bacterioplankton in lakes and rivers distributed globally [5, 59]. Within-lake horizontal heterogeneity of bacterioplankton community composition in Taihu and seasonal patterns of bacterial community structure in Meiliang Bay have been investigated [55, 56]. The results indicated that Actinobacteria was the most commonly represented phylum in the bacterioplankton communities, accounting for 44.8% and 59.4% in Meiliang Bay and East Taihu clone libraries, respectively [55]. However, our results demonstrate a substantial difference between OA-associated and free-living bacterial communities at both the phylum and subphylum levels. Only 29 clones (4.6%) were affiliated with Actinobacteria in the OA-associated bacterial clone library. Allgaier and Grossart [3] compared free-living and particle-associated bacterial communities in four lakes of Northeastern Germany by FISH, and their results showed that the majority of Actinobacteria are free-living and only few are attached. This is consistent with our results.
Generally, the origin, size, and composition of organic aggregates have a pronounced effect on the bacterial colonization. Although many sequences were similar to those found in the previously studied pelagic freshwater habitat, our data showed that nearly half of the clones obtained grouped with the sequences from other habitats (Fig. 4). To better understand the habitats of the new phylotypes, we searched for the closest published sequences on GenBank from aquatic or terrestrial environmental samples. Using these data, we found that about 55% of total clones from the OA-associated bacterial libraries were closely related (88–100% sequence similarity) organisms or clones from pelagic freshwater environments (Fig. 6). Among these pelagic freshwater bacteria, about 19% were from Taihu, which were identified by Wu et al. [55, 56]. In East Taihu, these sequences were most similar to sequences recovered from pelagic habitats and represented 78% of the total OA-associated bacteria. In other locations, however, pelagic-associated bacteria accounted for less than 51%. Meanwhile, the relative proportion of sediment-associated bacteria in East Taihu was the smallest. The observation of these sequences recovered from multiple habitat types indicates potential microbial exchange between OA and the surrounding water. The observed release of amino acids into the surrounding water and the high calculated detachment rates of aggregate-associated bacteria [20, 27, 51] support our hypothesis that there is substantial exchange between the two communities. Soil bacteria constituted the next largest group in Meiliang Bay, Lake Center, and East Taihu, whereas sludge bacteria account for the second largest group (20%) in the River Mouth (Fig. 6). The high proportion of sludge bacteria in the River Mouth library may have resulted from Liangxi River discharge. Previous studies have demonstrated significant influences of allochthonous bacteria on bacterioplankton diversity in lakes [8, 55]. The presence of clones in each OA-associated bacterial library closely related to clones or isolates from soils and sludge may indicate the import of allochthonous bacteria associated with detritus from terrestrial environments.
Proportion of habitat affiliations of the sequences based on comparison of our 16S rRNA clone library sequences with the closest relatives from the GenBank. The left bars give the frequency distribution for the sample River Mouth (A), Meiliang Bay (B), Lake Center (C), East Taihu (D); the bar labeled Total gives the frequency distribution for all samples. Habitats affiliated with anaerobic swine lagoon, subsurface aquifer, wastewater, rumen fluid, human gut, and fuel cell anode as well are included in Others
The high inter-habitat variability of OABC might be governed by differences in environmental conditions between the sites. The result of CCA illustrates that the trophic state was the most important environmental factor in structuring the OABC (Fig. 5). In freshwater systems, increasing nutrient loading causes shifts in food web structure, increased intensity of destructive algal blooms, and the disruption of important ecosystem functions, and all responses are tightly linked [54, 55, 57]. Heterotrophic bacteria respond to nutrient enrichment promptly, resulting in changes in microbial species and community composition.
The different composition of OABC in this study (Table 3 and Fig. 1) is the result of the trophic gradient and the food web dynamics. In East Taihu, the dominant (78%) heterotrophic bacterial sequences were associated with pelagic habitats (Fig. 6); this may be related to low nutrient levels, abundance of macrophytes, and scarcity of OA. Submersed macrophytes caused an immobilization of the sediment, and, due to the lack of sediment resuspension, the water column in the macrophyte-dominated parts contained a much lower concentration of OA [55]. Our sampling procedure integrated over many different particle types with different origins. The chemical composition of OA from different sites was quite different, e.g., the ratio of the OA-associated TN to the TN of the sampled water was much higher (by twofold to fourfold) in Meiliang Bay and Lake Center compared to the other two sites (Table 1). Trophic states might structure OABC by way of influencing the chemical composition of OA. In Meiliang Bay and Lake Center, we found substantial amounts of brown senescent and dead algal material in the water column. The decomposed algae are a good source of OA. And this source of organic matter can be utilized by specific groups of OA-associated bacteria. The relationship between nutrient states, macrophytes, organic aggregate concentrations, cyanobacterial blooms, and composition of OABC should be examined in more detail.
In conclusion, this is among the first reports on microscopic aggregate-associated bacterial diversity in a large shallow aquatic system. Our results demonstrate that OA microhabitats harbor diverse bacterial communities which differ from planktonic ones in Lake Taihu. Phylogenetically, clone libraries of noncyanobacterial OA-associated bacteria were dominated by β-proteobacteria, Bacteroidetes, and α-proteobacteria sequences, but, at individual sites, these proportions varied. Horizontal heterogeneity of OABC existed between habitats and the relative high proportion of Bacteroidetes associated with algae-derived aggregates suggests that the trophic state and the physicochemical properties of OA play a key role in sustaining OABC structures. Comparative statistical analyses of the habitats of OA-associated bacteria highlight the potential ecological importance of the exchange between OABC and the surrounding planktonic community. Furthermore, microorganisms from terrestrial and sediment habitats may be an important component of OA.
References
Alldredge AL (1976) Discarded appendicularian houses as sources of food, surface habitats, and particulate organic matter in planktonic environments. Limnol Oceanogr 21:14–23
Alldredge AL, Silver MW (1988) Characteristics, dynamics and significance of marine snow. Prog Oceanog 20:41–82
Allgaier M, Grossart H-P (2006) Diversity and seasonal dynamics of Actinobacteria populations in four lakes in northeastern Germany. Appl Environ Microbiol 72:3489–3497
Allgaier M, Grossart H-P (2006) Seasonal dynamics and phylogenetic diversity of free-living and particle-associated bacterial communities in four lakes in northeastern Germany. Aquat Microb Ecol 45:115–128
Allgaier M, Bruckner S, Jaspers E, Grossart H-P (2007) Intra- and inter-lake variability of free-living and particle-associated Actinobacteria communities. Environ Microbiol 9:2728–2741
Azam F, Richard AL (2001) Sea snow microcosms. Nature 414:495–498
Böckelmann U, Manz W, Neu TR, Szewzyk U (2000) Characterization of the microbial community of lotic organic aggregates (‘river snow’) in the Elbe River of Germany by cultivation and molecular methods. FEMS Microbiol Ecol 33:157–170
Bergström AK, Jansson M (2000) Bacterioplankton production in humic Lake Örträsket in relation to input of bacterial cells and input of allochthonous organic carbon. Microb Ecol 39:101–115
Caron DA, Davis PG, Madin LP, Sieburth JM (1982) Heterotrophic bacteria and bacterivorous protozoa in oceanic macroaggregates. Sciences 218:795–797
Chao A (1984) Nonparametric estimation of the number of classes in a population. Scand J Statist 11:265–270
Chao A, Chazdon RL, Colwell RK, Shen T-J (2005) A new statistical approach for assessing similarity of species composition with incidence and abundance data. Ecol Lett 8:148–159
Chen Y, Qin B, Teubner K, Dokulil MT (2003) Long-term dynamics of phytoplankton assemblages: Microcystis-domination in Lake Taihu, a large shallow lake in China. J Plankton Res 25:445–453
Crump BC, Hobbie JE (2005) Synchrony and seasonality in bacterioplankton communities of two temperate rivers. Limnol Oceanogr 50:1718–1729
DeLong EF, Franks DG, Alldredge AL (1993) Phylogenetic diversity of aggregate-attached vs. free living marine bacterial assemblages. Limnol Oceanogr 38:924–934
Eiler A, Bertilsson S (2004) Composition of freshwater bacterial communities associated with cyanobacterial blooms in four Swedish lakes. Environ Microbiol 6:1228–1243
Eiler A, Olsson JA, Bertilsson S (2006) Diurnal variations in the auto- and heterotrophic activity of cyanobacterial phycospheres (Gloeotrichia echinulata) and the identity of attached bacteria. Freshw Biol 51:298–311
Glöckner FO, Fuchs BM, Amann R (1999) Bacterioplankton compositions of lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl Environ Microbiol 65:3721–3726
Grossart H-P, Simon M (1993) Limnetic macroscopic organic aggregates (lake snow): occurrence, characteristics, and microbial dynamics in Lake Constance. Limnol Oceanogr 38:532–546
Grossart H-P, Simon M (1998) Bacterial colonization and microbial decomposition of limnetic organic aggregates (lake snow). Aquat Microb Ecol 15:127–140
Grossart H-P, Hietanen S, Ploug H (2003) Microbial dynamics on diatom aggregates in Øresund, Denmark. Mar Ecol Prog Ser 249:69–78
Grossart H-P, Tang KW, Kiorboe T, Ploug H (2007) Comparison of cell-specific activity between free-living and attached bacteria using isolates and natural assemblages. FEMS Microbiol Lett 266:194–200
Hahn MW (2003) Isolation of strains belonging to the cosmopolitan Polynucleobacter necessarius cluster from freshwater habitats located in three climatic zones. Appl Environ Microbiol 69:5248–5254
Jin XC, Tu QY (eds) (1990) In: The standard methods for observation and analysis of lake eutrophication. 2nd edn. China Environmental Science Press, Beijing, pp 138–272 (in Chinese)
Kemp PF, Aller JY (2004) Estimating prokaryotic diversity: when are 16S rDNA libraries large enough? Limnol Oceanogr: Methods 2:114–125
Kepner R, Pratt JR (1994) Use of fluorochromes for direct enumeration of total bacteria in environmental samples: past and present. Microbiol Rev 58:603–615
Kiørboe T, Jackson GA (2001) Marine snow, organic solute plumes, and optimal chemosensory behavior of bacteria. Limnol Oceanogr 46:1309–1318
Kiørboe T, Grossart H-P, Ploug H, Tang K (2002) Mechanisms and rates of bacterial colonization of sinking aggregates. Appl Environ Microbiol 68:3996–4006
Kirchman DL (2002) The ecology of Cytophaga–Flavobacteria in aquatic environments. FEMS Microbiol Ecol 39:91–100
Maidak BL, Cole JR, Lilburn TG, Parker CT Jr, Saxman PR, Stredwick JM, Garrity GM, Li B, Olsen GJ, Pramanik S, Schmidt TM, Tiedje JM (2000) The RDP (Ribosomal Database Project) continues. Nucleic Acids Res 28:173–174
Maruyama T, Kato K, Yokoyama A, Tanaka T, Hiraishi A, Park HD (2003) Dynamics of microcystin-degrading bacteria in mucilage of Microcystis. Microb Ecol 46:279–288
McCarthy MJ, Lavrentyev PJ, Yang L, Zhang L, Chen Y, Qin B, Gardner WS (2007) Nitrogen dynamics and microbial food web structure during a summer cyanobacterial bloom in a subtropical, shallow, well-mixed, eutrophic lake (Lake Taihu, China). Hydrobiologia 581:195–207
Newton RJ, Kent AD, Triplett EW, McMahon KD (2006) Microbial community dynamics in a humic lake: differential persistence of common freshwater phylotypes. Environ Microbiol 8:956–970
Paerl HW (1974) Bacterial uptake of dissolved organic matter in relation to detrital aggregation in marine and freshwater systems. Limnol Oceanogr 19:966–972
Paerl HW, Prufert LE (1987) Oxygen-poor microzones as potential sites of microbial N2 fixation in nitrogen-depleted aerobic marine waters. Appl Environ Microbiol 53:1078–1087
Paerl HW, Shimp SL (1973) Preparation of filtered plankton and detritus for study with scanning electron microscopy. Limnol Oceanogr 18:802–805
Qin BQ, Xu PZ, Wu QL, Luo LC, Zhang YL (2007) Environmental issues of Lake Taihu, China. Hydrobiologia 581:3–14
Rashidan K, Bird D (2001) Role of predatory bacteria in the termination of a cyanobacterial bloom. Microb Ecol 41:97–105
Rath J, Wu KY, Herndl GJ, DeLong EF (1998) High phylogenetic diversity in a marine-snow-associated bacterial assemblage. Aquat Microb Ecol 14:261–269
Riemann L, Winding A (2001) Community dynamics of free-living and particle-associated bacterial assemblages during a freshwater phytoplankton bloom. Microb Ecol 42:274–285
Rzhetsky A, Nei M (1992) A simple method for estimating and testing minimum-evolution trees. Mol Biol Evol 9:945–967
Schweitzer B, Huber I, Amann R, Ludwig W, Simon M (2001) α- and β-proteobacteria control the consumption and release of amino acids on lake snow aggregates. Appl Environ Microbiol 67:632–645
Selje N, Simon M (2003) Composition and dynamics of particle-associated and free-living bacterial communities in the Weser estuary, Germany. Aquat Microb Ecol 30:221–237
Simon M, Grossart H-P, Schweitzer B, Ploug H (2002) Microbial ecology of organic aggregates in aquatic ecosystems. Aquat Microb Ecol 28:175–211
Smith DC, Simon M, Alldredge AL, Azam F (1992) Intense hydrolytic enzyme activity on marine aggregates and implications for rapid particle dissolution. Nature 359:139–141
Tamura K, Nei M, Kumar S (2004) Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci 101:11030–11035
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Vannini C, Pockl M, Petroni G, Wu QL, Lang E, Stackebrandt E, Schrallhammer M, Richardson PM, Hahn MW (2007) Endosymbiosis in statu nascendi: close phylogenetic relationship between obligately endosymbiotic and obligately free-living Polynucleobacter strains (Betaproteobacteria). Environ Microbiol 9:347–359
Velji MI, Albright LJ (1993) Improved sample preparation for enumeration of aggregated aquatic substrate bacteria. In: Kemp PF, F. Sherr B, Sherr EB, Cole JJ (eds) Handbook of methods in aquatic microbial ecology. Lewis, Boca Raton, pp 139–142
Warnecke F, Amann R, Pernthaler J (2004) Actinobacterial 16S rRNA genes from freshwater habitats cluster in four distinct lineages. Environ Microbiol 6:242–253
Weiss P, Schweitzer B, Amann R, Simon M (1996) Identification in situ and dynamics of bacteria on limnetic organic aggregates (lake snow). Appl Environ Microbiol 62:1998–2005
Worm J, Søndergaard M (1998) Dynamics of heterotrophic bacteria attached to Microcystis spp. (Cyanobacteria). Aquat Microb Ecol 14:19–28
Wu QL, Hahn MW (2006) Differences in structure and dynamics of Polynucleobacter communities in a temperate and a subtropical lake, revealed at three phylogenetic levels. FEMS Microbiol Ecol 57:67–79
Wu QL, Hahn MW (2006) High predictability of the seasonal dynamics of a species-like Polynucleobacter population in a freshwater lake. Environ Microbiol 8:1660–1666
Wu Q, Chen Y, Xu K, Liu Z, Hahn M (2007) Intra-habitat heterogeneity of microbial food web structure under the regime of eutrophication and sediment resuspension in the large subtropical shallow Lake Taihu, China. Hydrobiologia 581:241–254
Wu QL, Zwart G, Wu J, Kamst-van Agterveld MP, Liu S, Hahn MW (2007) Submersed macrophytes play a key role in structuring bacterioplankton community composition in the large, shallow, subtropical Taihu Lake, China. Environ Microbiol 9:2765–2774
Wu X, Xi W, Ye W, Yang H (2007) Bacterial community composition of a shallow hypertrophic freshwater lake in China, revealed by 16S rRNA gene sequences. FEMS Microbiol Ecol 61:85–96
Xing P, Kong F (2007) Intra-habitat heterogeneity of environmental factors regulating bacterioplankton community composition in Lake Taihu, China. Aquat Microb Ecol 48:113–122
Zhou J, Bruns MA, Tiedje JM (1996) DNA recovery from soils of diverse composition. Appl Environ Microbiol 62:316–322
Zwart G, Crump BC, Agterveld MPK-v, Hagen F, Han S-K (2002) Typical freshwater bacteria: an analysis of available 16S rRNA gene sequences from plankton of lakes and rivers. Aquat Microb Ecol 28:141–155
Acknowledgements
We thank Dr. Qinglong Wu, Dr. Meijun Chen, and Dr. Peng Xing for providing valuable advice. Dr. Guangwei Zhu and Shurong Qian provided much appreciated field support on sample collections. Taihu Laboratory for Lake Ecosystem Research, Chinese Academy of Sciences is thanked for providing data on chemistry and phytoplankton. We also thank Guijuan Xie and Yi Zhang for experimental assistance. We especially thank Prof. Hans W. Paerl for his careful review and insightful comments on an earlier version of the manuscript. This research was supported by the National Natural Science Foundation of China (grant no. 40573062) and the Knowledge Innovation Program of the Chinese Academy of Sciences (grant no. kzcx2-yw-419). Finally, we thank the anonymous reviewers and the editor for their thoughtful and constructive comments.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the Electronic supplementary material.
Fig. S1
(PDF 637.42 KB)
Rights and permissions
About this article
Cite this article
Tang, X., Gao, G., Qin, B. et al. Characterization of Bacterial Communities Associated with Organic Aggregates in a Large, Shallow, Eutrophic Freshwater Lake (Lake Taihu, China). Microb Ecol 58, 307–322 (2009). https://doi.org/10.1007/s00248-008-9482-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-008-9482-8